Abstract
At the high concentrations used in medicine, antibiotics exert strong selection on bacterial populations for the evolution of resistance. However, these lethal concentrations may not be representative of the concentrations bacteria face in soil, a recognition that has led to questions of the role of antibiotics in soil environments as well as the dynamics of resistance evolution during sublethal challenge. Here we examine the evolution of resistance to sub-minimal inhibitory concentrations (sub-MIC) of streptomycin in the filamentous soil bacterium Streptomyces coelicolor. First, we show that spontaneous resistance to streptomycin causes an average fitness deficit of ~21% in the absence of drugs; however, these costs are eliminated at concentrations as low as 1/10 the MIC of susceptible strains. Using experimental evolution, we next show that resistance to >MIC levels of streptomycin readily evolves when bacteria are exposed to sub-MIC doses for 500 generations. Furthermore, the resistant clones that evolved at sub-MIC streptomycin concentrations carry no fitness cost. Whole-genome analyses reveal that evolved resistant clones fixed some of the same mutations as those isolated at high drug concentrations; however, all evolved clones carry additional mutations and some fixed mutations that either compensate for costly resistance or have no associated fitness costs. Our results broaden the conditions under which resistance can evolve in nature and suggest that rather than low-concentration antibiotics acting as signals, resistance evolves in response to antibiotics used as weapons.
Introduction
Because of their lethal effects on target bacteria, antibiotics exert strong natural selection on bacterial populations for the evolution of resistance (Andersson and Levin, 1999). At the high concentrations used in clinical environments, antibiotic-resistant clones can rapidly increase in frequency because these strains gain an absolute advantage compared with their susceptible counterparts (Baquero et al., 2009). However, it is likely that these high concentrations, above the so-called mutant selection window (Drusano, 2004), represent an extreme of the drug concentrations bacteria naturally experience (Davies et al., 2006). Drug concentrations within patients can vary markedly through time and across body sites because of the difference in drug penetrance, excretion or metabolism (Negri et al., 2000; Gullberg et al., 2011). Equally, in the natural environment, where bacteria are exposed to antibiotics from anthropogenic sources as well as endogenous antibiotics produced by bacteria and fungi, bacteria may experience a broad range of drug concentrations (Thiele-Bruhn, 2003; Andersson and Hughes, 2014). For example, exposure to anthropogenic sources of antibiotics will be greatest near the point of contamination and declines with distance from this source. And although the overall drug concentrations because of endogenous sources are likely low (Yim et al., 2006), gradients in concentrations are anticipated as a function of the distance from these antibiotic-producing microbes. While decades of research have unraveled the dynamics of the evolution of antibiotic resistance at high drug concentrations, scarcely little is understood of the emergence of resistance at the low concentrations that are more reflective of natural values (Hughes and Andersson, 2012). What are the dynamics of resistance evolution at low antibiotic concentrations outside the traditional mutant selective window? And if resistance evolves, is it associated with the same pleiotropic costs borne by clones that evolve resistance after exposure to high drug concentrations? Here we address these questions with a focus on the evolution of streptomycin resistance in the environmental bacterium Streptomyces coelicolor.
Streptomycetes produce a wide range of natural products, including some 50% of all known antibiotics (Hopwood, 2007; Barka et al., 2016), and are also well-known environmental reservoirs of anti-microbial resistance (D’Costa et al., 2006b); they are therefore ideal organisms for this study.
Pharmacodynamic models assume that drug-resistant mutants are selected when antibiotic concentrations fall into a specific range known as the mutant selection window (Drusano, 2004; Drlica and Zhao, 2007; Andersson and Hughes, 2014). This traditional selective window encompasses the antibiotic concentrations between the minimal inhibitory concentration (MIC) of the susceptible strain and the MIC of the resistant strain (Drlica and Zhao, 2007). However, while this model correctly identifies the MIC as the threshold where resistant cells persist and susceptible cells die, it fails to account for the fact that below the MIC the two cell types are not otherwise competitively equivalent (Andersson and Hughes, 2014). Indeed, susceptible cells can be significantly harmed by non-lethal, sub-MIC, antibiotics and these negative effects on growth can markedly increase the range of drug concentrations where resistant cells are selected (Gullberg et al., 2011). Of equal importance, the antibiotic concentration where resistance evolves can have crucial implications for the type of resistance that evolves (Baquero, 2001; Andersson and Hughes, 2014).
While antibiotic resistance that evolves at high concentrations often has a significant cost in terms of bacterial fitness (Andersson and Levin, 1999), recent studies have predicted that this cost will not be evident for resistance that emerges at low drug concentrations (Gullberg et al., 2011; Andersson and Hughes, 2014; Chow et al., 2015). The reasons for this can be intuitively explained as follows: while resistant cells above the MIC gain an absolute fitness advantage against susceptible strains, below the MIC, resistant cells and susceptible cells will compete with one another. Accordingly, the success of resistant strains below the MIC will be determined both by their ability to withstand the effects of drug exposure and also their intrinsic competitiveness relative to susceptible cells. Strains with costly resistance may therefore fail to outcompete susceptible strains, whereas strains with no-cost resistance will thrive. As a consequence of these lower costs, it is furthermore predicted that resistance that evolves at sub-MIC antibiotic concentrations will persist when growing in environments without drugs, whereas strains with costly resistance may be outcompeted (Andersson and Hughes, 2011).
Our aims here are to quantify the concentration-dependent fitness effects of spontaneous streptomycin resistance in S. coelicolor. Streptomycin is an aminoglycoside antibiotic that is produced in the soil by the natural antibiotic producer Streptomyces griseus (Schatz et al., 1944); although difficult to quantify directly, it is believed that streptomycin concentrations in soil are extremely low, raising questions about the role of this antibiotic in nature for the bacteria that produce it (Laskaris et al., 2011). It has even been argued that because antibiotics at such low, non-lethal, concentrations are insufficient to select for resistance, these secondary metabolites are better viewed as signals than as weapons (Yim et al., 2006, 2007; Davies and Davies, 2010). The results of the present work fail to support this perspective. We first show that the frequency of streptomycin resistance among natural bacterial isolates is relatively high. Next, we show that while resistance that evolves at high concentrations of antibiotics is highly costly, resistance evolving at sub-MIC drug concentrations over 500 generations is cost-free. We discuss the implications of these results for understanding the evolution and persistence of resistant bacterial strains in nature and also for understanding the roles of antibiotics in natural environments.
Materials and methods
Bacterial strains and culturing conditions
Two S. coelicolor strains were used in this study: S. coelicolor A3(2) M145 (designated WT) and S. coelicolor A3(2) M145 Apra, an isogenic strain carrying an integrated pSET152 plasmid conferring apramycin resistance (designated WTApr). The MIC of streptomycin for both ancestral strains is 2 μg ml−1, indicating that there is no cross-resistance between apramycin and streptomycin (methods for MIC determination are outlined below). Strains were routinely grown at 30 °C on Soy Flour Mannitol Agar (SFM) containing 20 g Soy Flour (Biofresh Belgium, Onze-Lieve-Vrouw-Waver, Belgium), 20 g Mannitol (Merck KGaA, Darmstadt, Germany) and 15 g agar (Hispanagar, Burgos, Spain) per liter (pH 7.2–7.4). To generate high-density spore stocks, plates were uniformly spread with 50 μl of spore-containing solution. After 3–4 days of growth, spores were harvested with a cotton disc soaked in 3 ml of 30% glycerol, and then spores were extracted from the cotton by passing the liquid through an 18 g syringe to remove the vegetative mycelium. Resulting spore stocks were titred and stored at −20 °C. Growth rates were estimated on SFM plates by inoculating plates with ~105 spores and then harvesting after 3 and 4 days of growth. This resulted in ~1.67 × 109 and 5.97 × 109 spores, respectively, corresponding to 14 and 16 elapsed generations in total.
MIC testing
The MIC for streptomycin of laboratory isolates was determined according to the EUCAST (European Committee of Anti-microbial Susceptibility Testing) protocol (Kahlmeter et al., 2003). MICs were estimated by spotting ~104 spores on SFM plates containing 0, 2, 4, 6, 8, 12, 16, 24, 32, 48, 64, 92, 128, 192 and 256 μg ml−1 streptomycin sulfate (Sigma, St Louis, MO, USA). Plates were incubated at 30 °C for 4 days. The MIC was set to the lowest concentration of antibiotic yielding no visible growth. To investigate the level of streptomycin resistance in nature, we determined the MIC of a collection of 85 Streptomyces strains isolated from soil collected from the Himalaya in Nepal and Qinling Mountains in China (Zhu et al., 2014). MICs were estimated as described above by spotting 1 μl of a 100-fold diluted spore stock.
Spontaneous streptomycin resistance
Spontaneous streptomycin-resistant clones were isolated from the WT strain by plating 109 spores onto SFM agar containing 2, 4, 8 or 16 μg ml−1 streptomycin. After 2–3 days of growth, random single colonies were selected from independent plates from each streptomycin concentration and then restreaked onto a plate containing the same concentration of streptomycin as the selection plate. Spore stocks of these single colonies were collected as outlined above and stored at −20 °C.
Experimental evolution at sub-MIC streptomycin
To investigate the evolution and costs of streptomycin resistance at sub-MIC concentrations of streptomycin, we serially transferred six replicate populations for ~500 generations on plates containing 0.2 μg ml−1 streptomycin. This value corresponds to the minimum estimate of the minimal selective concentration (MSC) for spontaneous resistant clones and is ~1/10 the MIC of the susceptible parent strain. Replicate populations, initiated from independent colonies, were grown for either 3 (14 generations) or 4 days (16 generations), after which spores were harvested as above, and then replated at a density of ~105 spores per plate. Experimental populations were stored at −20 °C after every transfer. After ~332 generations, replicates of all six populations were in addition serially transferred to plates containing 0.4 μg ml−1 streptomycin, leading to a total of 12 populations. To quantify the evolution of streptomycin resistance through time, we plated 105 spores of all evolved populations at 50-generation intervals onto SFM supplemented with 2 μg ml−1 of streptomycin. Resistant colonies were scored after 6 days of growth. After ~500 generations a single random resistant colony was isolated from each 0.2 μg ml−1 population to be used to quantify the fitness of evolved resistant clones. This same clone was subsequently sequenced.
Fitness assays
To assess the fitness of the spontaneous and evolved streptomycin-resistant strains, we carried out head-to-head competition experiments between evolved clones and ancestral clones that were differentially marked with an apramycin resistance cassette (Lenski et al., 1991). Costs of resistance were quantified by competing strains in the absence of streptomycin, whereas the MSC of resistant clones was determined by competing strains in the presence of 0, 0.125, 0.25, 0.5 and 1.0 μg ml−1 streptomycin (susceptible clones at or above the MIC were fully displaced). Competition assays were initiated by mixing strains 1:1 and then plating 105 total spores onto SFM at the indicated streptomycin concentration. To determine the fraction of the inoculum that was apramycin resistant or sensitive, we simultaneously plated a 10−3 dilution of this mix on SFM and SFM containing 50 μg ml−1 apramycin sulfate (Duchefa Biochemie, Haarlem, The Netherlands). After 4 days of growth at 30 °C, the plates were harvested and the number of each competitor quantified following plating on SFM agar plates with or without 50 μg ml−1 apramycin. Control assays between WT and WTApr ancestral clones were used to correct for any fitness effects associated with the apramycin marker. Following Lenski et al. (1991), relative fitness was calculated as the ratio of the Malthusian parameters of both strains: 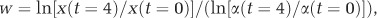 , where x is the competing streptomycin-resistant strain and α is the wild-type or ancestral control strain and t is the time in days of growth after inoculation. For determination of the MSC, the selection rate constant (r) was used to define relative fitness, where instead of the ratio, we calculated the difference in the Malthusian parameters of both strains (Travisano and Lenski, 1996). Selection rate constant was used to control for the fact that under antibiotic exposure one or both competing clones may decline in density during the course of the assay. The MSC was estimated as the antibiotic concentration where both strains have equal selection rate constants (Gullberg et al., 2011).
, where x is the competing streptomycin-resistant strain and α is the wild-type or ancestral control strain and t is the time in days of growth after inoculation. For determination of the MSC, the selection rate constant (r) was used to define relative fitness, where instead of the ratio, we calculated the difference in the Malthusian parameters of both strains (Travisano and Lenski, 1996). Selection rate constant was used to control for the fact that under antibiotic exposure one or both competing clones may decline in density during the course of the assay. The MSC was estimated as the antibiotic concentration where both strains have equal selection rate constants (Gullberg et al., 2011).
DNA extraction and sequencing
Streptomycetes to be sequenced were grown in liquid culture containing 50% YEME (yeast extract/malt extract)/50% TSBS (trypticase soy broth-starch) with 5 mm MgCl2 and 0.5% glycine incubated at 30 °C and shaken at 250 r.p.m. for 2 days. After centrifugation, the pellet was resuspended in TEG (Tris buffer, EDTA and glucose) buffer with 1.5 mg ml−1 lysozyme, and after 1 h of incubation at 30 °C, the reaction was stopped by adding 0.5 volume of 2 m NaCl. DNA was extracted using standard phenol–chloroform extraction, followed by DNA precipitation and washing in isopropanol and 96% ethanol. Dried DNA was resuspended in MQ water and then treated with 50 μg ml−1 of RNase and incubated at 37 °C for 1 h. Following RNase treatement, the mixture was purified and cleaned as above, after which the purified DNA was washed with 70% ethanol and resuspended in MQ water. The genomes of the spontaneous and evolved clones as well as those of their ancestral strains were sequenced on the Illumina HiSeq4000 (Illumina, Singapore, Malaysia) with paired-end 150 bp reads at the Leiden Genome Technology Center (LGTC). All samples were prepped with an amplification-free prep (KAPA Hyper Kit, Kapa Biosystems, Cape Town, South Africa) after Covaris shearing of the DNA.
Sequence analysis
All genomes were assembled to the S. coelicolor A3(2) genome sequence available from the NCBI database (http://www.ncbi.nlm.nih.gov/assembly/GCF_000203835.1/) using Geneious 9.1.4. The ‘Find variation/single-nucleotide polymorphism (SNP)’ tool in Geneious was used to identify SNPs and indels with a minimum sequencing coverage of 10 and a variant frequency of at least 50%. Unique mutations in the spontaneous and evolved resistant strains were identified by direct comparison with the ancestral strains.
Results
Streptomycin resistance among natural isolates
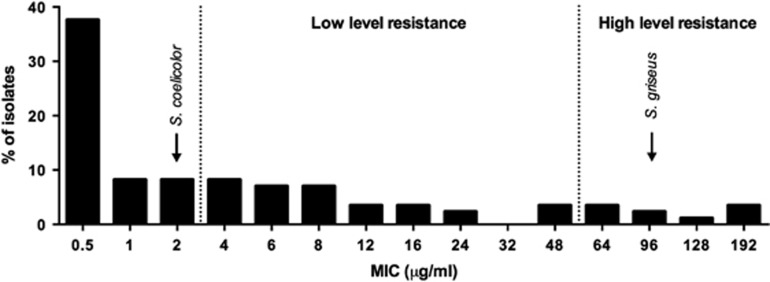
To assess the level of streptomycin resistance among streptomycetes in nature, we tested the MICs of 85 natural Streptomyces strains originally isolated from the Himalaya and Qinling Mountains (Zhu et al., 2014; Figure 1). In accordance with literature estimates, we found resistance (MIC ⩾4 μg ml−1 streptomycin) in a substantial fraction of these strains (46%) with low-level resistance being more prevalent than high-level resistance (MIC ⩾64 μg ml−1 streptomycin; Tolba et al., 2002). This survey confirms that streptomycin resistance is common among streptomycetes in nature and raises questions about the benefits of streptomycin resistance at the presumably low streptomycin concentrations in the soil. Here we use the well-characterized lab strain S. coelicolor M145 that, with an MIC of 2 μg ml−1 streptomycin, has negligible resistance to streptomycin, to study the costs and benefits of streptomycin resistance.
Figure 1.
Streptomycin resistance of a collection of 85 natural Streptomyces isolates. MICs of S. coelicolor and S. griseus are indicated in the figure.
Spontaneous streptomycin resistance
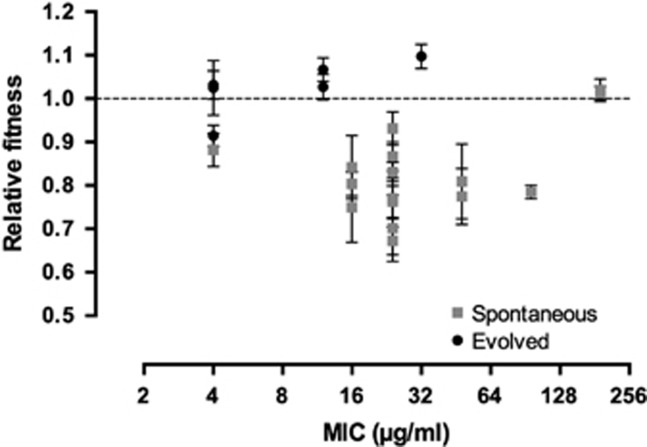
To gain insight into fitness effects of streptomycin resistance, we selected 16 independent isolates that were resistant to >2 μg ml−1 streptomycin (the MIC of the susceptible WT parent strain; Table 1). The resultant clones had MICs ranging from 4 to 192 μg ml−1 streptomycin (Figure 2). Competition experiments between these resistant clones and their susceptible parent in a drug-free environment revealed that although there is significant heterogeneity in the cost of resistance (analysis of variance: F15=2.92, P=0.002), 12 of 16 resistant strains were significantly less fit than the parent, with an average cost of ~21% (mean±s.e.m.=0.79±0.018). Notably, two highly resistant clones with MICs of 196 μg ml−1 streptomycin appeared to have no evident costs of resistance (P>0.05 for both clones). Across all mutants with significant costs, we found that there was no significant relationship between MIC and fitness (P>0.05).
Table 1. Strains used in this study.
| Strain | Streptomycin concentration (μg ml−1) used for selection | MIC (μg ml−1) | Relative fitness in the absence of streptomycin |
|---|---|---|---|
| Ancestral WT | — | 2 | 1 |
| Ancestral WTApr | — | 2 | — |
| S1 | 2 | 16 | 0.749635 |
| S2 | 2 | 16 | 0.804028 |
| S3 | 2 | 16 | 0.841599 |
| S4 | 2 | 24 | 0.831249 |
| S5 | 2 | 4 | 0.881947 |
| S6 | 4 | 24 | 0.672712 |
| S7 | 4 | 24 | 0.701725 |
| S8 | 4 | 24 | 0.931398 |
| S9 | 8 | 24 | 0.762897 |
| S10 | 8 | 24 | 0.769794 |
| S11 | 8 | 48 | 0.774609 |
| S12 | 8 | 24 | 0.866925 |
| S13 | 16 | 192 | 1.019433 |
| S14 | 16 | 192 | 1.013181 |
| S15 | 16 | 96 | 0.784886 |
| S16 | 16 | 48 | 0.809275 |
| WT1 | 0.2 | 4 | 1.025088 |
| WT2 | 0.2 | 12 | 1.027081 |
| WT3 | 0.2 | 4 | 0.914401 |
| WTApr1 | 0.2 | 12 | 1.066676 |
| WTApr2 | 0.2 | 4 | 1.031835 |
| WTApr3 | 0.2 | 32 | 1.097033 |
Abbreviation: MIC, minimal inhibitory concentration.
Figure 2.
Relative fitness in the absence of streptomycin as a function of the MIC for the spontaneous and evolved streptomycin-resistant mutants. Error bars represent standard error of the mean.
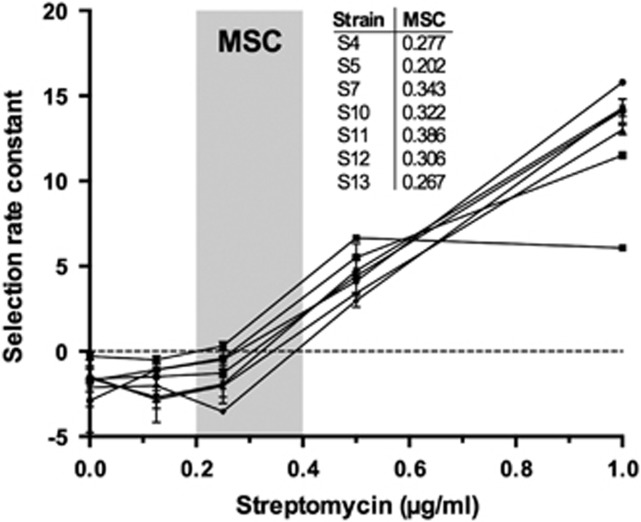
To estimate the MSC, we carried out competition experiments for a subset of clones across the breadth of streptomycin MIC at increasing streptomycin concentrations and determined the MSC as the antibiotic concentration where the fitness of the susceptible and resistant strain are equal. Figure 3 shows the change in fitness as a function of streptomycin concentration for seven strains, from which we draw two conclusions. First, the fitness of each strain is strongly dependent on the drug concentration to which it is exposed during competition; as anticipated, fitness is lowest in the absence of drugs, but increases sharply with small increases in the concentration of streptomycin. Second, there is variation in the MSC of different clones; the lowest MSC we measured (0.202) corresponds to ~1/10 the MIC of streptomycin against the susceptible parent strain, while the highest value (0.386) corresponds to ~1/5 the MIC. These data led to the prediction that selection of de novo resistance should be possible at concentrations significantly less than the MIC of wild-type cells.
Figure 3.
Selection rate constants as a function of the streptomycin concentration for a subset of spontaneous mutants. Error bars represent standard error of the mean.
Evolution of resistance at sub-MIC concentrations of streptomycin
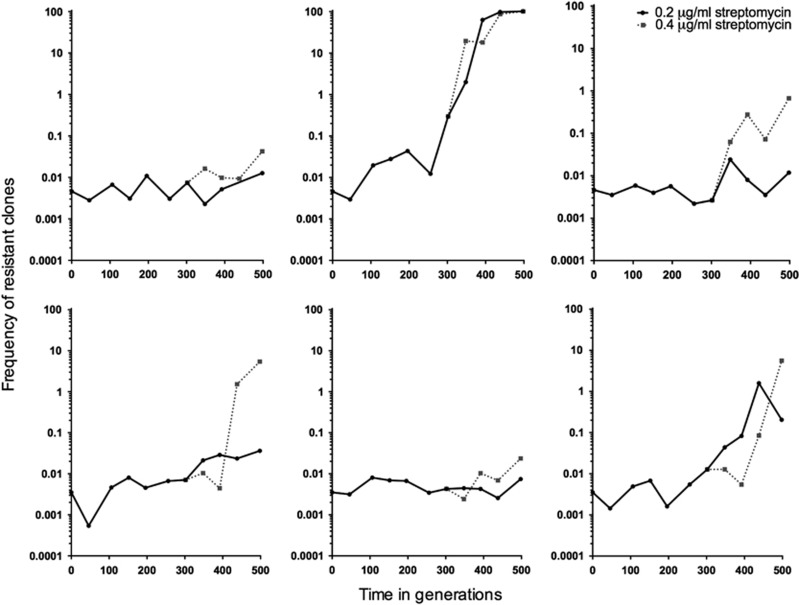
Having shown that antibiotic-resistant clones gain significant fitness benefits even at low antibiotic concentrations, we next sought to determine if these same low concentrations could select for de novo resistance. We further aimed to quantify the spectrum of fitness costs of evolved resistant strains, as these are predicted to be lower than the costs of spontaneous resistance. We serially transferred six replicate populations on media containing 0.2 μg ml−1 streptomycin, which corresponds to 1/10 of the MIC of the susceptible parent strain. As shown in Figure 4, while the frequency of resistant clones increased by at least 10-fold in three populations, with fixation of resistance in one of the populations, the remaining three populations remained static. We considered two alternative explanations for the apparent absence of resistance in these populations: either resistance mutations had not yet arisen, or alternatively, mutants were present but they were only slowly increasing because of limited benefits at the streptomycin concentrations they faced. To distinguish these possibilities we doubled the drug concentration to 0.4 μg ml−1 after ~300 generations and then continued transferring these six new populations in parallel with the original replicates. Consistent with the idea that resistant clones were present, but only slowly increasing, we observed a rapid and significant overall increase in the fraction of resistant cells in these supplemented populations as compared with those evolved at the lower concentration (paired t-test, d.f.=5, P=0.028). We confirmed the evolution of de novo streptomycin resistance by measuring the MIC of random clones isolated from evolved populations; clones from all six populations evolved at 0.2 μg ml−1 streptomycin had MIC >2 μg ml−1 (Figure 2).
Figure 4.
The frequency through time of strains resistant to 2 μg ml−1 streptomycin in populations evolved for 500 generations in the presence of 0.2 or 0.4 μg ml−1 (started at ~332 generations from the 0.2 μg ml−1 population) streptomycin. Resistance was estimated approximately every 50 generations.
Evolution of drug resistance below the MIC is predicted to enrich for strains with reduced fitness costs of resistance. This is because resistant strains must still compete with susceptible strains that are inhibited, but not killed, by the antibiotic. To test this prediction, we measured the fitness of random resistant clones in the absence of streptomycin that were isolated from the final time point of all replicated populations evolved at 0.2 μg ml−1 streptomycin. As shown in Figure 2, the fitness of evolved resistant clones is significantly different from the spectrum of fitness effects of spontaneous mutants (generalized linear mixed models, P<0.001). While one of six clones does have fitness costs, the fitness of the remaining five populations is either higher than or indistinguishable from 1. Overall, in contrast to the significant ~21% cost of spontaneous resistance, clones that evolved resistance after exposure to sub-MIC streptomycin for 500 generations had an average fitness benefit of ~3%, which did not differ significantly from 1 (Figure 2). In summary, strains of S. coelicolor evolving at sub-MIC streptomycin can evolve high levels of resistance while simultaneously avoiding the costs associated with this phenotype.
Genetics of resistance
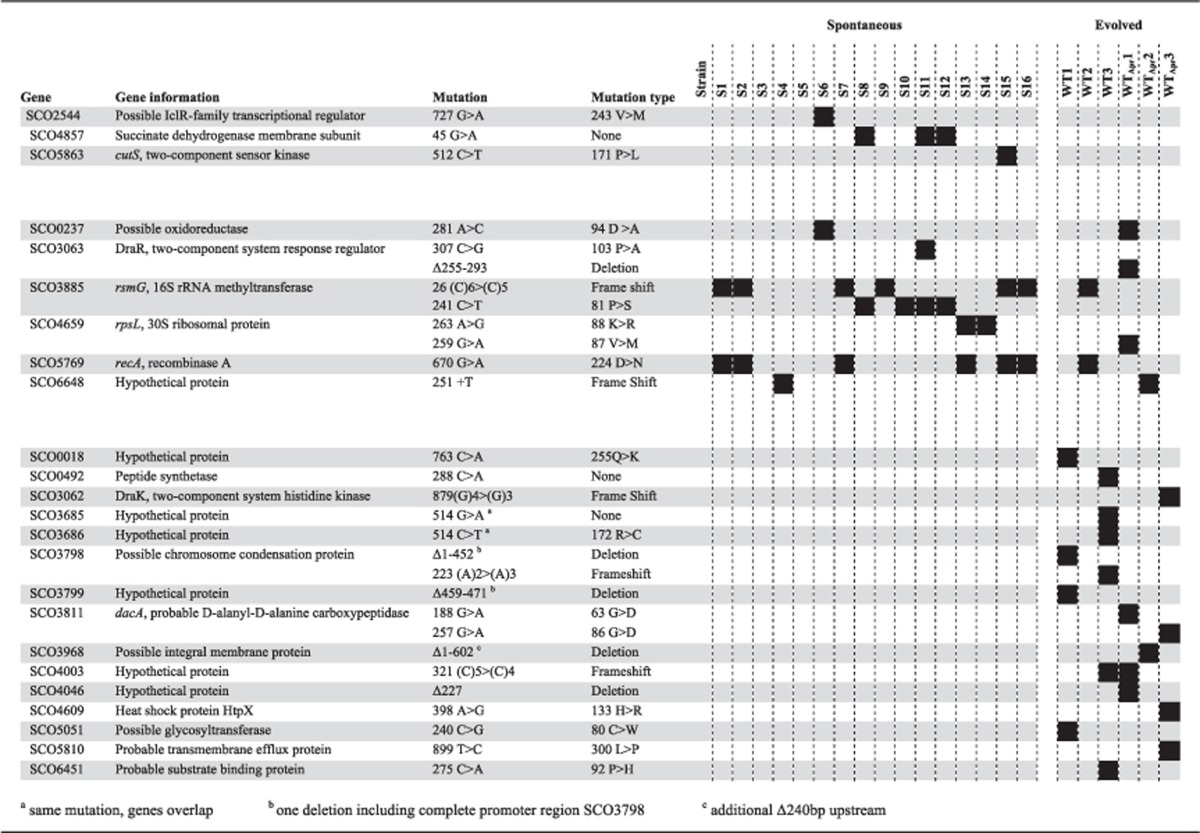
To gain insight into the mechanisms of resistance, we sequenced the genomes of ancestral and resistant strains. Across all resistant strains, we identified a total of 93 mutations: 4 synonymous substitutions, 27 nonsynonymous substitutions, 3 insertions, 14 deletions (11 single bp deletions) and 45 intergenic mutations. Consistent with extensive convergence across clones, these 93 mutations mapped to only 24 genes (Table 2) and 20 intergenic regions (Supplementary Table S1). On average, we identified 3.1 mutations in the spontaneous mutants, with an average of 1.6 mutations in genes and 1.4 mutations in intergenic regions. As the evolved clones were exposed to sub-MIC levels of streptomycin for 500 generations, it is not surprising that we found significantly more mutations in this set, with an average of 7.4 mutations per clone (3.7 mutations in genes and 3.7 in intergenic regions).
Table 2. Mutations in genes in the spontaneous and evolved clones.
As the spontaneous mutants show significant fitness defects, we hypothesized that the mutations identified in this set will correspond to costly resistance mutations. The evolved clones, by contrast, did not show any fitness deficits. This could be explained in two ways: either they fixed costly resistance mutations, and also acquired compensatory mutations during their 500 generation evolution, or they fixed new intrinsically cost-free resistance mutations. Three out of six evolved clones share mutations in genes with spontaneous resistant clones. Taken together with our finding that these evolved clones bear no fitness cost, this result suggests that they have acquired additional mutations that compensate for the fitness costs of resistance. For the other three evolved clones, two share mutations in intergenic regions with one spontaneous resistant clone, whereas the third has no mutations in common with any spontaneous clone. This suggests that these clones may carry a distinct spectrum of resistance mutations that comprise novel resistance mutations together with compensatory mutations, or that these mutations are intrinsically cost-free.
Parallel mutation fixation was observed for 10 genes. Six of these genes were mutated both in spontaneous and evolved mutants, strongly suggesting that they are associated with costly streptomycin resistance. Mutations in two of these genes, rsmG and rpsL, are known to confer low- (Nishimura et al., 2007a) and high-level (Hosaka et al., 2009) streptomycin resistance in S. coelicolor, respectively. Fourteen strains showed a mutation in either gene, whereas no strains were mutated in both genes. Eleven strains (10 spontaneous and one evolved), with MICs ranging from 12 to 96 μg ml−1, were found to have a mutation in rsmG, which encodes an rRNA methyltransferase that methylates base G527 in the 16S rRNA (Nishimura et al., 2007b). Seven of these carried the same effective lesion in a homopolymeric tract of five cytosine residues in this gene (26 (C)6>(C)5), resulting in a frameshift mutation that leads to an early stop codon (Nishimura et al., 2007a; Tanaka et al., 2009), whereas the other four show the same nonsynonymous substitution. Three clones are mutated in rpsL, encoding ribosomal protein S12; one evolved clone with an MIC of 12 μg ml−1 and two spontaneous clones with an MIC of 192 μg ml−1, the latter two carrying the same 88K>R mutation that is known to cause high-level resistance (Tanaka et al., 2009). Interestingly, these two spontaneous mutants (S13 and S14) are highly resistant to streptomycin, yet neither bears a cost of resistance.
Although these are the only genes known to cause streptomycin resistance in streptomycetes, the fact that parallel mutations were fixed elsewhere suggests that these mutations may be causally associated with streptomycin resistance. An interesting case can be made for the two-component system consisting of response regulator DraR (SCO3063) and sensory kinase DraK (SCO3062). While two strains (one spontaneous and one evolved) showed a different mutation in the gene for DraR, another strain was mutated in the gene for DraK. The DraR two-component system has been shown to be involved in the regulation of antibiotic production in S. coelicolor and the structural configuration of the extracellular signal domain of DraK is pH-dependent, but its ligand is not known (Yu et al., 2012; Yeo et al., 2013). Surprisingly, seven resistant strains have the same mutation in recA, encoding recombinase A that is involved in the homologous recombination of single-stranded DNA. This mutation always co-occurs with a mutation in rsmG or rpsL; however, when comparing strains that do not have this additional mutation in recA, we do not see a difference in MIC or fitness, implying that it may not be involved in streptomycin resistance or compensatory mechanisms. Other parallel mutations occurring both in spontaneous and evolved strains were located in a possible oxidoreductase and another hypothetical protein.
Within the mutations appearing only in the evolved clones, there are three cases of parallelism. A possible chromosome condensation protein is mutated in both of the evolved strains that do not share any mutations with the spontaneous mutants, making it a likely candidate for conferring streptomycin resistance. Two evolved clones are mutated in dacA, which encodes a d-alanyl-d-alanine carboxypeptidase, an enzyme belonging to the group of penicillin binding proteins involved in cell wall synthesis. Notably, we identified a mutation in the promoter region of the same gene in a third evolved clone, 101 bp upstream of the predicted translational start site. The third parallel mutation is located in a hypothetical protein. Furthermore, we identified mutations in 12 more genes that were only mutated in evolved clones, none of which were shared with the spontaneous resistant isolates.
Discussion
Despite the appropriate emphasis on the clinical crisis in antibiotic resistance, it is also important to recognize that antibiotic resistance is a natural phenomenon that long predates the modern selective pressure of antibiotic use by man (D’Costa et al., 2011). Genes for antibiotic resistance are commonly found in nature (Allen et al., 2010), even in pristine environments untouched by human influence (D’Costa et al., 2006a; Allen et al., 2009); however, very little is understood about the processes by which antibiotic resistance arises in these conditions. This has led to questions about the role of antibiotics in soil, where their concentrations are believed to be extremely low, as well as the role of resistance at these sublethal concentrations (Yim et al., 2006; Aminov, 2009). Here we focus on the evolution of antibiotic resistance in the soil bacterium S. coelicolor in response to streptomycin, an antibiotic produced by S. griseus. We first show that streptomycin resistance among natural Streptomyces isolates is widespread, with ~50% of strains reaching an MIC greater than S. coelicolor. Next, we show that costly antibiotic resistance can be offset at very low streptomycin concentrations; drug concentrations of antibiotics as low as 1/10 the MIC of susceptible strains are sufficient to provide direct fitness benefits for resistant strains. Using experimental evolution, we next find that resistant strains readily evolved during 500 generations of evolution at very low concentrations and furthermore that these evolved mutants do not carry a fitness cost, in striking contrast to strains that evolved spontaneous resistance with fitness costs of more than 20%. Finally, whole-genome sequencing revealed that some sub-MIC evolved mutants acquired the same spectrum of mutations as resistant clones isolated at concentrations above the MIC, while also carrying additional mutations that could compensate for the initial cost of resistance. The other clones evolved at these subinhibitory concentrations of streptomycin contained a distinct spectrum of mutations that could confer resistance at no cost. Although at present we cannot clearly distinguish these possibilities, this will be possible in future work using directed mutagenesis in isogenic clones.
There are several important implications of these results. First, consistent with the results of Gullberg et al. (2011), our data clarify that antibiotics do not need to reach lethal concentrations to exert pronounced effects on resistance evolution. Even if susceptible cells are not obviously inhibited by sub-MIC antibiotics, their growth rates are diminished and this provides a broad range of opportunities for resistant cells to increase in frequency (Negri et al., 2000; Gullberg et al., 2011; Jorgensen et al., 2013). This has clear relevance to the evolution of resistance in soil, where antibiotic concentrations due to endogenous production by microorganisms, both bacteria and fungi, are believed to be typically too low to inhibit competing susceptible strains (Davies et al., 2006; Raaijmakers and Mazzola, 2012; Andersson and Hughes, 2014). Thus, even if antibiotics are produced at sublethal levels, a suggestion requiring further study, they can nevertheless strongly and directly select for the emergence of resistant cells. Accordingly, emphasis on the MIC of bacteria is likely to be misguided for understanding the roles of both antibiotic production and resistance in soil; instead, emphasis should be reoriented to the MSC to determine boundary conditions for the emergence of resistant isolates.
Second, our results have clear implications for the persistence of resistant strains. Resistant bacteria that were isolated following exposure to lethal streptomycin concentrations were burdened with significant fitness costs, an outcome widely observed across species (Melnyk et al., 2014). One possibility is that these effects are caused by resistance mutations, for example, in rpsL or rsmG, that lead to hyperaccurate protein translation and therefore slower growth (Ruusala et al., 1984; Kurland, 1992). Alternatively, and specific to Streptomyces, streptomycin resistance can in some cases lead to hyperproduction of antibiotics (Hosaka et al., 2009; Tanaka et al., 2009; Wu et al., 2015), although we did not observe any increased susceptibility of our ancestral strain of S. coelicolor to any of the evolved strains. In contrast to bacteria that were selected at high streptomycin concentrations (Kurland, 1992; Melnyk et al., 2014), S. coelicolor clones that evolved resistance at sub-MIC doses for 500 generations did not show a fitness cost. From an environmental standpoint, this suggests that resistance evolving at sub-MIC antibiotics in soil will persist in the face of competition with susceptible cells, whereas cells that bear the significant fitness costs of spontaneous resistance would be predicted to decline (Andersson and Hughes, 2011, 2014). Consistent with this, and as corroborated here, streptomycin resistance is commonly found in nature, with low-level resistance being more prevalent than high-level resistance (Tolba et al., 2002; van Overbeek et al., 2002). Although there are many potential reasons for this, including high densities of S. griseus that are naturally resistant to their own antibiotic (Laskaris et al., 2011), resistance in other species may arise because of the direct benefits resistance provides. From a clinical standpoint, cost-free mutations emerging at sub-MIC antibiotic concentrations are problematic because this could serve to reduce the reversibility of resistance, a potential that relies on durable fitness costs in resistant isolates (Andersson and Hughes, 2011). Certainly, infectious bacteria face a range of antibiotic doses during treatment (Baquero et al., 1998; Negri et al., 2000); if this influences the types of resistance mutations that arise and fix, and in particular their costs, it will be necessary to take this into consideration during the development of treatment protocols.
Third, our results suggest that resistance mutations selected at sub-MIC concentrations could differ from those arising above the MIC. While mutations in genes rsmG and rpsL, known to be associated with streptomycin resistance, were identified in 12 out of 16 spontaneous and 2 out of 6 evolved clones, the resistance mechanisms in the other clones remain to be elucidated. Many of the mutations/mutated genes occur in parallel, suggesting that they are directly involved in streptomycin resistance or potentially that these mutations influence the costs of resistance. For example, the DraR-K two-component system is mutated in several lineages. Various two-component systems have been implicated in the control of antibiotic production (van Wezel and McDowall, 2011), but as far as we are aware none have been specifically tied to resistance in the absence of the related biosynthetic gene cluster. Further research into the DraR-K response regulon is required to shed light on this important phenomenon. Another intriguing parallel mutation is located in recA and was found in seven sequenced strains. As a disruption of recA in S. coelicolor increases genetic instability (Huang and Chen, 2006), it is possible that this mutation increases the likelihood for subsequent resistance evolution. Despite these cases of parallelism, many evolved lineages carry unique mutations in hypothetical genes or intergenic regions. Moreover, there is little overlap between mutations found in sub-MIC evolved lineages and those selected for spontaneous resistance at higher drug concentrations. This indicates that many routes and mechanisms towards drug resistance are unknown. Also, it may indicate that studying antibiotic resistance at lethal doses provides only part of the spectrum of resistance mutations. At present, the role these mutations play in resistance is unknown; however, these are strong candidates for testing in future work. In addition, these mutations clarify the value of using experimental evolution at sub-MIC drug concentrations to elucidate novel modes of resistance. Finally, we note that in 2 of 16 spontaneously resistant lineages, we failed to identify any mutations at all, whereas two strains that were completely isogenic according to our sequencing data (S2 and S16) demonstrated a threefold MIC difference. Although our sequencing coverage was high in these clones, the Streptomyces chromosome is very GC rich (>70% G+C content), making assembly challenging and rendering certain regions difficult to sequence. Additionally, short-read sequencing may fail to capture duplications that could be highly relevant for resistance evolution (Sandegren and Andersson, 2009). Longer-read sequencing platforms should hopefully address these problems in this system in the future.
While antibiotics have been traditionally considered as interbacterial weapons (Raaijmakers and Mazzola, 2012), their role has been re-examined in the past few decades in light of results showing that cells respond to sub-MIC antibiotics with broad and diverse changes to gene expression and cellular physiology (Davies et al., 2006). By this new view, antibiotics are not weapons but instead are reinterpreted as signals, whereas resistance is understood to modify signal strength (Yim et al., 2006; Aminov, 2009). We recently cast doubt on this reinterpretation in studies showing that social interactions among competing Streptomycetes had a marked influence on antibiotic production (Abrudan et al., 2015), a result consistent with their likely role as intermicrobial weapons. The present work supports this view. In short, irrespective of any other effects sub-MIC antibiotics have on cells, these low concentrations are sufficient to both inhibit competing susceptible cells and to provide sufficient natural selection to enrich for resistance.
Several questions nevertheless remain from this study. First, we lack a clear understanding of the effective concentrations of streptomycin in soil. While concentrations are often claimed to be low, little direct evidence supports this possibility, and local concentrations may in fact be high. Moreover, it remains unclear how antibiotic concentrations in soil are influenced by the physicochemical properties of soils together with the role of other intermicrobial dynamics that influence antibiotic production. It therefore remains a key goal to extend this work to more natural microcosms that include structured soil, as well as including competition with the natural streptomycin producer S. griseus. Second, it remains unclear how cost-free antibiotic-resistant clones arise at sub-MIC streptomycin concentrations. Our genome sequencing has identified several putatively causal mutations for resistance in two well-studied genes; moreover, it has suggested candidate genes that could either compensate for costs of resistance or alternatively could represent entirely new suites of resistance mechanisms that are intrinsically cost-free. This needs to be followed with more mechanistic studies to determine the precise functional role of these mutations. Finally, it will be important to extend our analyses to the evolution of resistance in natural environments influenced by anthropogenic antibiotic pollution (Thiele-Bruhn, 2003; Andersson and Hughes, 2014). Natural reservoirs for resistance can transfer genes for resistance to clinically relevant pathogens (Forsberg et al., 2012); if these mechanisms are enriched for low-cost resistance mutations, then this has profound potential consequences for the distribution and persistence of resistance types among infectious bacteria.
Acknowledgments
We appreciate the helpful comments of Angus Buckling and two anonymous reviewers on an earlier version of this manuscript. This work was financially supported by a grant from the Dutch National Science Foundation (NWO) (to DER). and by a grant from the China Scholarship Council (CSC) (to ZZ). Additional support was provided by the UK Biotechnology and Biological Sciences Research Council (BB/J006009/1) (to DER) and Ian S Roberts (University of Manchester).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Abrudan MI, Smakman F, Grimbergen AJ, Westhoff S, Miller EL, van Wezel GP et al. (2015). Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc Natl Acad Sci USA 112: 11054–11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8: 251–259. [DOI] [PubMed] [Google Scholar]
- Allen HK, Moe La, Rodbumrer J, Gaarder A, Handelsman J. (2009). Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J 3: 243–251. [DOI] [PubMed] [Google Scholar]
- Aminov RI. (2009). The role of antibiotics and antibiotic resistance in nature. Environ Microbiol 11: 2970–2988. [DOI] [PubMed] [Google Scholar]
- Andersson DI, Hughes D. (2011). Persistence of antibiotic resistance in bacterial populations. FEMS Microbiol Rev 35: 901–911. [DOI] [PubMed] [Google Scholar]
- Andersson DI, Hughes D. (2014). Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12: 465–478. [DOI] [PubMed] [Google Scholar]
- Andersson DI, Levin BR. (1999). The biological cost of antibiotic resistance. Curr Opin Microbiol 2: 489–493. [DOI] [PubMed] [Google Scholar]
- Baquero F. (2001). Low-level antibacterial resistance: a gateway to clinical resistance. Drug Resist Updat 4: 93–105. [DOI] [PubMed] [Google Scholar]
- Baquero F, Alvarez-Ortega C, Martinez JL. (2009). Ecology and evolution of antibiotic resistance. Environ Microbiol Rep 1: 469–476. [DOI] [PubMed] [Google Scholar]
- Baquero F, Negri M, Morosini M-I, Blazquez J. (1998). Antibiotic-selective environments. Clin Infect Dis 27: 5–11. [DOI] [PubMed] [Google Scholar]
- Barka EA, Vatsa P, Sanchez L, Gaveau-vaillant N, Jacquard C, Klenk H-P et al. (2016). Taxonomy, physiology, and natural products of actinobacteria. Microbiol Mol Biol Rev 80: 1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L, Waldron L, Gillings MR. (2015). Potential impacts of aquatic pollutants: sub-clinical antibiotic concentrations induce genome changes and promote antibiotic resistance. Front Microbiol 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C et al. (2011). Antibiotic resistance is ancient. Nature 477: 457–461. [DOI] [PubMed] [Google Scholar]
- D’Costa VM, McGrann K, Hughes DW, Wright GD. (2006. a). Sampling the antibiotic resistome. Science 311: 374–377. [DOI] [PubMed] [Google Scholar]
- D’Costa VM, McGrann KM, Hughes DW, Wright GD. (2006. b). Sampling the antibiotic resistome. Science 311: 374–377. [DOI] [PubMed] [Google Scholar]
- Davies J, Davies D. (2010). Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74: 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Spiegelman GB, Yim G. (2006). The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol 9: 445–453. [DOI] [PubMed] [Google Scholar]
- Drlica K, Zhao X. (2007). Mutant selection window hypothesis updated. Clin Infect Dis 44: 681–688. [DOI] [PubMed] [Google Scholar]
- Drusano GL. (2004). Antimicrobial pharmacodynamics: critical interactions of ‘bug and drug’. Nat Rev Microbiol 2: 289–300. [DOI] [PubMed] [Google Scholar]
- Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337: 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D et al. (2011). Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 7: e1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood DA. (2007) Streptomyces in Nature and Medicine: The Antibiotic Makers. Oxford University Press: New York, NY, USA. [Google Scholar]
- Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, Kodani S et al. (2009). Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol 27: 462–464. [DOI] [PubMed] [Google Scholar]
- Huang TW, Chen CW. (2006). A recA null mutation may be generated in Streptomyces coelicolor. J Bacteriol 188: 6771–6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D, Andersson DI. (2012). Selection of resistance at lethal and non-lethal antibiotic concentrations. Curr Opin Microbiol 15: 555–560. [DOI] [PubMed] [Google Scholar]
- Jorgensen KM, Wassermann T, Jensen PO, Hengzuang W, Molin S, Hoiby N et al. (2013). Sublethal ciprofloxacin treatment leads to rapid development of high-level ciprofloxacin resistance during long-term experimental evolution of Pseudomonas aeruginosa. Antimicrob Agents Chemother 57: 4215–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlmeter G, Brown DFJ, Goldstein FW, MacGowan AP, Mouton JW, Osterlund A et al. (2003). European harmonization of MIC breakpoints for antimicrobial susceptibility testing of bacteria. J Antimicrob Chemother 52: 145–148. [DOI] [PubMed] [Google Scholar]
- Kurland CG. (1992). Translational accuracy and the fitness of bacteria. Annu Rev Genet 26: 29–50. [DOI] [PubMed] [Google Scholar]
- Laskaris P, Gaze WH, Wellington EMH. (2011) Environmental reservoirs of resistance genes in antibiotic-producing bacteria and their possible impact on the evolution of antibiotic resistance. In: Keen PL, Montforts MHMM (eds). Antimicrobial Resistance in the Environment. Wiley: New York, NY, pp 73–92. [Google Scholar]
- Lenski R, Rose M, Simpson S, Tadler S. (1991). Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat 138: 1315–1341. [Google Scholar]
- Melnyk AH, Wong A, Kassen R. (2014). The fitness costs of antibiotic resistance mutations. Evol Appl 8: 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri MC, Lipsitch M, Blázquez J, Levin BR, Baquero F. (2000). Concentration-dependent selection of small phenotypic differences in TEM beta-lactamase-mediated antibiotic resistance. Antimicrob Agents Chemother 44: 2485–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Hosaka T, Tokuyama S, Okamoto S, Ochi K. (2007. a). Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2). J Bacteriol 189: 3876–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Johansen SK, Inaoka T, Hosaka T, Tokuyama S, Tahara Y et al. (2007. b). Identification of the RsmG methyltransferase target as 16S rRNA nucleotide G527 and characterization of Bacillus subtilis rsmG mutants. J Bacteriol 189: 6068–6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek LS, Wellington EMH, Egan S, Smalla K, Heuer H, Collard JM et al. (2002). Prevalence of streptomycin-resistance genes in bacterial populations in European habitats. (Special issue: The contribution of mobile genetic elements in bacterial adaptability and diversity.). FEMS Microbiol Ecol 42: 277–288 53 ref. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JM, Mazzola M. (2012). Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu Rev Phytopathol 50: 403–424. [DOI] [PubMed] [Google Scholar]
- Ruusala T, Andersson D, Ehrenberg M, Kurland CG. (1984). Hyper-accurate ribosomes inhibit growth. EMBO J 3: 2575–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandegren L, Andersson DI. (2009). Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat Rev Microbiol 7: 578–588. [DOI] [PubMed] [Google Scholar]
- Schatz A, Bugie E, Waksman SA. (1944). Streptomycin, a substance exhibiting antibiotic activity against Gram-positive and Gram-negative bacteria. Proc Soc Exp Biol Med 55: 66–69. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Komatsu M, Okamoto S, Tokuyama S, Kaji A, Ikeda H et al. (2009). Antibiotic overproduction by rpsL and rsmG mutants of various actinomycetes. Appl Environ Microbiol 75: 4919–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele-Bruhn S. (2003). Pharmaceutical antibiotic compounds in soils—a review. J Plant Nutr Soil Sci 166: 145–167. [Google Scholar]
- Tolba S, Egan S, Kallifidas D, Wellington EMH. (2002). Distribution of streptomycin resistance and biosynthesis genes in streptomycetes recovered from different soil sites. FEMS Microbiol Ecol 42: 269–276. [DOI] [PubMed] [Google Scholar]
- Travisano M, Lenski RE. (1996). Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics 143: 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezel GP, McDowall KJ. (2011). The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep 28: 1311. [DOI] [PubMed] [Google Scholar]
- Wu C, Du C, Gubbens J, Choi YH, van Wezel GP. (2015). Metabolomics-driven discovery of a prenylated isatin antibiotic produced by streptomyces species MBT28. J Nat Prod 78: 2355–2363. [DOI] [PubMed] [Google Scholar]
- Yeo KJ, Kim EH, Hwang E, Han Y-H, Eo Y, Kim HJ et al. (2013). pH-dependent structural change of the extracellular sensor domain of the DraK histidine kinase from Streptomyces coelicolor. Biochem Biophys Res Commun 431: 554–559. [DOI] [PubMed] [Google Scholar]
- Yim G, Wang HH, Davies J. (2006). The truth about antibiotics. Int J Med Microbiol 296: 163–170. [DOI] [PubMed] [Google Scholar]
- Yim G, Wang HH, Davies J. (2007). Antibiotics as signalling molecules. Philos Trans R Soc Lond Ser B 362: 1195–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Zhu H, Dang F, Zhang W, Qin Z, Yang S et al. (2012). Differential regulation of antibiotic biosynthesis by DraR-K, a novel two-component system in Streptomyces coelicolor. Mol Microbiol 85: 535–556. [DOI] [PubMed] [Google Scholar]
- Zhu H, Swierstra J, Wu C, Girard G, Choi YH, van Wamel W et al. (2014). Eliciting antibiotics active against the ESKAPE pathogens in a collection of actinomycetes isolated from mountain soils. Microbiology 160: 1714–1725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.