Abstract
Inherited eye disorders have a large clinical and genetic heterogeneity, which makes genetic diagnosis cumbersome. An exome-sequencing approach was developed in which data analysis was divided into two steps: the vision gene panel and exome analysis. In the vision gene panel analysis, variants in genes known to cause inherited eye disorders were assessed for pathogenicity. If no causative variants were detected and when the patient consented, the entire exome data was analyzed. A total of 266 Dutch patients with different types of inherited eye disorders, including inherited retinal dystrophies, cataract, developmental eye disorders and optic atrophy, were investigated. In the vision gene panel analysis (likely), causative variants were detected in 49% and in the exome analysis in an additional 2% of the patients. The highest detection rate of (likely) causative variants was in patients with inherited retinal dystrophies, for instance a yield of 63% in patients with retinitis pigmentosa. In patients with developmental eye defects, cataract and optic atrophy, the detection rate was 50, 33 and 17%, respectively. An exome-sequencing approach enables a genetic diagnosis in patients with different types of inherited eye disorders using one test. The exome approach has the same detection rate as targeted panel sequencing tests, but offers a number of advantages. For instance, the vision gene panel can be frequently and easily updated with additional (novel) eye disorder genes. Determination of the genetic diagnosis improved the clinical diagnosis, regarding the assessment of the inheritance pattern as well as future disease perspective.
Introduction
Inherited eye disorders are an important cause of visual impairment in children and working age people in the Western world.1, 2 Within the spectrum of inherited eye disorders, a large clinical heterogeneity is observed. Among the most frequently observed hereditary eye diseases are inherited retinal dystrophies (IRDs), with an incidence of approximately 2.5 in 10 000.1, 2, 3 In patients with IRDs rod photoreceptors, cone photoreceptors and/or retinal pigment epithelial cells degenerate due to monogenic inheritance of highly penetrant disease-causing genetic variants.4 Using clinical features, IRDs are divided into a wide array of clinical subtypes, such as retinitis pigmentosa, Leber congenital amaurosis and so on.4 Another frequent hereditary eye disorder is congenital and childhood cataract with a cumulative incidence of 3.5 per 10 000 at age 15 years.1, 2, 5 Only half of the congenital and childhood cataract has a genetic origin.6 Developmental eye defects and optic atrophy are less frequent among the inherited eye disorders.1 The clinical heterogeneity of inherited eye disorders is even more complex, as they can manifest as an isolated eye disorder or can be part of a syndrome with involvement of additional organs.4 For instance, patients with Bardet–Biedl syndrome have, besides retinal dystrophy, also skeletal, genitourinary and neurological symptoms.7
Determination of the genetic cause of inherited eye disorders has a significant clinical relevance. In Western countries, most patients are the only affected individual in their family, making it impossible to determine the exact inheritance pattern and recurrence risk in offspring. With a genetic diagnosis, accurate genetic counseling can be provided and prenatal testing can be offered.8 Furthermore, for patients to become eligible for genetic therapies that target specific disease genes, their genetic disease cause must be known.9 Moreover, in multiple syndromes the manifestation of the eye disorder is the first detectable symptom, for instance in the metabolic syndrome galactokinase deficiency.10 Genetic testing can give a diagnosis at an early stage and, if available, preventive measures and treatment can start before other symptoms have developed.11
Within the different inherited eye disorders, a large genetic heterogeneity is present. Variants in more than 200 genes have been described to cause IRDs and variants in more than 100 genes have been associated with cataract.11, 12 Because in many eye disorders there is no single gene that accounts for disease in the majority of patients, the detection of the underlying genetic defect by Sanger sequencing is time consuming and expensive and is often not feasible. Since the introduction of next-generation sequencing, many different targeted gene panel tests that sequence a large set of disease-associated genes in one experiment have been developed for specific inherited eye disorders.13 The targeted sequencing tests have a far higher diagnostic yield compared with previous genetic tests, such as Sanger sequence.11, 13, 14 These tests, however, target only a subset of the inherited eye disorders and therefore for each disease category a gene panel test needs to be developed; moreover, for every update the test needs to be validated and patients need to be retested. In contrast, exome sequencing allows the determination of a genetic diagnosis for all types of inherited eye disorders in one single test and when a new panel is implemented, a bioinformatics re-analysis of the already available data is sufficient. The aim of this study was to determine whether exome sequencing in a diagnostic setting is a good method to provide a genetic diagnosis, and to assess the detection rate of causative variants in Dutch patients with inherited eye disorders.
Subjects and methods
Subjects and clinical evaluation
This study was approved by the institutional review boards of the Radboud University Medical Center and adhered to the tenets of the Declaration of Helsinki. Informed consent was obtained from all the participants. A cohort of 266 patients with visual impairment diagnosed by different medical centers in the Netherlands were included in this study. The specific eye disorders were diagnosed by expert ophthalmologists based on appropriate clinical examinations.
The exome results of the patient with unique identifier (UID)5085,15 and the causative variants of the affected brother of UID4785 have already been published.16
Exome sequencing
Sequencing of the exome was performed after the participants were counseled by a clinical geneticist at one of the medical university centers located in the Netherlands. DNA was extracted from peripheral blood using standard procedures. In all 266 cases, exome enrichment was performed using Agilent's SureSelect XT Human All Exon V4 (Agilent Technologies, Santa Clara, CA, USA). For 48 cases, next-generation sequencing was performed using a 5500xl SOLiD System (Life Technologies) and data analysis was performed as described previously.17 For 218 cases, next-generation sequencing was performed using a Illumina HiSeq2000TM sequencer at BGI-Europe (Copenhagen, Denmark). Read alignment to the human reference genome (GrCH37/hg19) and variant calling was performed at BGI using BWA and GATK software, respectively. For all cases, variant annotation was performed using a custom designed in-house annotation and variant prioritization pipeline. Copy number analysis was performed in all cases using CoNIFER 0.2.0 software and annotation was performed using an in-house strategy.
Exome analysis
The analysis of the exome data was divided into two steps: the vision gene panel analysis and the exome analysis. In the vision gene panel analysis, an in silico enrichment of genes associated with genetic eye disorders was performed. This vision gene panel was updated three times during the 3-year period of this study. Fifteen cases were analyzed with the v120612 version consisting of 164 genes, 22 cases were analyzed with version v220313 consisting of 194 genes, 196 cases were analyzed with version v200614 consisting of 273 genes and 33 cases were analyzed with version v141114 consisting of 286 genes (Supplementary Tables 1 and 2). In the vision gene panel analysis, variants in the panel genes with a frequency below 5% in dbSNP and below 1% in an in-house database (consisting of exome frequency data of mostly Dutch individuals) and within the exon or within intronic position −20 till +8 were interpreted and classified based on existing guidelines for variant classification that were determined by the Association for Clinical Genetic Science (classification range from class 1 benign till class 5 pathogenic).18, 19 In the second step, the exome analysis, all genes were analyzed. Protein truncating variants, missense variants with a PhyloP above or equal to 3.5 and canonical splice site variants with a frequency below 5% in dbSNP and below 1% in an in-house database were interpreted and classified in the same way as in the vision gene panel analysis. In both steps of the analysis, cases were grouped into distinct groups based on the variant classification, segregation analysis and associated phenotype known from literature. Three groups were distinguished: cases with causative variant(s), cases with likely causative variant(s) and cases in whom no causative variants were detected. Cases were determined to have causative variant(s) when (i) the detected variant arose de novo, (ii) the detected variant(s) segregated in the family and the affected gene matched the described phenotype or (iii) when no segregation was performed at least one of the detected variants was a published pathogenic, a protein truncating or canonical splice site variant and the affected gene matched the described phenotype. Cases were determined to have likely causative variant(s) when the affected gene matched the described phenotype, no segregation was performed and when a novel missense variant was present in a gene causing autosomal dominant or X-linked inheritance, or when two novel heterozygous missense variants or a novel homozygous missense variant were present in a gene causing autosomal recessive inheritance. Cases were determined to have no causative variants when (i) no pathogenic variants were detected, (ii) one heterozygous variant was detected in a gene in which disease-causing variants are associated with autosomal recessive inherited disease or (iii) variant(s) did not segregate in the family. When no causative variant(s) were detected in the vision gene panel analysis and when the patient consented for this analysis, the second step, the exome analysis, was performed. The disease-causing variants were submitted to ClinVar (submission ID: SUB1936409).
Sanger sequencing and MLPA
All reported single-nucleotide variants have been validated by Sanger sequencing. Primers were developed using Primer3 software. Primers and PCR conditions are available upon request. In 28 cases in whom only one (likely) pathogenic variant was detected in a gene that is causing a recessive inherited eye disorder all coding exons with flanking introns and the splice sites of all noncoding exons were Sanger sequenced. When pathogenic intronic and/or copy number variants were described for the gene, Sanger sequencing of the specific intronic regions (ABCA4 intron 31 and 36, CEP290 intron 26 or USH2A intron 40), a deletion PCR (ABCA4 exon 20-22 deletion or USH2A exon 38-56 deletion) and/or MLPA (ABCA4 MRC-Holland kit P151-B1, EYS MRC-Holland kit P328-A1, or USH2A: MRC-Holland kits P361-A1 and P362-A1) were also performed. Copy number variants detected in the exome data were validated by MLPA (PANK2 MRC-Holland kit P120-B1, PAX6 MRC-Holland kit P219-B2 or USH2A MRC-Holland kit P361-A1 and P362-A2), deletion PCR (CLN3 exon 7-8 deletion) or genome-wide SNP array (PDE6B).
Results
Patients
In this study, 266 Dutch probands with visual impairment, who were referred for diagnostic exome sequencing in 2012, 2013 and 2014 to the Department of Human Genetics of the Radboud University Medical Center, were included. The patients had different types of inherited eye disorders, with IRD as the most common (~90% of the cases; Figure 1a). Furthermore, the cohort contained patients with cataract, optic atrophy and developmental eye defects (Figure 1a). In 12% of the cases, the eye disorder was part of a syndrome, for instance Usher syndrome or Bardet–Biedl syndrome (Supplementary Table 2). In most cases, the family had only one affected individual (simplex cases), while in 13% an autosomal recessive, in 12% an autosomal dominant and in 3% an X-linked inheritance was predicted based on the family pedigree. In 12% of the cases the parents were consanguineous and in these families 24 cases were simplex and eight had multiple affected siblings (Supplementary Table 2). In 63% of the cases, one or more genetic tests were performed previously (Supplementary Table 2).
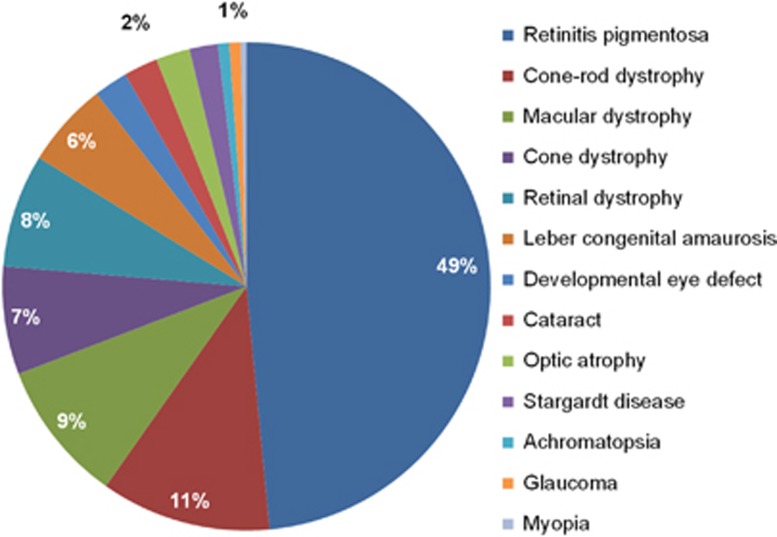
Figure 1.
Clinical details of the 266 Dutch patients with visual impairment. In our cohort of patients with visual impairment 13 different (sub)types of inherited eye disorders were clinically diagnosed. Most cases had an IRD, including 129 patients (49%) with retinitis pigmentosa, 44 patients (16%) with cone or macular dystrophy, 30 patients (11%) with cone–rod dystrophy and 15 patients (6%) with Leber congenital amaurosis. Furthermore, six patients with cataract, six patients with developmental eye disorders, including patients with aniridia, coloboma, microcornea and micro- or nanopthalmos and six patients with optic atrophy were included. The group of retinal dystrophies contained unspecified retinal dystrophies (n=20) and rare forms of retinal dystrophies, including patients with benign concentric annular macular dystrophy, bradyopsia, congenital stationary night blindness, cystoid macular dystrophy, enhanced S-cone syndrome, foveal hypoplasia or night blindness with retinal detachment.
Exome sequencing provides more than 20 × coverage in most inherited eye disease genes
The enrichment of the exome was not tailored specifically towards inherited eye disease genes. To investigate whether the 298 genes of the vision panels are well covered, we determined for each vision panel gene the percentage of base pairs in the gene with an average coverage of at least 20 ×, only including regions targeted for enrichment (Supplementary Figures 1a and b). On the SOLiD platform, the majority of genes in the vision panel genes achieved a mean coverage above 20 × in 91 till 95% of the targeted gene region, while for the HiSeq platform this was in 96 till 100% of the targeted gene region (Figure 2, Supplementary Figures 1a and b).
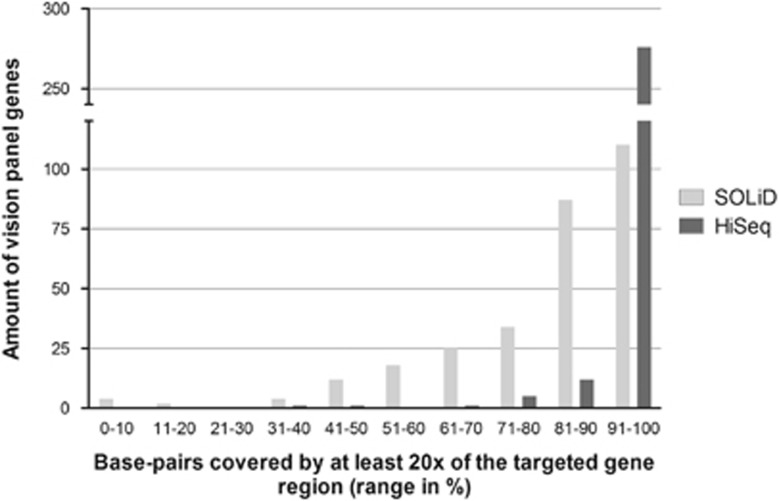
Figure 2.
Mean percentage base pairs covered by at least 20 × of the vision panel genes. For each of the 298 vision panel genes, the mean percentage of base pairs covered by at least 20 × of the targeted gene region was calculated for all 48 samples on the SOLiD and for all 218 cases run on the HiSeq platform. On both platforms, most genes had a mean percentage of base pairs covered by at least 20 × in the range of 91 till 100%. However, if this range is further dissected, the SOLiD platform had most genes in the range of 91 till 95%, while for the HiSeq platform this was in the range of 96 till 100%. Furthermore, the HiSeq platform had only three genes with a mean percentage of base pairs covered by at least 20 × below 70%, while for the SOLiD platform this was the case in 65 genes.
To investigate the extent of possible false-negative results, we determined for all 298 panel genes which exons had an average coverage below 10 × in 75% or more base pairs, including the regions that are not targeted for enrichment. The majority of exons with a coverage below 10 × were not targeted for enrichment. Most of these exons were noncoding, but also 35 exons with coding sequence were not targeted (Supplementary Table 3). For instance, all exons of the OPN1LW and OPN1MW gene were not targeted, because of their high homology. In 11 of the noncoding and 20 of the coding exons, disease-causing variants have been described (Supplementary Table 3). Furthermore, several exons were not efficiently enriched; on the SOLiD platform, 136 targeted exons, and on the HiSeq platform, 15 targeted exons had a coverage below 10 × in 75% of the base pairs. In approximately half of these exons, pathogenic disease-causing variants have been described (Supplementary Table 3).
Vision gene panel analysis determined (likely) causative variants in 49% of the cases
In the first step of analysis of the exome data, the vision gene panel analysis, variants in genes associated with inherited eye disorders were analyzed. Most genes in the vision gene panel cause syndromic or non-syndromic IRDs, but also genes associated with cataract, optic atrophy and developmental eye disease are present (Figure 3a). In 122 cases (46%) causative variant(s), in nine cases (3%) likely causative variant(s), and in 135 cases (51%) no causative variants were detected (Supplementary Table 4). In seven cases, copy number analysis of the exome data detected a causative duplication or deletion (Supplementary Table 4). In 11 of the 122 cases in whom a (likely) genetic cause was detected, the second allele was determined using a second genetic test (Supplementary Table 4). These results are discussed in more detail in the next paragraph. Three cases had (likely) causative variants in two genes, that might have a synergistic effect (Supplementary Table 4).
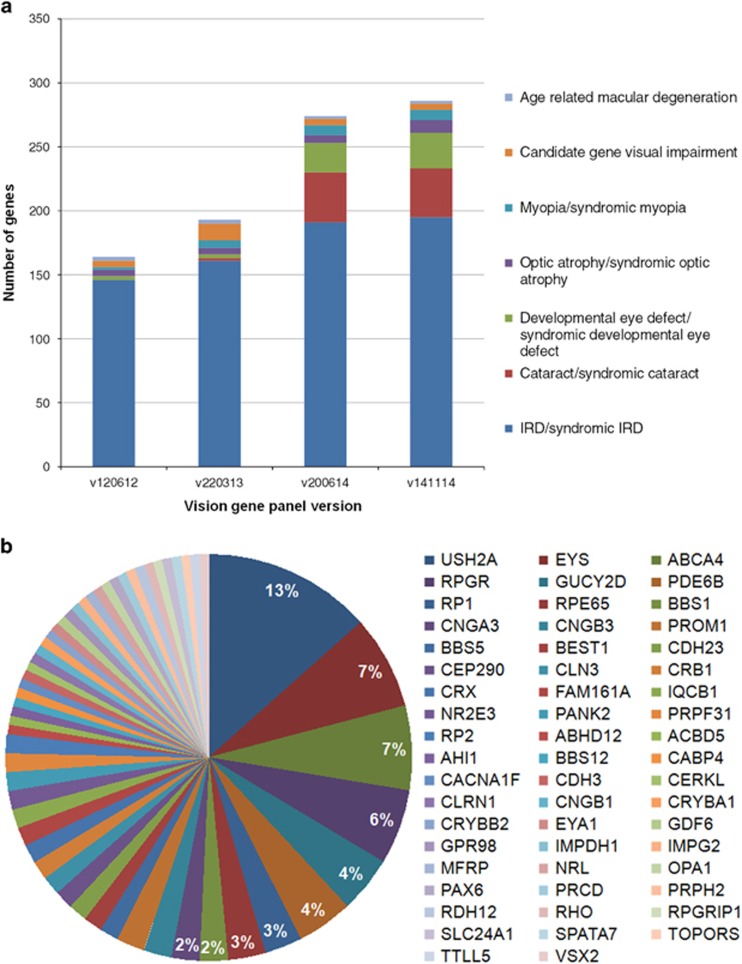
Figure 3.
Vision gene panel revisions and genes carrying variants in the vision gene panel analysis. (a) The genes present in the vision gene panel were grouped into seven main types of inherited visual impairment. In the 3-year time period of this study, the vision gene panel, used in the first step of analysis, was updated three times. The initial version of the panel consisted mostly of genes known to cause syndromic and/or non-syndromic IRD. In the second update also genes known to cause syndromic and/or non-syndromic cataract or developmental eye defects were added. Furthermore, in each update, recently published inherited eye disease genes were added. (b) In the vision gene panel analysis (likely) causative variants were detected in 56 different inherited eye disease genes. USH2A, EYS, ABCA4 and RPGR carried most often the causative variant and taken together they are the responsible causative gene in more than a quarter of the cases in whom a genetic cause was detected. The other genes were implicated to cause disease in 4% or less of the cases.
In 53 cases, the DNA of affected and/or unaffected family member(s) was tested for segregation of the (likely) causative variant(s) in the proband. In seven of these cases, the variant did not segregate within the family (Table 1). In four simplex cases, a heterozygous variant in a gene involved in autosomal dominant inheritance was also present in one of the unaffected parents (BEST1, SNRNP200 and TEAD1) and in three cases the variant(s) was/were absent in an affected family member (BBS5, CRB1 and ZNF644; Table 1).
Table 1. Variants determined to be non-causative after analysis of affected and/or unaffected family members.
| Detected variant(s) | |||||||
|---|---|---|---|---|---|---|---|
| UID | Eye phenotype | Gene | Allele 1 | Detected in | Result segregation analysis | ||
| 1385 | Cone–rod dystrophy | BBS5 | NM_152384.2 | c.92T>C c.92T>C | p.(Ile31Thr) p.(Ile31Thr) | Blind panel analysis | Affected brother has the variant heterozygous |
| 3026 | Macular dystrophy | BEST1 | NM_001139443.1 | c.1223C>T | p.(Pro408Leu) | Blind panel analysis | Unaffected mother has the variant |
| 8732 | Macular dystrophy | CRB1 | NM_201253.2 | c.1023T>A c.3713_3716dup | p.(Asn341Lys) p.(Cys1240fs) | Blind panel analysis | Affected sister carries both variants, affected brother doesn't carry any of the variants |
| 6217 | Retinitis pigmentosa | SNRNP200 | NM_014014.4 | c.4835C>T | p.(Thr1612Met) | Blind panel analysis | Unaffected father has the variant |
| 8681 | Cone–rod dystrophy | SNRNP200 | NM_014014.4 | c.5644C>A | p.(Pro1882Thr) | Blind panel analysis | Unaffected mother has the variant |
| 9722 | Macular dystrophy | TEAD1 | NM_021961.5 | c.59G>A | p.(Ser20Asn) | Blind panel analysis | Unaffected father has the variant |
| 8239 | Iris and choroid coloboma | TFAP2A | NM_003220.2 | c.723del | p.(Glu242fs) | Exome analysis | Affected brother, unaffected mother and unaffected grandfather have the variant |
| 1728 | Benign concentric annular macular dystrophy | ZNF644 | NM_201269.1 | c.3031C>G | p.(Gln1011Glu) | Blind panel analysis | Affected father does not have the variant |
In the vision gene panel analysis (likely) causative variants were identified in 56 different genes (Figure 3b). USH2A was most often the causative gene, while 31 genes were causative in single cases (Figure 3b). In the 56 genes, 178 different variants were detected (including variants identified in a second genetic test). Of the total 81 (46%) novel detected variants, 47 were either protein truncating or canonical splice site variants (Supplementary Table 4). The majority of variants (73%) was detected once, only three variants were detected four times, namely BBS1 c.1169T>G, CRX c.205C>T and USH2A c.2276G>T.
A subsequent genetic test identified the second causative allele in 11 of 32 tested cases
In 46 cases, the vision gene panel analysis detected one heterozygous variant in a gene that is associated with an autosomal recessive inherited eye disorder (Table 2). In 11 of 32 cases, subsequent test(s) identified a second pathogenic variant (Table 2). Interestingly, in these 11 cases, the variant allele detected in the exome analysis was either a known pathogenic variant, a protein truncating variant or a canonical splice site variant (Table 2). In six cases, a (likely) pathogenic deep-intronic variant (ABCA4 or CEP290) was identified. These variants were not present in the exome data, because deep-intronic regions are not enriched. In four cases, a deletion or duplication of a small region (50 bp or 1 exon) of either the EYS or the PDE6B gene was identified. Copy number analysis performed on the exome data can only detect deletions and duplications of at least three consecutive exons20 and, therefore, these four small deletions were not identified. In the last case, the second allele was a canonical splice site variant, not detected in the exome data due to low coverage of the region in this case (nine reads of which only two reads contained the variant nucleotide).
Table 2. Cases with a single heterozygous (likely) causative variant in a gene causing recessive disease.
| Detected variant(s) | ||||||
|---|---|---|---|---|---|---|
| UID | Eye phenotype | Gene | Transcript ID | Allele | Result additional genetic test(s) | |
| 7762 | Stargardt disease | ABCA4 | NM_000350.2 | c.1822T>A | p.(Phe608Ile)a | Deep-intronic variant c.5196+1137G>A detected |
| 4459 | Stargardt disease | ABCA4 | NM_000350.2 | c.3113C>T | p.(Ala1038Val)a | Deep-intronic variant c.4539+2001G>A detected |
| 4913 | Retinitis pigmentosa | ABCA4 | NM_000350.2 | c.3113C>T | p.(Ala1038Val)a | No second variant |
| 0732 | Retinitis pigmentosa | ABCA4 | NM_000350.2 | c.4771G>A | p.(Gly1591Arg)a | No second variant |
| 4795Z | Cone dystrophy | ABCA4 | NM_000350.2 | c.5056G>A | p.(Val1686Met)a | No second variant |
| 4182 | Stargardt disease | ABCA4 | NM_000350.2 | c.5584+6T>C | r.(spl?)a | Deep-intronic variant c.4539+2001G>A detected |
| 2851 | Cone–rod dystrophy | ABCA4 | NM_000350.2 | c.5882G>A | p.(Gly1961Glu)a | ND |
| 4261 | Macular dystrophy | ABCA4 | NM_000350.2 | c.6148G>C | p.(Val2050Leu)a | No second variant |
| 0889 | Stargardt disease | ABCA4 | NM_000350.2 | c.634C>T | p.(Arg212Cys)a | Deep-intronic variant c.4539+2001G>A detected |
| 3013 | Retinitis pigmentosa | ALMS1 | NM_015120.4 | c.6571_6574del | p.(Ser2191fs) | ND |
| USH2A | NM_206933.2 | c.11815G>A | p.(Glu3939Lys)a | ND | ||
| 3988 | Leber congenital amaurosis | BBS1 | NM_024649.4 | c.1131_1135del | p.(Cys377fs)a | No second variant |
| 5017 | Retinitis pigmentosa | BBS5 | NM_152384.2 | c.551A>G | p.(Asn184Ser)a | No second variant |
| 3366 | Retinitis pigmentosa | C2ORF71 | NM_001029883.1 | c.530C>T | p.(Pro177Leu) | No second variant |
| CNGB1 | NM_001297.4 | c.2360A>G | p.(Tyr787Cys) | No second variant | ||
| 3803 | Leber congenital amaurosis | CEP290 RPE65 | NM_025114.3 NM_000329.2 | c.1079G>A c.676G>A | p.(Arg360Gln) p.(Val226Ile) | No second variant No second variant |
| 5502 | Leber congenital amaurosis | CEP290 | NM_025114.3 | c.4960C>T | p.(Gln1654*) | Deep-intronic variant c.2991+1655A>G detected |
| 8389 | Retinitis pigmentosa | CEP290 | NM_025114.3 | c.5649dup | p.(Leu1884fs)a | Deep-intronic variant c.2991+1655A>G detected |
| 6858 | Retinal dystrophy | CEP290 | NM_025114.3 | c.6547C>T | p.(His2183Tyr) | No second variant |
| 8120 | Macular dystrophy | CNGA3 | NM_001298.2 | c.1618G>A | p.(Val540Ile)a | ND |
| CDHR1 | NM_033100.2 | c.783G>A | r.(spl?)a | ND | ||
| 0308 | Retinitis pigmentosa | EYS | NM_001142800.1 | c.1161del | p.(Lys387fs) | Copy number variant c.(2137+1_2138-1)_(2259+1_2260-1)dup detected |
| 2400 | Retinitis pigmentosa | EYS | NM_001142800.1 | c.5167_5168del | p.(Leu1723fs) | Copy number variant c.(6424+1_6425-1)_(6571+1_6572-1)del detected |
| 3982 | Retinitis pigmentosa | EYS | NM_001142800.1 | c.5167_5168del | p.(Leu1723fs) | Copy number variant c.(3243+1_3244-1)_(3443+1_3444-1)dup detected |
| 1073 | Cone dystrophy | KCNV2 | NM_133497.3 | c.820G>A | p.(Val274Met) | No second variant |
| 5428 | Retinal dystrophy | LEPREL1 | NM_018192.3 | c.1906C>T | p.(Pro636Ser) | No second variant |
| 3854 | Cone–rod dystrophy | MAK | NM_001242957 | c.941C>T | p.(Pro314Leu) | ND |
| 5615 | Cone dystrophy | MFRP | NM_031433.3 | c.855T>A | p.(Cys285*) | No second variant |
| 2666 | Developmental eye defect | MKS1 | NM_017777.3 | c.857A>G | p.(Asp286Gly)a | No second variant |
| 4368 | Macular dystrophy | NMNAT1 | NM_022787.3 | c.769G>A | p.(Glu257Lys)a | No second variant |
| 9731 | Cone–rod dystrophy | NR2E3 | NM_014249.2 | c.724_725del | p.(Ser242fs) | Second variant c.119-2A>C detected |
| 4795B | Retinal dystrophy | OCA2 | NM_000275.2 | c.1025A>G | p.(Tyr342Cys)a | ND |
| 0235 | Retinitis pigmentosa | PDE6B | NM_000283.3 | c.(?_-53)_(*785_?)del | p.0 | No second variant |
| 6317 | Retinitis pigmentosa | PDE6B | NM_000283.3 | c.1210A>G | p.(Arg404Gly) | ND |
| 6488 | Retinitis pigmentosa | PDE6B | NM_000283.3 | c.2193+1G>A | r.(spl?)a | Copy number variant c.1923_1971delinsTCTGGGTA detected |
| 8843 | Retinitis pigmentosa | PDE6B | NM_000283.3 | c.2503+5G>C | r.(spl?)a | ND |
| 7248 | Cone dystrophy | RLBP1 | NM_000326.4 | c.454G>A | p.(Asp152Asn) | ND |
| 8369 | Retinitis pigmentosa | RP1 | NM_006269.1 | c.2005G>A | p.(Ala669Thr)a | ND |
| 2438 | Retinitis pigmentosa | RP1 | NM_006269.1 | c.368_369dup | p.(Pro124fs)a | No second variant |
| 4698 | Retinitis pigmentosa | RP1 | NM_006269.1 | c.368_369dup | p.(Pro124fs)a | ND |
| 9807 | Retinitis pigmentosa | RP1 | NM_006269.1 | c.4961C>T | p.(Ser1654Phe) | ND |
| MAK | NM_005906.4 | c.383C>A | p.(Pro128Gln) | ND | ||
| 0459 | Retinitis pigmentosa | RP1L1 | NM_178857.5 | c.1153C>T | p.(Arg385Trp) | ND |
| 9088 | Retinitis pigmentosa | USH1G | NM_173477.4 | c.83C>T | p.(Pro28Leu) | No second variant |
| 1428 | Retinitis pigmentosa | USH2A | NM_206933.2 | c.10510C>G | p.(Pro3504Ala)a | No second variant |
| 2681 | Retinitis pigmentosa | USH2A | NM_206933.2 | c.12823T>A | p.(Ser4275Thr) | ND |
| 0731 | Cone–rod dystrophy | USH2A | NM_206933.2 | c.15427C>T | p.(Arg5143Cys)a | ND |
| 8964 | Retinitis pigmentosa | USH2A | NM_206933.2 | c.1582G>A | p.(Asp528Asn) | No second variant |
| 8481 | Retinitis pigmentosa | USH2A | NM_206933.2 | c.5516T>A | p.(Val1839Glu) | No second variant |
| CDH23 | NM_022124.5 | c.5563C>T | p.(Pro1855Ser) | No second variant | ||
| 9593 | Retinitis pigmentosa | USH2A | NM_206933.2 | c.5698T>G | p.(Cys1900Gly) | No second variant |
Abbreviation: ND, not determined.
Variant already described to cause an inherited eye disorder.
Analysis of the exome determined a (likely) genetic cause in five additional cases
When no causative variants were identified in the vision gene panel analysis, the analysis of the complete exome data was performed as a second step. As there is a possibility of approximately 1% to have an unsolicited finding in this analysis,21 for instance a disease-causing BRCA1 variant, this second step was only performed in patients who consented for this analysis. Consent to analyze the complete exome was given by 79% of patients (106 of the 135 cases) with a negative vision gene panel result. In this additional analysis, in three cases (3%) causative variants and in three cases (3%) likely causative variants were detected (Supplementary Table 4). Variants were identified in four different genes. Disease-causing variants in two of these genes, POC1B and MFSD8, were recently published to cause cone–rod dystrophy and macular dystrophy, respectively.22, 23 Consequently, these genes were not yet in the vision gene panel version used for these cases. MFSD8 was added to vision gene panel v200614 and POC1B to v141114. Likely disease-causing variants were detected in CEP83 in a patient with apparent non-syndromic RP, while two of her siblings have Senior–Loken syndrome. The last case had a de novo variant in the KIF21A gene. Disease-causing variants in KIF21A cause a syndrome in which the muscular symptoms are most apparent and this gene is therefore not in the vision gene panel. In one case, it was concluded that the frameshift variant in TFAP2A was not the cause of the disease, because it was also present in the unaffected mother and grandfather (Table 1).
The diagnostic yield is highest in cases with IRD
Taking the detection rate of causative variants of the vision gene panel and exome analysis together, we determined (likely) causative variants in 52% of the visual impairment cases. As already described in literature, the detection rate differed considerably between the different (sub)types of inherited eye disorders. The different subtypes of IRDs had the highest detection rate. In all patients with achromatopsia and Stargardt disease (likely) causative variants were identified, though for both a small number of cases were tested (Table 3). Of the IRDs of which more than 10 cases were tested, Leber congenital amaurosis and retinitis pigmentosa had the highest diagnostic yield, 60% and 63% respectively, and macular dystrophy and retinal dystrophy (containing rare forms of retinal dystrophies and unspecified retinal dystrophies) had the lowest detection rate of 28% and 25%, respectively (Table 3). Cases with developmental eye defects, cataract and optic atrophy had a detection rate of 50%, 33% and 17%, respectively (Table 3).
Table 3. Detection rate of (likely) causative variants per visual impairment (sub)type.
| (Sub)type of visual impairment | Total number of cases | Percentage of cases with (likely) causative variants (%) |
|---|---|---|
| Achromatopsia | 2 | 100 |
| Stargardt disease | 5 | 100 |
| Retinitis pigmentosa | 129 | 63 |
| Leber congenital amaurosis | 15 | 60 |
| Developmental eye defect | 6 | 50 |
| Cone dystrophy | 19 | 47 |
| Cone–rod dystrophy | 30 | 43 |
| Cataract | 6 | 33 |
| Macular dystrophy | 25 | 28 |
| Retinal dystrophy | 20 | 25 |
| Optic atrophy | 6 | 17 |
| Glaucoma | 2 | 0 |
| Myopia | 1 | 0 |
| Total | 266 | 52 |
Genetic diagnosis redefined the inheritance pattern and clinical diagnosis
The great majority of our patients were the only affected individual in their family, and in these simplex cases the inheritance pattern is unclear without a genetic diagnosis. Of the simplex cases in whom (likely) causative variants were identified, 88% had autosomal recessive, 6% had autosomal dominant and 6% had X-linked inheritance. The genetic diagnosis also redefined the inheritance pattern in cases in whom an inheritance pattern was predicted based on the family pedigree. Three families, in which autosomal dominant inheritance was predicted, actually had X-linked inheritance (RPGR), one family with predicted autosomal recessive inheritance, the genetic diagnosis determined dominant inheritance (the EYA1 variant was also present in the mother with a very mild phenotype), and last, one family with predicted X-linked inheritance showed an autosomal recessive inheritance after genetic diagnosis (USH2A).
The genetic diagnosis also redefined the clinical phenotype in three cases. Case UID3855 with cone–rod dystrophy and case UID4088 with retinitis pigmentosa had no clear signs of any additional clinical symptoms except for a slight motor developmental delay in UID4088. Exome sequencing identified in both cases causative variants in the PANK2 gene that causes NBIA1 syndrome, which is also known to express a broad range of neurological symptoms. In patients with PANK2 variants, retinal dystrophy is one of the first symptoms, and in both cases the neurological symptoms of NBIA1 syndrome presented in a later stage. The same holds true for case UID0504 with retinitis pigmentosa, in whom causative variants were detected in CLN3, which causes the progressive neurodegenerative disease neuronal ceroid lipofuscinosis. The patient developed the neurological symptoms later on.
Discussion
We performed a single genetic test, exome sequencing, in a diagnostic setting to identify disease-causing variants in 266 patients with different types of inherited eye disorders. Using this single test, a (likely) genetic cause was determined in 52% of the cases.
In our approach, the exome data analysis was divided into two parts, the vision gene panel and the exome analysis. This two-step analysis has two major advantages. First, the variant analysis was optimized for the requirements of each step. Within the vision gene panel analysis, variant frequency was the only exclusion criteria before variants were assessed for pathogenicity, while in the exome analysis additional criteria are used to keep the amount of variants that need to be assessed within workable range. Second, for the exome analysis step, patients must consent for unsolicited findings, such as disease-causing BRCA1 variants. In the cohort, 21% of the cases did not consent and if the complete exome would be analyzed directly, this test would not be suitable for at most a quarter of the patients.
Exome sequencing was performed on two different platforms, SOLiD and HiSeq, and both provide a coverage above 20 × for most vision panel genes. Yet the HiSeq platform outperformed the SOLiD platform significantly on uniformity and depth of coverage. Nevertheless, there were regions of the vision panel genes that were persistently not covered in our cohort. First, some regions of the vision panel genes were difficult to enrich. Huang et al. determined that next-generation sequencing has the potential to miss causative variants in the RPGR gene, accounting for 70–75% of X-linked retinitis pigmentosa cases.24 In the currently described exome approach, most of the RPGR gene had a coverage above 20 ×, but approximately 30% of the RPGR-ORF15 exon 15, a mutation hotspot region, had less than 10 × coverage. Nevertheless, the RPGR gene was the fourth most frequently mutated gene in our cohort, demonstrating that our approach is able to detect causative RPGR variants. In general, newer versions of the enrichment kit and longer read lengths could overcome enrichment difficulties. For instance, the OPN1LW and OPN1MW genes are enriched in Agilent enrichment kit V5, while this was not the case in V4 used in this study. Second, via an exome-sequencing approach, deep-intronic and small copy number variations (~50 bp till 2 exons) are not detected. In our cohort, an additional genetic test identified a deep-intronic variant (n=6) or a small copy number variant (n=4) as the second causative variant. We therefore conclude that in our exome-sequencing approach, an additional genetic test to identify variants in non-covered regions should be performed, when a single heterozygous pathogenic variant is detected in a gene associated with autosomal recessive inheritance. Unfortunately, patients in whom all causative variants are in poorly/non-covered region will not be genetically solved using the currently described exome approach. To provide the best care for patients with hereditary eye disorders, we developed a diagnostic flow-scheme to perform specific genetic tests, either as pre-screen, parallel to exome sequencing, or after a negative exome result (Figure 4). In two cases of our cohort, a subsequent genetic test identified the causative variant. In UID5265 with LCA, the deep-intronic c.2991+1655A>G in CEP290 was homozygously present and in UID8627 with cone–rod dystrophy the complete OPN1MW gene was deleted. In two of the 13 simplex males with retinitis pigmentosa in whom the exome analysis was negative, RPGR was Sanger sequenced and no causative variants were detected. Each diagnostic laboratory needs to decide whether a pre-screen (as suggested in the flow-scheme) or simultaneous exome sequencing and testing of intronic disease-causing variants/non-covered regions is the most cost-effective approach with a reasonable turnaround time in their situation.
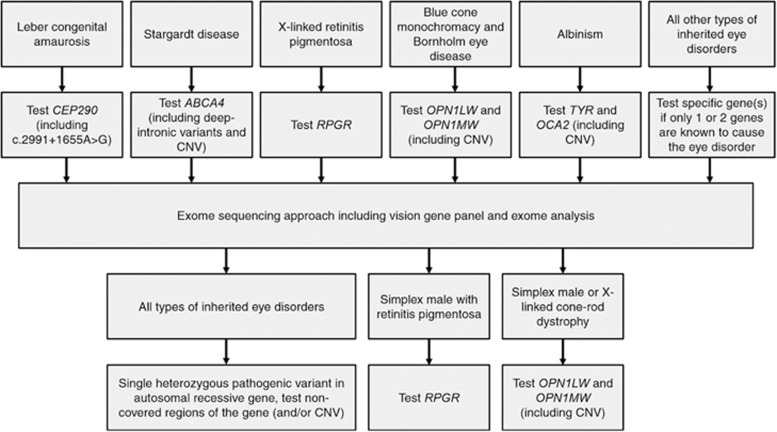
Figure 4.
Flow-scheme of genetic tests that should be performed in patients with inherited eye disorders. In our exome approach, some regions of genes associated with inherited eye disorders have consistently low or no coverage. For instance, regions with high homology, GC-rich regions and repetitive regions are difficult to enrich. Moreover, deep-intronic and small copy number variations are not detected via an exome-sequencing approach. To provide the best care, a diagnostic flow-scheme specific for our exome approach was developed, to provide an overview in which types of inherited eye disorders a genetic test should be performed as a pre-screen or after a negative exome result. In patients with Leber congenital amaurosis, Stargardt disease, X-linked retinitis pigmentosa, blue cone monochromacy, Bornholm eye disease and albinism, a genetic pre-screen of one or two specific genes is indicated. In simplex male patients with retinitis pigmentosa or cone–rod dystrophy and in patients with X-linked cone–rod dystrophy, a genetic follow-up test of one or two specific genes is indicated after a negative exome result. In patients in whom a single heterozygous pathogenic variant is detected in a gene associated with autosomal recessive inheritance, it is indicated to test the non-covered regions of that gene.
The vision gene panel analysis detected (likely) causative variants in 49% of the visual impairment cases. It has to be noted that 63% of our cases had pre-screening of one or more genetic tests, and when exome sequencing is performed as the first test the diagnostic yield will most likely further increase with 5 till 15%. Furthermore, we hypothesize that several of the 14 cases with one heterozygous variant in a gene involved in a recessive disease would have been solved when genetic follow-up was performed. In the vision gene panel analysis, variants were identified in 56 different genes, of which 31 genes were causative in single cases, and 46% of the identified variants were novel. This strengthens the hypothesis that a diagnostic test for inherited eye disorders has to have a broad genetic range and should not be restricted to known disease-causing variants. In the exome analysis, (likely) causative variants were detected in an additional 2% of the patients. In four of the six cases, the causative variants resided in a recently published gene (MFSD8 and POC1B) associated with inherited eye disorder, indicating the importance to update gene panels for inherited eye disorders in a regular manner. Our vision gene panel can be adjusted without changing the actual test itself and in this study the panel was updated three times. For targeted gene-sequencing techniques, a frequent update is far more complicated, because new probes/primers have to be developed and the enrichment has to be optimized for every update. In addition, in contrast to targeted gene sequencing, exome sequencing enables data to be periodically reanalyzed with a newer version of the vision gene panel, making it possible to determine the genetic cause in a novel eye disease gene within the already available data.
The past few years, multiple targeted gene panel tests have been developed. The targeted gene panels for subtypes of IRDs have a detection rate of approximately 50–70%.25 In our exome approach, most of the IRD subtypes have a detection rate within this range. Only the macular dystrophy and retinal dystrophy cases (containing rare forms of retinal dystrophies and unspecified retinal dystrophies) had a far lower detection rate of 28% and 25%, respectively. For the macular dystrophy, this was most likely due to the fact that almost all cases were prescreened for ABCA4 and PRPH2, the most frequently mutated genes in macular dystrophy. For retinal dystrophy, this was most likely due to the fact that many of the disease-causing genes are not yet known.
Only 50% of the congenital and childhood cataract is caused due to a monogenetic cause. Interestingly, in our cohort only in the cases with multiple affected family members a genetic cause was identified. Moreover, Gillespie et al. had a detection rate of 75% using a targeted gene sequencing method for congenital and childhood cataract.11 Indicating that strict patient inclusion criteria can increase the diagnostic yield in patients with childhood and congenital cataract.
As shown in the current and previous exome studies, even if the complete exome is analyzed the underlying genetic defect is not detected in all patients. Apart from the fact that there might be patients in whom the eye disorder is actually not genetic, there are several reasons for missing the causative variants in hereditary cases. This is, for instance, because in an increasing number of genes pathogenic variants are described to be present in noncoding gene regions. These variants are not present in exome-sequencing data, but will be detected when whole genome sequencing is introduced into patient care. Another important reason is that not all inherited eye disorder genes are known. It is difficult to identify candidate genes for inherited eye disorders as the genes with the highest disease-causing variant load have already been discovered. When a candidate gene is found to be mutated in a single family or even in a simplex case, the involvement of this gene in the inherited eye disorder must be further investigated. Besides gaining more insight in the role of this gene in the eye by functional testing and/or animal models, variant screening of the gene in large cohorts of unsolved patients with an overlapping phenotype is important. As there is an increasing number of laboratories performing exome sequencing, data sharing would facilitate the detection of novel inherited eye disorder genes.
In 48% of the simplex cases, (likely) causative variant(s) were detected and therefore in these patients the inheritance pattern and recurrence risk could be determined. Furthermore, the frequency of inheritance patterns determined in the cases in which a genetic cause was identified, can be used as an empirical risk in the genetic counseling of simplex cases in whom no genetic cause is known. Remarkably, in 7% of the cases in whom an X-linked, autosomal recessive or autosomal dominant inheritance pattern was predicted based on the family pedigree, the genetic diagnosis determined a different inheritance pattern. Besides the inheritance pattern, the genetic diagnosis also redefined the clinical diagnosis in three cases. All cases were thought to have a non-syndromic eye disorder, but genetic analysis revealed the presence of a syndromic disease, either NBIA1 or neuronal ceroid lipofuscinosis. Unfortunately, no treatment is available for these syndromes, but the early discovery was used to inform the patients and their families about the prognosis of these relatively severe syndromes. This demonstrates the importance of a thorough pre-test counseling even when the analysis is restricted to the vision gene panel.
In summary, our exome analysis approach for inherited eye disorders is a good diagnostic test to determine the genetic diagnosis in patients with inherited eye disorders and offers a number of advantages over targeted panel sequencing. Moreover, determining the genetic cause in patients with inherited eye disorders complements clinical diagnosis both on inheritance pattern as well as future disease prospective.
Acknowledgments
We thank Ilse de Wijs, Marleen van Bers and Jolanda Kooistra for the analysis of the exome data and Sanger sequence validation, Monique Losekoot for CLN3 validation and Krysta Voesenek for uploading disease-causing variants to ClinVar.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Kocur I, Resnikoff S: Visual impairment and blindness in Europe and their prevention. Br J Ophthalmol 2002; 86: 716–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger RP, Fimmers R, Holz FG, Scholl HP: Incidence of blindness and severe visual impairment in Germany: projections for 2030. Invest Ophthalmol Vis Sci 2011; 52: 4381–4389. [DOI] [PubMed] [Google Scholar]
- Ayuso C, Millan JM: Retinitis pigmentosa and allied conditions today: a paradigm of translational research. Genome Med 2010; 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahel JA, Marazova K, Audo I: Clinical characteristics and current therapies for inherited retinal degenerations. Cold Spring Harb Perspect Med 2015; 5: a017111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahi JS, Dezateux C, , British Congenital Cataract Interest G: Measuring and interpreting the incidence of congenital ocular anomalies: lessons from a national study of congenital cataract in the UK. Invest Ophthalmol Vis Sci 2001; 42: 1444–1448. [PubMed] [Google Scholar]
- Francis PJ, Moore AT: Genetics of childhood cataract. Curr Opin Ophthalmol 2004; 15: 10–15. [DOI] [PubMed] [Google Scholar]
- Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA: New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet 1999; 36: 437–446. [PMC free article] [PubMed] [Google Scholar]
- Wiggs JL, Pierce EA: Genetic testing for inherited eye disease: who benefits? JAMA Ophthalmol 2013; 131: 1265–1266. [DOI] [PubMed] [Google Scholar]
- Gillespie RL, Hall G, Black GC: Genetic testing for inherited ocular disease: delivering on the promise at last? Clin Exp Ophthalmol 2014; 42: 65–77. [DOI] [PubMed] [Google Scholar]
- Bosch AM, Bakker HD, van Gennip AH, van Kempen JV, Wanders RJ, Wijburg FA: Clinical features of galactokinase deficiency: a review of the literature. J Inherit Metab Dis 2002; 25: 629–634. [DOI] [PubMed] [Google Scholar]
- Gillespie RL, O'Sullivan J, Ashworth J et al: Personalized diagnosis and management of congenital cataract by next-generation sequencing. Ophthalmology 2014; 121: 2124–2137 e2121-2122. [DOI] [PubMed] [Google Scholar]
- Daiger SP, Sullivan LS, Bowne SJ, Rossiter BJF: Retnet, 2016, Available at http://sph.uth.edu/RetNet/ acessed on 13 June 2016.
- Lee K, Garg S: Navigating the current landscape of clinical genetic testing for inherited retinal dystrophies. Genet Med 2015; 17: 245–252. [DOI] [PubMed] [Google Scholar]
- Prokudin I, Simons C, Grigg JR et al: Exome sequencing in developmental eye disease leads to identification of causal variants in GJA8, CRYGC, PAX6 and CYP1B1. Eur J Hum Genet 2014; 22: 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi KM, Avila-Fernandez A, van Huet RA et al: Exome sequencing extends the phenotypic spectrum for ABHD12 mutations: from syndromic to nonsyndromic retinal degeneration. Ophthalmology 2014; 121: 1620–1627. [DOI] [PubMed] [Google Scholar]
- Neveling K, Collin RW, Gilissen C et al: Next-generation genetic testing for retinitis pigmentosa. Hum Mutat 2012; 33: 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K, Feenstra I, Gilissen C et al: A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat 2013; 34: 1721–1726. [DOI] [PubMed] [Google Scholar]
- Wallis Y, Payne S, McAnulty C et al: Practice Guidelines for the Evaluation of Pathogenicity and the Reporting of Sequence Variants in Clinical Molecular Genetics. ACGS/VGKL 2013, Available at http://www.acgs.uk.com/media/774853/evaluation_and_reporting_of_sequence_variants_bpgs_june_2013_-_finalpdf.pdf.
- Plon SE, Eccles DM, Easton D et al: Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 2008; 29: 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfundt R, Del Rosario M, Vissers LE et al: Detection of clinically relevant copy number variants by exome sequencing in a large cohort of genetic disorders. Genet Med 2016, e-pub ahead of print 27 October 2016doi:10.1038/gim.2016.163. [DOI] [PMC free article] [PubMed]
- Green RC, Berg JS, Grody WW et al: ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013; 15: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosing S, Lamers IJ, de Vrieze E et al: Disruption of the basal body protein POC1B results in autosomal-recessive cone-rod dystrophy. Am J Hum Genet 2014; 95: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosing S, van den Born LI, Sangermano R et al: Mutations in MFSD8, encoding a lysosomal membrane protein, are associated with nonsyndromic autosomal recessive macular dystrophy. Ophthalmology 2015; 122: 170–179. [DOI] [PubMed] [Google Scholar]
- Huang XF, Wu J, Lv JN, Zhang X, Jin ZB: Identification of false-negative mutations missed by next-generation sequencing in retinitis pigmentosa patients: a complementary approach to clinical genetic diagnostic testing. Genet Med 2015; 17: 307–311. [DOI] [PubMed] [Google Scholar]
- Combs R, McAllister M, Payne K et al: Understanding the impact of genetic testing for inherited retinal dystrophy. Eur J Hum Genet 2013; 21: 1209–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.