Abstract
Nitrification, the microbial oxidation of ammonia to nitrate via nitrite, occurs in a wide range of acidic soils. However, the ammonia-oxidizing bacteria (AOB) that have been isolated from soil to date are acid-sensitive. Here we report the isolation and characterization of an acid-adapted AOB from an acidic agricultural soil. The isolated AOB, strain TAO100, is classified within the Gammaproteobacteria based on phylogenetic characteristics. TAO100 can grow in the pH range of 5–7.5 and survive in highly acidic conditions until pH 2 by forming cell aggregates. Whereas all known gammaproteobacterial AOB (γ-AOB) species, which have been isolated from marine and saline aquatic environments, are halophiles, TAO100 is not phenotypically halophilic. Thus, TAO100 represents the first soil-originated and non-halophilic γ-AOB. The TAO100 genome is considerably smaller than those of other γ-AOB and lacks several genes associated with salt tolerance which are unnecessary for survival in soil. The ammonia monooxygenase subunit A gene of TAO100 and its transcript are higher in abundance than those of ammonia-oxidizing archaea and betaproteobacterial AOB in the strongly acidic soil. These results indicate that TAO100 plays an important role in the nitrification of acidic soils. Based on these results, we propose TAO100 as a novel species of a new genus, Candidatus Nitrosoglobus terrae.
Introduction
Extensive applications of nitrogen fertilizers have enhanced nitrification activity, resulting in the pollution of surface and ground waters and emission of the greenhouse gas nitrous oxide (Schlesinger, 2009; Wrage et al., 2001). Ammonia oxidation, the first and rate-limiting step of nitrification, is carried out in soil by ammonia oxidizing bacteria (AOB) (Kowalchuk and Stephen, 2001), ammonia-oxidizing archaea (AOA) (Leininger et al., 2006) and complete ammonia oxidizing (comammox) bacteria (Daims et al., 2015; van Kessel et al., 2015). AOB, of which sequences have been found suggesting that they may be involved in ammonia oxidation in soil (Di et al., 2009; Jia and Conrad, 2009), have been shown to be acid-sensitive and do not grow below pH 6.0 in pure culture (Jiang and Bakken, 1999). However, high-nitrification rates have been found in a wide variety of acidic soils with pH below 5.5 (Booth et al., 2005). Thus, the microbiological basis of ammonia oxidation in acidic soil remained unclear until the recent discovery of AOA (Konneke et al., 2005; Leininger et al., 2006; Tourna et al., 2011). Indeed, AOA are more abundant than AOB in many acidic soils (Zhang et al., 2012), and acidophilic AOA, Nitrosotalea devanaterra and Nitrosotalea species, have been isolated from acidic soil and characterized (Lehtovirta-Morley et al., 2011, 2014). These observations strongly support the hypothesis that AOA are more important than AOB in nitrification in acidic soil.
AOB have, however, been identified as the dominant ammonia oxidizers in several acidic soils (Long et al., 2012; Petersen et al., 2012; Wertz et al., 2012). A few AOB have been isolated from acidic soils with a pH around 4 (Jiang and Bakken, 1999) although they have been found to be acid sensitive in pure culture. Ammonia oxidation by these acid-sensitive AOB in acidic soils might be explained by several hypothetical mechanisms (De Boer and Kowalchuk, 2001) including pH-neutral micro-sites, urea hydrolysis (Burton and Prosser, 2001; De Boer et al., 1989), and acid resistant biofilm and aggregate formation (Allison and Prosser, 1993; De Boer et al., 1991). Conversely, we previously reported the potential presence of acid-tolerant AOB in acidic soil from tea fields, where nitrification occurs at high levels despite the soil having a pH of about 3 (Hayatsu, 1993; Hayatsu and Kosuge, 1993). These studies suggested the existence of acid adapted AOB and their contribution to nitrification in acidic soil. Therefore, AOB cannot be excluded from the organisms that mediate nitrification in acidic soil, and the discovery of new acid-tolerant AOB is anticipated.
The aim of the current study was to isolate and characterize acid-adapted AOB from the acidic soil of tea fields and to demonstrate the contribution of the isolated AOB to ammonia oxidation activity in the tea field soil. In addition, we aimed to provide genomic level evidence for the taxonomic and evolutionary relationships between isolated acid-adapted AOB and known related AOB.
Materials and methods
Soil samples
Soil samples, classified as Andosol, were obtained in July 2012 from tea field plots at Kanaya Tea Research Station, Institute of Fruit Tree and Tea Science, NARO, in Japan. These plots are fertilized every year to study the effects of N, P, K and Ca on tea plants (Table 1). N, P, K and Ca were supplied as ammonium sulfate, superphosphate, potassium sulfate and dolomite (CaCO3·MgCO3), respectively. Soil samples were collected as cores from 10-cm deep and immediately stored at −80 °C until molecular analyses or at 4 °C until potential nitrification rate analyses.
Table 1. Characteristics of soils used in the study.
| Treatmenta | Soil pH | Total C (%) | Total N (%) | PARb (nmole−1h−1g−1) |
Fertilization (kgha−1y−1)c |
|
|---|---|---|---|---|---|---|
| N input | Ca (dolomite) | |||||
| Standard | 3.36 (0.05) c | 16.86 (1.32) b | 1.23 (0.13) b | 45.91 (5.74) a | 400 | 1000 |
| High Ca | 4.74 (0.10) d | 11.93 (0.13) a | 0.84 (0.04) a | 253.66 (5.10) c | 400 | 3000 |
| No Ca | 2.89 (0.05) a | 19.47 (0.59) c | 1.54 (0.06) d | 55.84 (4.80) a | 400 | 0 |
| High N | 3.09 (0.04) b | 17.82 (0.37) b | 1.52 (0.06) c | 180.18 (17.90) b | 1200 | 1000 |
s.d. are given in parentheses. Values within the same column followed by the same letter are not significantly different at P<0.05.
Treatments: standard fertilization (Standard), high-rate Ca fertilizer input (high Ca), no Ca fertilizer input (no Ca) and high-rate N fertilizer input (high N).
PAR, potential ammonia oxidation rate (NO2−1 nmole−1 h−1 g−1 dry soil).
Dolomite and ammonium sulfate were added to the fields as sources of Ca and N, respectively.
Potential ammonia oxidation rate (PAR)
Potential rates of ammonia oxidation were measured as rates of nitrite accumulation in the presence of chlorate, which inhibits nitrite oxidation (Belser and Mays, 1980). Slurries were obtained by mixing 2.5 g soil sample with 10 ml of 1 mM potassium phosphate buffer (pH 7) containing 10 mM (NH4)2SO4 and 10 mM KClO3. Subsequently, the slurries were incubated for several hours at 25 °C with shaking at 150 r.p.m. Aliquots of 1 ml were drawn at 0 and 4 h and centrifuged at 10000g for 10 min at 4 °C. The nitrite concentration in the supernatant was determined calorimetrically via diazotization (Keeney and Nelson, 1982). PAR were calculated as nmol nitrite per hour per g of dried soil. The obtained potential rates, which were measured at pH 7, may not reflect actual nitrification rates in acidic soil at in situ pH. However, potential nitrification rate assessment at this pH value has been utilized in many studies, including those on nitrification in acidic soil (for example, Peterson et al., 2012; Wertz et al., 2012). We used the potential rate to facilitate comparisons with other studies.
Enrichment of ammonia oxidizing bacteria
Soil samples with pH 2.9 and 3.1 were collected from no Ca and high N treatment plots, respectively (Table 1) and used to obtain enrichment cultures. The initial cultures were obtained by suspending 10 g soil in 100 ml Schmidt AOB media (Schmidt and Belser, 1994) containing 100 mM ammonium sulfate at pH 5.5. These cultures were grown at 25 °C with shaking at 110 r.p.m. until high-rates of ammonia oxidation were observed. The cultures were then diluted 10-fold and grown further. Multiple cycles of dilution-to-extinction were performed until no culturable heterotrophic microorganisms were detected. Contaminating microorganisms were assessed by 10-fold diluted nutrient agar. We designed 16S rRNA-specific fluorescent probes for fluorescence in situ hybridization (FISH) analysis and a polymerase chain reaction (PCR) primer set (Supplementary Table S1) for enriched ammonia oxidizing bacteria using retrieved 16S rRNA gene sequences of DNA fragments amplified from the enriched culture using the Bacteria-specific primers 27F and 1492r (Lane, 1991). The final culture purity was evaluated by FISH (Supplementary Methods) and quantitative PCR (qPCR) for the 16S rRNA gene using the designed FISH probe and PCR primer set, respectively. Total bacterial 16S rRNA gene copy number was quantified by qPCR using a Bacteria-specific primer set. AOA ammonia monooxygenase sub-unit A (amoA) and betaproteobacterial AOB (βAOB)-amoA were checked by PCR using their specific primers. The primers and probes used in this study are listed in Supplementary Table S1.
Quantification of amoA genes
Total DNA was extracted from the enriched cultures of ammonia oxidizing bacteria designated as TAO100 by using the Fast DNA spin kit for bacteria (Qbiogene, Inc., Irvine, CA, USA), and the number of copies of the TAO100 amoA gene was quantified by real-time PCR using the SYBR Premix Ex Taq kit (TaKaRa Bio, Inc., Shiga, Japan). We designed a PCR primer set, TAO-amoAF and TAO-amoAR (Supplementary Table S1). TAO100 amoA was amplified using the following cycling conditions: 94 °C for 2 min; 40 cycles of 30 s at 94 °C, 30 s at 58 °C, 45 s at 72 °C; and assessment at 72 °C. A standard curve was constructed using the TAO100 amoA DNA fragment amplified by PCR that had been cloned into pGEM. The detection limit was ~2.5 × 105 copies ml−1 of culture.
Phylogenetic analysis
Genomic DNA was extracted from TAO100 cells using a Fast DNA spin kit for bacteria. Bacterial 16S rRNA genes were PCR-amplified using the bacterial 27F and 1492r primer set. The TAO100 amoA gene was amplified using TAO-amoAF and TAO-amoAR. The PCR products were sequenced using ABI Prism BigDye Terminator cycle sequencing ready reaction kits and an ABI Prism 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA). The phylogenetic analyses were performed using the MEGA 6 software program (Tamura et al., 2013). Average nucleotide identity (ANI) values for 3 whole-genome sequences of strains of the genera Nitrosococcus including N. oceani ATCC 19707, N. halophilus NC4 and N. watosoni C-113, were determined using a web-based service (http://enve-omics.ce.gatech.edu/ani/).
Growth experiments
Ten milliliters of cultures enriched in TAO100 were inoculated into 100 ml Schmidt AOB media modified as described below. Cultures were incubated at 25 °C with rotary shaking at 110 r.p.m. and 1 ml samples were drawn periodically. Subsequently, nitrite and/or ammonium in the supernatant were determined colorimetrically using diazotization and indophenol blue (Keeney and Nelson, 1982), respectively. To assess the effect of pH on growth, media were buffered with 80 mM 2-morpholinoethanesulfonic acid (MES) to pH 5.0, 5.5, 6.0, 6.5 and 7.0 or with 80 mM 4-(2-hydroxyethyl)-1-piperazine-ethanesulphonic acid (HEPES) to pH 7.0, 7.5 and 8.0. To assess the effects of NaCl, media were buffered with 80 mM MES to pH 6.0 and supplemented with 50, 100, 200, 300, 400 and 500 mM NaCl. The effects of NH4+ were assessed in the media with 80 mM MES to pH 6.0 and supplemented with 50, 100, 200, 300, 400 and 500 mM NH4+.
Kinetics of ammonia oxidation
The half-saturation constant (Km) values of ammonia oxidation were determined by estimating the O2 consumption at 25 °C of a suspension of TAO100 cells in a stirred vessel with a Clark-type oxygen electrode. The vessel contained TAO100 cells in 1 ml buffer containing 50 mM MES (pH 6.0) and various concentrations of ammonium sulfate. Km values were calculated using a double-reciprocal plot using Sigma Plot 12.5 (Systat Software, San Jose, CA, USA).
Analysis of conserved chromosome structure and genome-wide phylogenetic analysis
Whole-genome sequencing and annotation of TAO100 were performed as described in Supplementary Methods. The results were used for further analysis. Orthologous groups among the TAO100 and three γ-AOB were constructed using DomClust (Uchiyama, 2006) on the Microbial Genome Database (MBGD) server (Uchiyama et al., 2013). An alignment of syntenically conserved regions, designated the ‘core genome structure,’ was subsequently constructed using CoreAligner (Uchiyama, 2008) based on a consensus chromosomal arrangement conserved among these orthologs. To infer the phylogenetic relationships among TAO100, the three γ-AOB, and nine related strains belonging to order Chromatiales (the specific strains are listed in Supplementary Methods), we first constructed the core genome structure among these genomes using CoreAligner. Among the resulting core genes, 245 were found to be conserved in all 13 strains with one-to-one correspondence. CLUSTALW (Larkin et al., 2007) was used to generate a multiple sequence alignment of these genes, from which blocks suitable for phylogenetic analyses were extracted using Gblocks program (Talavera and Castresana, 2007). The resulting sequence blocks were concatenated to construct an approximately maximum likelihood phylogenetic tree using FastTree (Price et al., 2010).
Evaluation of metabolic and physiological potential
Metabolic and physiological potential harbored in the TAO100 genome were investigated by the Metabolic And Physiological potentiaL Evaluator (MAPLE) (http://www.genome.jp/tools/maple/) to calculate the completion ratio of the functional modules defined by KEGG (Takami et al., 2012, 2016). The module completion ratio (MCR) was calculated based on a Boolean algebra-like equation according to a previously described procedure (Takami et al., 2012). The MCR pattern of the TAO100 was compared with those of other γ-AOB to characterize differences in the physiological potential.
Quantification of amoA genes and their transcripts in soil samples
DNA was extracted from 0.4 g of triplicate soil samples using Fast DNA Spin Kits for soil, as described in previous studies (Morimoto et al., 2011; Takada-Hoshino and Matsumoto, 2004). The extracted DNA was then purified using a Sepharose gel filtration kit (Roche Applied Science, Indianapolis, IN, USA) and an UltraClean 15 DNA Purification Kit (MOBIO Labs, Solana Beach, CA, USA). The abundances of amoA of AOA, β-AOB, and TAO100 were quantified by real-time PCR using the SYBR Premix Ex Taq kit with the primer sets of amoA19F (Leininger et al., 2006) and CrenamoA616r (Tourna et al., 2008), amoA1F (Rotthauwe et al., 1977) and amoA2R (Nicolaisen and Ramsing, 2002), and TAO-amoAF and TAO-amoAR (details in Supplementary Methods).
For transcript analysis, RNA was extracted from the soil as described in a previous report (Wang et al., 2012). A one-step reverse transcription (RT)-qPCR was performed using a QuantiTect SYBR Green RT-PCR Kit (Qiagen, Inc., Valencia, CA, USA) and the StepOnePlus Real-Time PCR System (Applied Biosystems) (details in Supplementary Methods).
Availability of sequences
Sequences obtained in this study were deposited in the DNA Data Bank of Japan (DDBJ) under the accession numbers AP014836, AP014837 and DRA003611.
Results and discussion
Enrichment of acid-tolerant AOB
We used soil samples with pH values of 2.9 and 3.1 (Table 1), which were obtained from experimental plots of tea fields treated with heavy nitrogen fertilization, to isolate acid-tolerant AOB. The amoA gene abundance of AOA and β-AOB was below the detection limit of qPCR although the soil exhibited high nitrification activities. We therefore speculated that unknown acid-tolerant or acidophilic ammonia oxidizers might be involved in nitrification of these tea field soils.
The culture media used for isolation was adjusted to pH 5.5 and supplemented with ammonium at concentrations higher than those used to isolate AOA or AOB in previous studies (Jiang and Bakken, 1999; Tourna et al., 2011; Bollmann et al., 2011). Higher levels of ammonium were used because acidification reduces the concentration of NH3, which is believed to be the preferred substrate (instead of NH4+) for ammonia monooxygenase (Suzuki et al., 1974). Cultures highly enriched in ammonia-oxidizing microorganisms were successfully obtained from the soil samples via serial dilution-to-extinction techniques. The final enrichment culture contained single morphotype large coccus cells. AOA amoA and β-AOB amoA genes were not detected in the culture by PCR with specific primers for these genes. Clones amplified with the bacterial 16S rRNA gene primers 27F and 1492r were of a single sequence type. The 16S rRNA gene sequence showed the highest similarity to those of Nitrosococcus genus. We evaluated the culture purity by FISH and qPCR analysis using fluorescent probes and a PCR primer set specific to the 16S rRNA gene of the enriched AOB designed strain TAO100. FISH analysis showed that the enriched cells comprised >99% of all 4,6-diamidino-2-phenylindole (DAPI) stained cells (Supplementary Figure S1). The qPCR analysis also showed that the 16S rRNA gene copy number of the enriched AOB was equal to that obtained for the total bacteria. Ammonia consumption, nitrite production and the amoA gene abundance were determined to confirm the autotrophic ammonia oxidation and growth of TAO100. Growth of TAO100 was accompanied by a near-stoichiometric conversion of ammonia to nitrite (Figure 1a). To confirm the expression of essential genes required for autotrophic bacterial ammonia oxidation, total mRNA was extracted and analyzed by pyrosequencing (Supplementary Methods). Supplementary Table S2 lists the 23 transcripts with the highest copy numbers in the enrichment culture of TAO100. Genes encoding ammonia monooxygenase, hydroxylamine dehydrogenase and ribulose 1,5-bisphosphate carboxylase oxygenase were abundantly expressed (Supplementary Table S2).
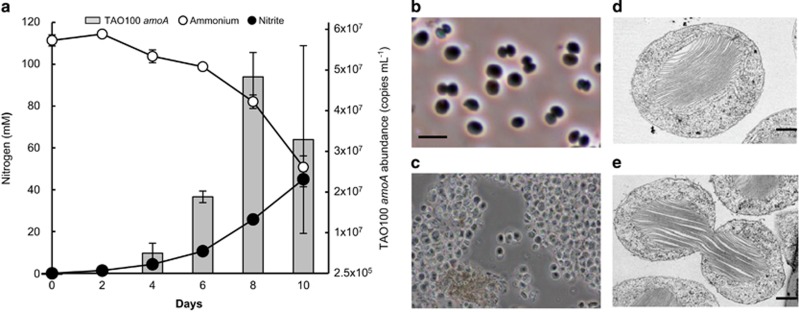
Figure 1.
Growth and morphology of TAO100. (a) Correlation of ammonia oxidation with growth of TAO100. TAO100 was grown in media at pH 6 and containing 100 mM ammonium. Error bars indicate s.d. and appear smaller than symbols in some cases. (b) Phase contrast photomicrograph. Scale bar, 5 μm. (c) Aggregates of TAO100 cells. Scale bar, 10 μm. (d, e) Thin-section electron micrographs of a cell (d) and paired cells (e). Scale bar, 0.5 μm. The method of electron microscopy was as described in Supplementary information.
Phylogenetic analysis and morphological and physiological characteristics
Phylogenetic analysis based on the 16S rRNA gene and AmoA amino acid sequences (Supplementary Figure S2) revealed that TAO100 clusters with γ-AOB of the genus Nitrosococcus, which includes three known species, N. oceani, N. halophilus and N. watosonii. However, the 16S rRNA and amoA gene sequences of TAO100 exhibited 94 and 70% (79% by amino acid sequence) similarity to those of the most closely related species of Nitrosococcus halophilus, respectively. The ANI value between TAO100 and these three species was 75%, which is far below the threshold for species demarcation (95–96%). These low percentages of 16S rRNA gene and amoA gene sequence similarities combined with the ANI value strongly suggest that TAO100 is phylogenetically distant from the species belonging to genus Nitrosococcus. AOB generally falls within the classes Betaproteobacteria or Gammaproteobacteria. Established β-AOB genera, such as Nitrosomonas and Nitrosospira, have been found in various environments such as soils, oceans, lakes and activated sludge (Prosser and Embley, 2002). The Nitrosospira genus has been shown to be the dominant functional AOB in agricultural and forest soils (Kowalchuk and Stephen, 2001). In contrast, γ-AOB of the Nitrosococcus genus thrive only in saline or hyper-saline aquatic environments such as oceans and salt lakes (Campbell et al., 2011). Thus, TAO100 is the first classified Gammaproteobacteria AOB to be isolated from soil.
TAO100 cells in liquid culture were found to be non-motile, spherical or ellipsoidal in shape, and occurred singly or in pairs with diameters ranging from 2 to 3 μm (Figure 1b), and aggregated cells were occasionally observed (Figure 1c). Each cell contained a centrally located extensive intracellular membrane system composed of a stack of flattened membrane vesicles (Figures 1d and e). These cell features are extremely similar to the morphological characteristics of the Nitrosococcus genus. TAO100 grew optimally at 25 °C and also grew at 15 °C, but no growth was observed at 5 or 40 °C.
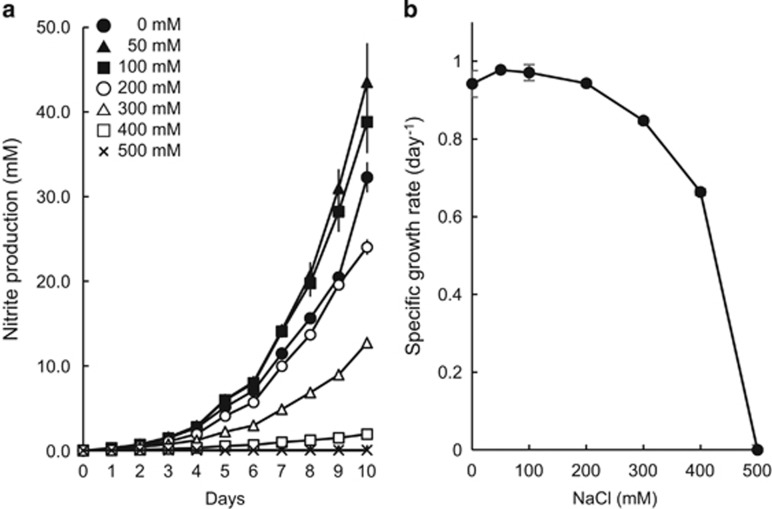
Previously isolated γ-AOB of the Nitrosococcus genus were reported to be obligatory halophiles and to require 500–700 mM NaCl for optimum growth (Campbell et al., 2011; Wang et al., 2016). Although TAO100 grew better at 50 mM NaCl than at 0 mM, TAO100 did not grow in NaCl concentrations greater than 400 mM (Figure 2), indicating that the strain is not halophilic.
Figure 2.
Effect of NaCl on nitrite production. (a) Nitrite production of TAO100 in media at pH 6 supplemented with 0–500 mM NaCl. Growth was estimated from nitrite production. (b) Specific growth rate from log-linear plots of nitrite during exponential growth. Error bars indicate s.d. from triplicate experiments and appear smaller than the symbols in some cases.
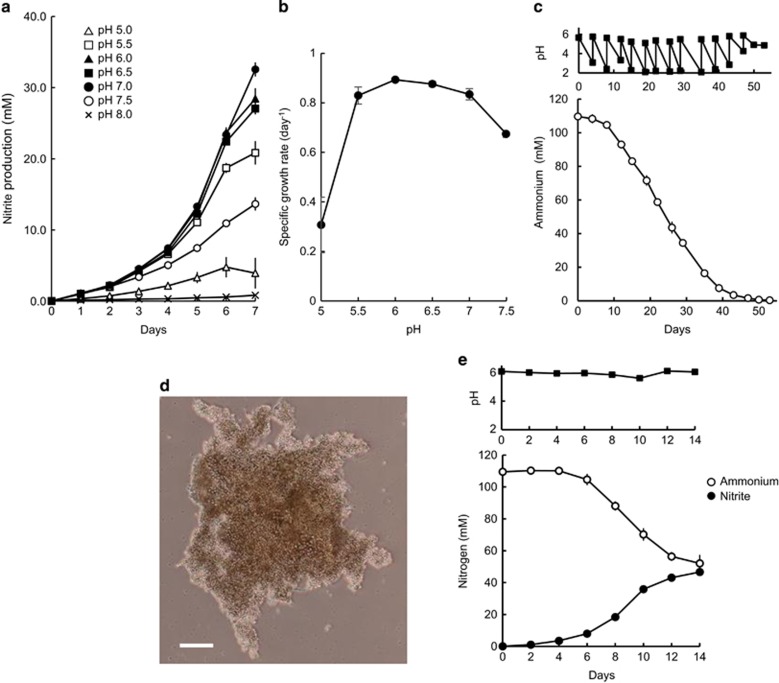
Growth of TAO100 was observed in the pH range of 5–7.5 in medium containing 100 mM ammonium. The optimal pH for growth was 6–6.5 and the specific growth rate was 0.89 day−1 at pH 6 (Figures 3a and b), which is equivalent to the generation time of 27.0 h. The pH of a vigorously growing culture decreased from 6 to ~2 when pH was not controlled (Figure 3c). After the acidified pH of the culture was adjusted to ~6, the cells continued to utilize ammonia. When this process was repeated several times, TAO100 formed visible, large cell aggregates (Figure 3d). When the culture containing these aggregates was inoculated into fresh medium at pH 6 (Figure 3e), TAO100 proliferated as a single cell, indicating that the aggregates themselves are not resistant, but the formation of aggregates might represent a mechanism enabling cells to cope with low pH. Similarly, aggregates formed in acidic conditions by Nitrosospira-like bacteria and Nitrosospira sp. AHB1 isolated from Dutch acid soils were able to nitrify at approximately pH 4 (De Boer et al., 1991). The lowest pH limit for the growth of TAO100 aggregates indicated that TAO100 is tolerant to more highly acidic conditions than β-AOB.
Figure 3.
Effect of pH on ammonia oxidation of TAO100. (a) Ammonia oxidation of TAO100 in media containing 100 mM ammonium at pH 5–8. (b) Specific growth rate from log-linear plots of nitrite during exponential growth. (c) Ammonia oxidation of TAO100 in medium without MES buffer in which pH was periodically adjusted to ~6. Growth was estimated by measuring ammonium oxidation to nitrite. Nitrite levels were not determined because nitric acid produced from ammonium was lost via evaporation under acidic conditions. (d) Visible cell aggregates formed in the acidified culture. Scale bar, 100 μm. (e) Growth of TAO100 in a medium with MES buffer in which pH was maintained continuously at ~6 after inoculation of the cell aggregates. Data were obtained from triplicate experiments. Error bars are s.d. and appear smaller than the symbols in some cases.
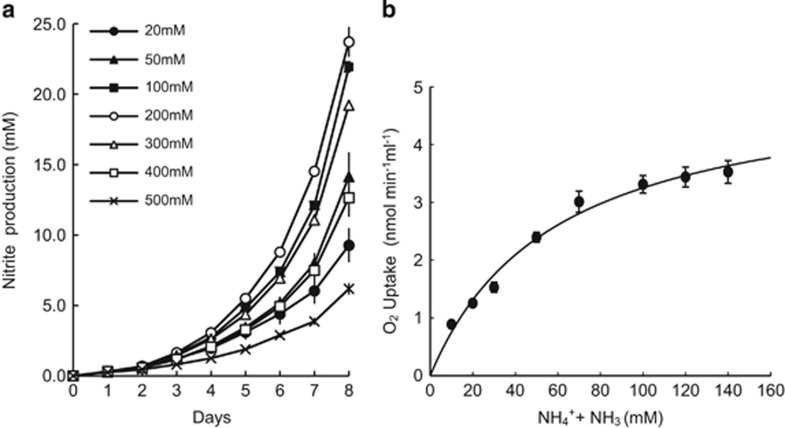
At pH 6, TAO100 grew optimally in ammonium concentrations between 100 and 200 mM (Figure 4a) but could tolerate concentrations up to 500 mM. Thus, TAO100 appears to require an unusually high concentration of ammonium, presumably because NH3 rather than NH4+ is the actual substrate for its ammonia monooxygenase, the concentration of which decreases with acidification. The Km for ammonia (NH3 + NH4+) oxidization was 58.5 mM at pH 6, at which the NH3 concentration was theoretically estimated to be 33.3 μM (Figure 4b). The estimated Km for NH3 was similar to that of Nitrosomonas europaea but higher than those of several Nitrosospira strains (Koper et al., 2010). Overall, TAO100 appeared to have adapted to the soil, which is highly acidic and contains high amounts of ammonium.
Figure 4.
Effect of ammonium concentration on ammonium oxidation to nitrite by TAO100. (a) Nitrite production by TAO100 in media at pH 6 supplemented in various concentrations of ammonium. Data are from triplicate experiments. Error bars indicate s.d. and are smaller than the symbols in some cases. (b) Michaelis–Menten plot of ammonium-dependent oxygen uptake. The Michaelis–Menten equation was used to calculate kinetic parameters. Km for ammonia (NH3+NH4+)=58.5 mM at pH 6, at which the NH3 concentration was theoretically estimated as 33.3 μM.
Genome characteristics
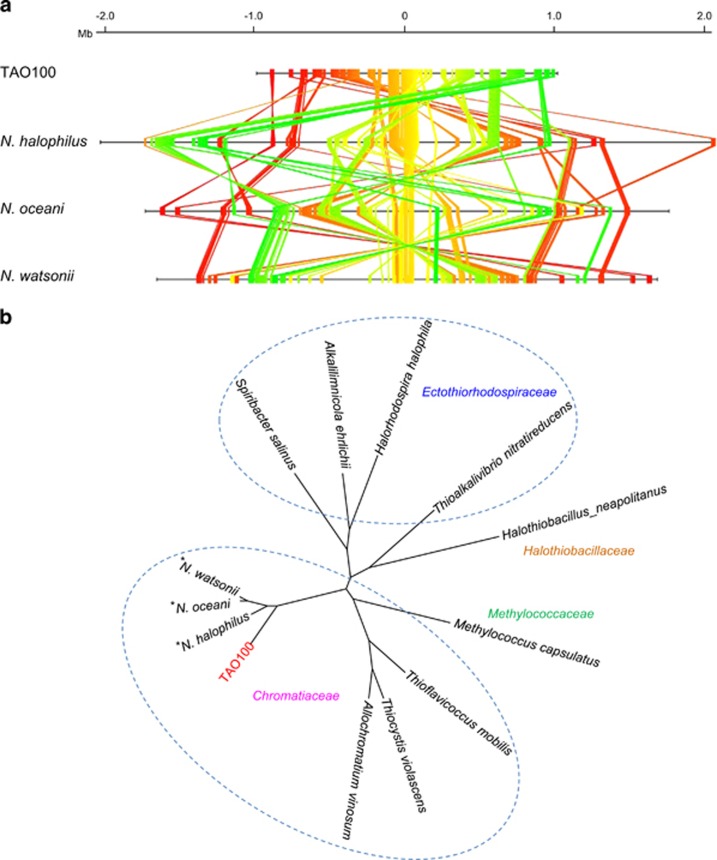
The TAO100 genome consists of a single circular chromosome (2011873 bp) and a plasmid (6441 bp) with mean G+C contents of 42.1% and 40.9%, respectively (Supplementary Figure S3). This chromosome is considerably smaller than the typical Nitrosococcus genus γ-AOB chromosome, which ranges from 3.33 to 4.08 Mb (Klotz and Stein, 2011). The G+C content of the TAO100 chromosome was lower than those of Nitrosococcus, which range from 50.1 to 51.6%. Of the 1839 protein-coding sequences (CDSs) identified in the chromosome, 946 genes (55%) were most similar to those of Nitrosococcus halophilus. In addition, 260 (15%) and 189 genes (11%) were most similar to CDSs in Nitrosococcus oceani and Nitrosococcus watsonii, respectively. We performed ortholog analyses among TAO100 and three other γ-AOB using the MBGD (Uchiyama et al., 2013) and identified 1,381 orthologous genes that were shared among the four strains (Supplementary Figure S4). The proportion of these shared orthologous genes in the TAO100 genome is considerably higher than that in other γ-AOB. Furthermore, we investigated the synteny (‘core genome structure’) conserved in at least two of the four species (Figure 5a; Supplementary Figure S5 and Table S3). The results indicated that TAO100 appears to have lost a number of genes from a common ancestor of γ-AOB through the evolutionary process. Finally, based on a genome-wide phylogenetic analysis using the alignment of concatenated sequences of 245 orthologs shared among TAO100 and 12 other related strains, we found that TAO100 is distinct from the Nitrosococcus genus despite the fact that TAO100 belongs to family Chromatiaceae (Figure 5b).
Figure 5.
Comparison of orthologous genes in TAO100 and related strains. (a) Location of the genes in the core genome structure conserved among TAO100 and three halophilic γ-AOB on each chromosome. The conserved chromosomal structure (‘core genome structure’) was constructed based on consensus arrangement of conserved orthologs. In the core genome structure, an ortholog group is denoted as a colored line. For simplicity, only universally conserved ortholog groups with one-to-one correspondence are presented. To visualize chromosomal rearrangement, a color gradation scale from red to yellow to green is assigned according to location on the TAO100 chromosome. Replication origins are located at the center. (b) Phylogenetic relationships of orthologs common to TAO100, three γ-AOB, and nine related strains from class Gammaproteobacteria. The tree was generated using the concatenated amino acid sequence of 245 orthologous genes conserved in all genomes. Asterisk indicates three halophilic γ-AOB belonging to the genus Nitrosococcus.
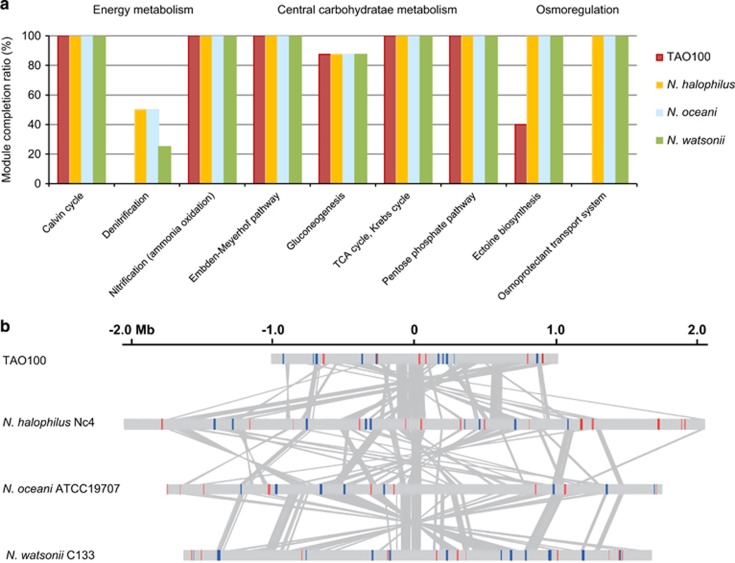
An analysis of the metabolic and physiological potential in the TAO100 genome using MAPLE indicated that the TAO100 genome possesses the elements necessary for establishment of complete pathways responsible for a chemolithoautotrophic lifestyle such as ammonia oxidation and the Calvin cycle (Figure 6a). The organization of the amo operon in TAO100 is identical to that of N. halophilus. On the other hand the gene for nitrite reductase (nirK) is missing from the TAO100 genome. The enzyme required for denitrification is found in all known AOB except Nitrosomonas communis Nm2 (Kozlowski et al., 2016). Kozlowski et al. (2014) proposed that the nirK of AOB might be primarily involved in an ammonia oxidation process rather than nitrite reduction during denitrification. Thus, a comparison of TAO100 with AOB strains having the nirK gene and with N. communis Nm2 might provide useful information to investigate the role of nirK in the ammonia oxidation process. Additionally, although all known AOB utilize polar flagella for motility, flagella-related genes were not identified in the TAO100 genome. Furthermore, ammonia transporters of the Rhesus clade are encoded in many AOB genomes (Offre et al., 2014); however, an ammonia transporter gene was not identified in the TAO100 genome. Similarly, N. oceani and Nitrosomonas eutropha genomes also do not encode known ammonia transporters (Klotz et al., 2006; Stein et al., 2007). TAO100 and these AOB might therefore primarily depend on a passive ammonia diffusion process across the membrane.
Figure 6.
Metabolic and physiological potential encoded in the TAO100 genome. (a) Comparison of MCR patterns in major pathway modules between TAO100 and other γ-AOB. Completion ratio patterns of the modules categorized into energy metabolism, central carbohydrate metabolism and osmoregulation were compared between TAO100 and other major γ-AOB. MCR of each module was calculated by using the MAPLE system. (b) Distribution of glycosyltransferase and cation transporter genes in TAO100 and three γ-AOB. Glycosyltransferase and cation transporter genes are denoted in blue and red, respectively. Orthologous genes found in all four strains are connected with gray lines.
In contrast to the halophilic γ-AOB genome, the TAO100 genome does not encode a gene set for ectoine synthesis or for osmoprotectant transport (Figure 6a), both of which confer salt tolerance (Ventosa et al., 1998). In addition, fewer genes were found to be associated with sodium transport, which would also likely confer halophilicity in the TAO100 genome. These genomic features are consistent with a nonhalophilic TAO100 classification, as indicated by the cultivation experiments. TAO100 therefore appears to have lost a number of genes critical for survival in oceans or salt lakes after diverging away from a common ancestor of halophilic γ-AOB. In contrast, the gene cluster encoding urea hydrolase is present in the TAO100 genome. This finding is consistent with the importance of ureolysis for survival in acidic soil (Burton and Prosser, 2001; De Boer et al., 1989). Cation transporter and glycosyltransferase genes, both potentially associated with acid tolerance (Krulwich et al., 2011; Stack et al., 2010), were also identified in the TAO100 genome (Figure 6b). Furthermore, glycosyltransferase has been implicated in exopolysaccharide synthesis and the formation of cell aggregates and biofilms, which confer stress tolerance including acid tolerance (Krulwich et al., 2011). However, because these genes have also been identified in the acid-sensitive, neutrophilic γ-AOB genome, further study is required to clarify the molecular mechanism of acid tolerance in TAO100.
Contribution of TAO100 to nitrification in tea field soil
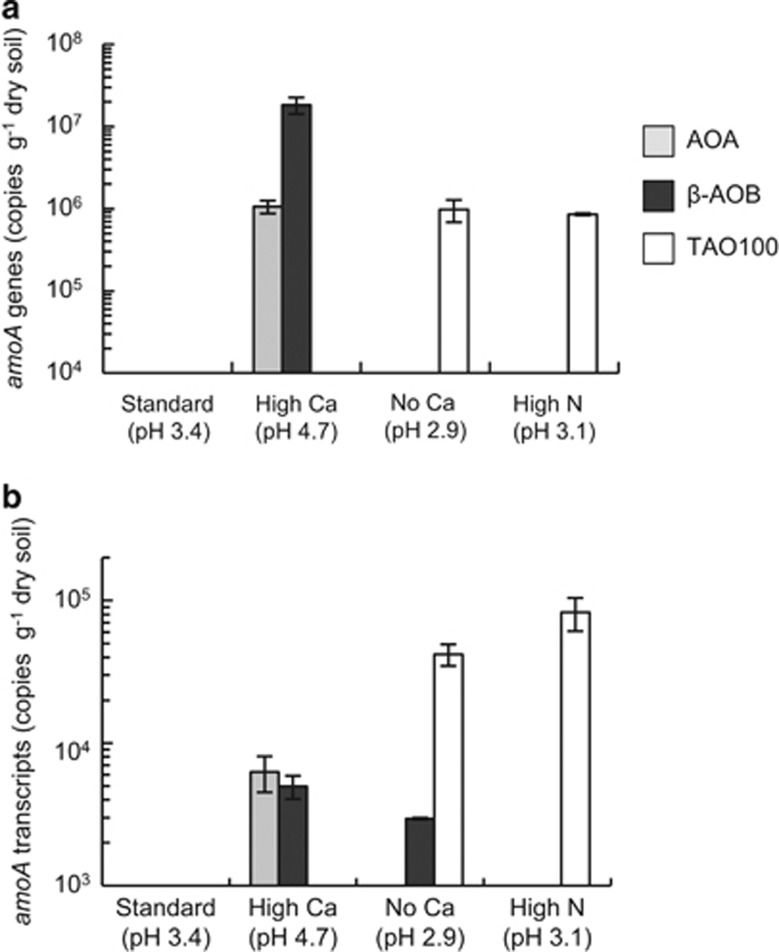
To evaluate the contribution of TAO100 to nitrification in the used soil samples (Table 1), the abundance of amoA genes and their transcript levels were measured in the soils by qPCR using primers specific for TAO100, β-AOB and AOA (Figure 7). In the pH 2.9 and 3.1 soil samples, the TAO100 amoA gene and its transcript were more abundant compared with those of AOA and β-AOB. In contrast, AOA and β-AOB amoA genes and their transcripts were more abundant in pH 4.7 soil. The results indicated that TAO100 may be the primary group of ammonia oxidizers in very acidic soil. High-nitrogen application and the strongly acidic conditions of tea field soils might therefore have resulted in preferential growth of TAO100. On the other hand, the soil pH values of high N and no Ca were lower than that of the limit of growth of TAO100 in pure culture. Further experimentation such as monitoring the growth of TAO100 and measuring the actual nitrification rates using the acidic soil microcosms would be required to elucidate the role and validate contribution of TAO100 to nitrification in acidic soils.
Figure 7.
Abundances of amoA genes and amoA transcripts in the used soil samples. (a) amoA gene abundances of AOA, β-AOB and TAO100 in the soil samples. (b) amoA transcript abundances of AOA, β-AOB and TAO100 in the soil samples. Abundance is expressed as gene and transcript copy numbers per g of dry soil. No column indicates that the copy numbers of amoA gene and its transcript were below the detection limit of the qPCR assays. Error bars represent standard errors for triplicate samples. Soil pH is indicated in parentheses. Standard, High N, High Ca and No Ca indicate the fertilization conditions of the sampling fields (Table 1).
We next screened 2212 whole genome shotgun sequencing data sets from soil, sediments and wastewater treatment plants available on the Metagenomics Rapid Annotation using Subsystem Technology (MG-RAST) server (Meyer et al., 2008) (Supplementary Methods). However no sequence read was affiliated with TAO100-amoA indicating that TAO100-amoA related γ-AOB amoA appeared to be absent or extremely rare in the soils used for the metagenomic analysis. Ultradeep sequencing is required to detect rare species in soil samples necessitating an estimated 50 Tbp for an individual gram of soil (Gans et al., 2005). As few metagenome data sets of acidic and high-ammonium content soils such as that of the studied tea field are available, further metagenomic surveys of acidic soil are required to evaluate the distribution of TAO-related γ-AOB in soil and other environments.
In summary, evidence from genomic, phenotypic and also ecological analyses indicate that strain TAO100 of γ-AOB is uniquely adapted to acidic conditions and could contribute significantly to nitrification in acidic soil. To date, no strains of γ-AOB except TAO100 have been isolated and characterized from terrestrial ecosytems. This study suggests that γ-AOB are more phylogenetically and physiologically diverse than previously observed. In support of this, recently, 16S rRNA gene sequences of Nitrosococcus genera were detected in moderately acidic pristine soil (Wang et al., 2015), indicating the existence and distribution of γ-AOB in soil ecosystems. As such, the isolation and characterization of γ-AOB from soil can provide useful information to further our understanding of nitrification in acidic soil.
The differentiation of TAO100 from its closely related genus Nitrosococcus was confirmed by phylogenetic analysis based on 16S rRNA gene and amoA gene sequences, genome-wide analyses, and physiological features. These results support the classification of TAO100 into a separate genus and species from Nitrosococcus. We propose the new name Nitrosoglobus terrae gen. nov., sp. nov., and the following candidate status.
Description of Candidatus Nitrosoglobus terrae
The candidate status of strain TAO100 comprises Nitrosoglobus terrae, (Ni.tro.so.glo’bus. N.L. adj.), with the following etymology: nitrosus, full of natron; here intended to mean nitrous; L. masc. n. globus, a ball; N.L. masc. n. Nitrosoglobus, the globus producing nitrite: ter’rae. L. gen. n. terrae, soil, where the type strain was isolated.
Cells are large cocci or very short rods with 2.0–3.0 μm diameters. Cells contained a centrally located and well-developed intracytoplasmic membrane system composed of a stack of flattened membrane vesicles. This species is an obligate chemolithotroph and oxidizes ammonia to nitrite. Cells are nonmotile and the genes involved in flagellum synthesis have not been identified. The optimum growth temperature is ~25 °C and growth is observed between 15 and 35 °C. The optimum pH for growth is between 6 and 6.5. The growth is confirmed acidic conditions with a pH range between 5 and 6. Aggregated cells are formed in strongly acidic conditions (pH 2–4). Cell aggregates are acid-resistant to pH 2. The optimum ammonium concentration for growth is 100 mM at pH 6. A urease operon was identified in the genome and cells have urease activity. Cell growth is better at 50 mM NaCl than at 0 mM and does not occur at concentrations over 400 mM NaCl. The GC content of the chromosome DNA is 42.1%.
Acknowledgments
We would like to thank the staff of the Comparative Genomics Laboratory at NIG for their support in genome sequencing. This work was financially supported by the Bio-oriented Technology Research Advancement Institution, Japan; the Funding Program for Next Generation World-Leading Researchers, Ministry of Education Culture, Sports, Science and Technology ‘Program to Disseminate Tenure Tracking System; KAKENHI Grant Numbers 26310315, 26292184 and 16K14874; and the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (28004A). We would also like to thank Dr Koji Mori (National Institute of Technology and Evaluation, Japan) for nomenclature advice.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Allison SM, Prosser JI. (1993). Ammonia oxidation at low pH by attached populations of nitrifying bacteria. Soil Biol Biochem 25: 935–941. [Google Scholar]
- Belser LW, Mays EL. (1980). Specific inhibition of nitrite oxidation by chlorate and its use in assessing nitrification in soils and sediments. Appl Environ Microbiol 39: 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollmann A, French E, Laanbroek HJ. (2011). Isolation, cultivation, and characterization of ammonia-oxidizing bacteria and archaea adapted to low ammonium concentrations. Methods Enzymol 486: 55–88. [DOI] [PubMed] [Google Scholar]
- Booth MS, Stark JM, Rastetter E. (2005). Controls on nitrogen cycling in terrestrial ecosystems: a synthetic analysis of literature data. Ecol Monogr 75: 139–157. [Google Scholar]
- Burton SAQ, Prosser JI. (2001). Autotrophic ammonia oxidation at low pH through urea hydrolysis. Appl Environ Microbiol 67: 2952–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MA, Chain PS, Dang H, Sheikh AF El, Norton JM, Ward NL et al. (2011). Nitrosococcus watsonii sp. nov., a new species of marine obligate ammonia-oxidizing bacteria that is not omnipresent in the world's oceans: calls to validate the names 'Nitrosococcus halophilus' and 'Nitrosomonas mobilis'. FEMS Microbiol Ecol 76: 39–48. [DOI] [PubMed] [Google Scholar]
- Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 52: 504–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer W, Duyts H, Laanbroek HJ. (1989). Urea stimulated autotrophic nitrification in suspensions of fertilized, acid heath soil. Soil Biol Biochem 21: 349–354. [Google Scholar]
- De Boer W, Gunnewiek PJ, Veenhuis M, Bock E, Laanbroek HJ. (1991). Nitrification at low pH by aggregated autotrophic bacteria. Appl Environ Microbiol 57: 3600–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer W, Kowalchuk GA. (2001). Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol Biochem 33: 853–866. [Google Scholar]
- Di HJ, Cameron KC, Shen JP, Winefield CS, O’Callaghan M, Bowatte S et al. (2009). Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2: 621–624. [Google Scholar]
- Gans J, Wolinsky M, Dunbar J. (2005). Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309: 1387–1390. [DOI] [PubMed] [Google Scholar]
- Hayatsu M. (1993). The lowest limit of pH for nitrification in tea soil and isolation of an acidophilic ammonia oxidizing bacterium. Soil Sci Plant Nut 39: 219–226. [Google Scholar]
- Hayatsu M, Kosuge N. (1993). Autotrophic nitrification in acid tea soils. Soil Sci Plant Nut 39: 209–217. [Google Scholar]
- Jia Z, Conrad R. (2009). Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol 11: 1658–1671. [DOI] [PubMed] [Google Scholar]
- Jiang QQ, Bakken LR. (1999). Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol Ecol 30: 171–186. [DOI] [PubMed] [Google Scholar]
- Keeney DR, Nelson DW. (1982) Nitrogen–inorganic forms. In: Page AL et al (ed.). Methods of Soil Analysis: Part 2. Agronomy Monogr. 9, 2nd ed. ASA and SSSA: Madison, WI, pp 643–687. [Google Scholar]
- Klotz MG, Arp DJ, Chain PSG, El-Sheikh AF, Hauser LJ, Hommes NG et al. (2006). Complete genome sequence of the marine, chemolithoautotrophic, ammonia-oxidizing bacterium Nitrosococcus oceani ATCC 19707. Appl Environ Microbiol 72: 6299–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz MG, Stein LY. (2011) Genomic of ammonia-oxidizing bacteria and insights into their evolution. In: Ward BB, Arp DJ, Klotz MG (eds). Nitrification. ASM Press: Washington DC, pp 57–94. [Google Scholar]
- Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546. [DOI] [PubMed] [Google Scholar]
- Koper TE, Stark JM, Habteselassie MY, Norton JM. (2010). Nitrification exhibits Haldane kinetics in an agricultural soil treated with ammonium sulfate or dairy-waste compost. FEMS Microbiol Ecol 74: 316–322. [DOI] [PubMed] [Google Scholar]
- Kowalchuk GA, Stephen JR. (2001). Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol 55: 485–529. [DOI] [PubMed] [Google Scholar]
- Kozlowski JA, Kits KD, Stein LY. (2016). Genome sequence of Nitrosomonas communis strain Nm2, a mesophilic ammonia-oxidizing bacterium isolated from Mediterranean soil. Genome Announc 4: e01541–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski JA, Price J, Stein LY. (2014). Revision of N2O-producing pathways in the ammonia-oxidizing bacterium Nitrosomonas europaea ATCC 19718. Appl Environ Microbiol 80: 4930–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krulwich TA, Sachs G, Padan E. (2011). Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9: 330–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ. (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (ed.). Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons: Chichester, UK, pp 115–175. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. (2011). Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci USA 108: 15892–15897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtovirta-Morley LE, Ge C, Ross J, Yao H, Nicol GW, Prosser JM. (2014). Characterisation of terrestrial acidophilic archaeal ammonia oxidisers and their inhibition and stimulation by organic compounds. FEMS Microbiol Ecol 89: 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809. [DOI] [PubMed] [Google Scholar]
- Long X, Chen CR, Xu ZH, Oren R, He JZ. (2012). Abundance and community structure of ammonia-oxidizing bacteria and archaea in a temperate forest ecosystem under ten-years elevated CO2. Soil Biol Biochem 46: 163–171. [Google Scholar]
- Meyer F, Paarman D, D’Souza M, Olson R, Glass EM, Kubal M et al. (2008). The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinf 9: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto S, Hayatsu M, Takada-Hoshino Y, Nagaoka K, Yamazaki M, Karasawa T et al. (2011). Quantitative analyses of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in fields with different soil types. Microbes Environ 26: 248–253. [DOI] [PubMed] [Google Scholar]
- Nicolaisen MH, Ramsing NB. (2002). Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J Microbiol Methods 50: 189–203. [DOI] [PubMed] [Google Scholar]
- Offre P, Kerou M, Spang A, Schleper C. (2014). Variability of the transporter gene complement in ammonia-oxidizing archaea. Trends Microbiol 22: 665–675. [DOI] [PubMed] [Google Scholar]
- Petersen DG, Blazewicz SJ, Firestone M, Herman DJ, Turetsky M, Waldrop M. (2012). Abundance of microbial genes associated with nitrogen cycling as indices of biogeochemical process rates across a vegetation gradient in Alaska. Environ Microbiol 14: 993–1008. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser JI, Embley TM. (2002). Cultivation-based and molecular approaches to characterization of terrestrial and aquatic nitrifiers. Antonie Van Leeuwenhoek 81: 165–179. [DOI] [PubMed] [Google Scholar]
- Rotthauwe JH, Witzel KP, Liesack W. (1977). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63: 4704–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger WH. (2009). On the fate of anthropogenic nitrogen. Proc Natl Acad Sci USA 106: 203–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EL, Belser LW. (1994) Autotrophic nitrifying bacteria. In: Weaver RW, Angle S, Bottomley P, Bezdicek D, Smith S, Tabatabai A et al (eds). Methods of Soil Analysis, Part 2, Microbiological and Biochemical Properties SSSA Book Series No. 5 Soil Science Society of America Inc.: Madison, WI, pp 159–177. [Google Scholar]
- Stack HM, Kearney N, Stanton C, Fitzgerald GF, Ross RP. (2010). Association of beta-glucan endogenous production with increased stress tolerance of intestinal lactobacilli. Appl Environ Microbiol 76: 500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LY, Daniel JA, Paul MB, Patrick SGC, Loren H, Mike SMJ et al. (2007). Whole-genome analysis of the ammonia-oxidizing bacterium, Nitrosomonas eutropha C91: implications for niche adaptation. Environ Microbiol 9: 2993–3007. [DOI] [PubMed] [Google Scholar]
- Suzuki I, Dular U, Kwok SC. (1974). Ammonia or ammonium ion as substrate for oxidation by Nitrosomonas europaea cells and extracts. J Bacteriol 120: 556–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada-Hoshino Y, Matsumoto N. (2004). An improved DNA extraction method using skim milk from soils that strongly adsorb DNA. Microbes Environ 19: 13–19. [Google Scholar]
- Takami H, Taniguchi T, Moriya Y, Kuwahara T, Kanehisa M, Goto S. (2012). Evaluation method for the potential functionome harbored in the genome and metagenome. BMC Genomics 13: 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami H, Taniguchi T, Arai W, Takemoto K, Moriya Y, Goto S. (2016). An automated systemfor evaluation of the potential functionome: MAPLE version 2.1.0. DNA Res 23: 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56: 564–577. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourna M, Freitag TE, Nicol GW, Prosser JI. (2008). Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol 10: 1357–1364. [DOI] [PubMed] [Google Scholar]
- Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA 108: 8420–8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama I. (2006). Hierarchical clustering algorithm for comprehensive orthologous-domain classification in multiple genomes. Nucleic Acids Res 34: 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama I. (2008). Multiple genome alignment for identifying the core structure among moderately related microbial genomes. BMC Genomics 9: 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama I, Mihara M, Nishide H, Chiba H. (2013). MBGD update 2013: the microbial genome database for exploring the diversity of microbial world. Nucleic Acids Res. 41: 631–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B et al. (2015). Complete nitrification by a single microorganism. Nature 52: 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventosa A, Nieto JJ, Oren A. (1998). Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev 62: 504–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang NF, Zhang T, Zhang F, Wang ET, He JF, Ding H et al. (2015). Diversity and structure of soil bacterial communities in the Fildes Region (maritime Antarctica) as revealed by 454 pyrosequencing. Front Microbiol 6: 1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nagaoka K, Hayatsu M, Sakai Y, Tago K, Asakawa S et al. (2012). A novel method for RNA extraction from Andosols using casein and its application to amoA gene expression study in soil. Appl Microbiol Biotechnol 96: 793–802. [DOI] [PubMed] [Google Scholar]
- Wang L, Lim CK, Dang H, Hanson TE, Klotz MG. (2016). D1FHS, the type strain of the ammonia-oxidizing bacterium Nitrosococcus wardiae spec. nov.: enrichment, isolation, phylogenetic, and growth physiological characterization. Front Microbiol 7: 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz S, Leigh AKK, Grayston SJ. (2012). Effects of long-term fertilization of forest soils on potential nitrification and on the abundance and community structure of ammonia oxidizers and nitrite oxidizers. FEMS Microbiol Ecol 79: 142–154. [DOI] [PubMed] [Google Scholar]
- Wrage N, Velthof GL, van Beusichem ML, Oenema O. (2001). Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol Biochem 33: 1723–1732. [Google Scholar]
- Zhang LM, Hu HW, Shen JP, He JZ. (2012). Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6: 1032–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.