Abstract
Explaining the enormous biodiversity observed in bacterial communities is challenging because ecological theory predicts that competition between species occupying the same niche should lead to the exclusion of less competitive community members. Competitive exclusion should be particularly strong when species compete for a single limiting resource or live in unstructured habitats that offer no refuge for weaker competitors. Here, we describe the ‘cheating effect’, a form of intra-specific competition that can counterbalance between-species competition, thereby fostering biodiversity in unstructured habitats. Using experimental communities consisting of the strong competitor Pseudomonas aeruginosa (PA) and its weaker counterpart Burkholderia cenocepacia (BC), we show that co-existence is impossible when the two species compete for a single limiting resource, iron. However, when introducing a PA cheating mutant, which specifically exploits the iron-scavenging siderophores produced by the PA wild type, we found that biodiversity was preserved under well-mixed conditions where PA cheats could outcompete the PA wild type. Cheating fosters biodiversity in our system because it creates strong intra-specific competition, which equalizes fitness differences between PA and BC. Our study identifies cheating – typically considered a destructive element – as a constructive force in shaping biodiversity.
Introduction
Most habitats on earth are populated by diverse bacterial communities (Curtis et al., 2002; Rusch et al., 2007; The Human Microbiome Project, 2012; Rinke et al., 2013; Sunagawa et al., 2015). Explaining the evolution and maintenance of this enormous bacterial biodiversity is challenging because competition between species occupying the same niche should often result in the exclusion of competitively inferior community members (Hardin, 1960; Becker et al., 2012; Foster and Bell, 2012). Ecological theory predicts that species can only co-exist if intra-specific competition exceeds inter-specific competition, a scenario that can apply if competing species differ in their resource use or are spatially separated from one another (Holt et al., 1994; Chesson, 2000; Lankau, 2009; Barabás et al., 2016). Previous experimental work has demonstrated that these two mechanisms indeed apply to microbial systems. In toxin-mediated interference competition, for instance, spatial structure can prevent competitive exclusion because it allows weaker community members to find refuge from their more competitive counterparts (Kerr et al., 2002; Kim et al., 2008; Vos et al., 2013). Moreover, it has been shown that evolving bacterial populations rapidly diversify, which promotes niche differentiation and reduces inter-specific resource competition (Rosenzweig et al., 1994; Rainey and Travisano, 1998; Brockhurst et al., 2006).
Here, we propose a novel mechanism of how increased intra- vs inter-specific competition can foster species co-existence, even under conditions where bacterial species compete for a single limiting resource in an unstructured, well-mixed habitat. In particular, we argue that a common form of intra-specific competition in bacteria is social cheating, where bacteria, secreting a group-beneficial compound, are exploited by ‘cheating’ variants of the same species (Rainey and Rainey, 2003; Griffin et al., 2004; Cordero et al., 2012; Raymond et al., 2012). Cheats are mutants that have lost the ability to produce the costly compound but still free ride on the compounds produced by others. Following the theoretical considerations and terminology of Chesson (2000) we predict that cheating has both equalizing and stabilizing fitness effects. If cheating occurs in the more competitive species then cheats equalize fitness differences between species, because they exclusively compromise the fitness of their own species. By doing so, they also increase intra-specific competition, creating a stabilizing fitness effect. Here, we empirically test these predictions and examine whether within-species cheating is a strong enough force to foster species co-existence and biodiversity.
We tested these predictions by constructing experimental bacterial communities consisting of Pseudomonas aeruginosa (PA) and Burkholderia cenocepacia (BC), two opportunistic human pathogens. These two species are notoriously hard to maintain in planktonic laboratory co-cultures because PA is a very strong competitor and is able to quickly suppress BC (Bragonzi et al., 2012; Costello et al., 2014). Although previous work examined the interactions between these two species because they can simultaneously be isolated from soils and infections (Weaver and Kolter, 2004; Lambiase et al., 2006), we here use them as a model system to test fundamental ecological theory. To match conditions where the competitive exclusion principle applies (Hardin, 1960), we created an environment where the two species compete for a single limiting resource – iron: a key essential nutrient, limiting bacterial growth both in natural environments (Hersman et al., 2000) and within hosts (Miethke and Marahiel, 2007). In order to scavenge iron, both species rely on the secretion of small peptide derivates (pyoverdine for PA and ornibactin for BC), called siderophores (Miethke and Marahiel, 2007; Tyrrell and Callaghan, 2015). Importantly, pyoverdine production in PA is a cooperative trait as secreted molecules can be shared as public goods within the Pseudomonas population (Griffin et al., 2004). Pyoverdine sharing selects for cheating mutants, which exploit the pyoverdine produced by others without contributing to it themselves (Harrison et al., 2008; Dumas and Kümmerli, 2012). Thus, to examine how cheating affects biodiversity, we introduced a cheating variant of PA into our community, which is unable to produce pyoverdine but possesses the receptor for its uptake.
We studied the competitive relationships and evolutionary dynamics between the PA wild type, the PA cheat and BC in pairwise combinations and in three-strain communities across a gradient of spatial structures, ranging from well-mixed (that is, low spatial structure) to viscous medium (that is, increased spatial structure). Consistent with the competitive exclusion principle, we found that the PA wild type completely displaced BC regardless of spatial structure. This dynamic changed when adding a PA cheat to the community, where, under low spatial structure, the PA cheat efficiently exploited the pyoverdine of the PA wild type, thereby weakening its competitive strength and fostering species co-existence. Conversely, increased spatial structure impeded biodiversity, as it prevented PA cheats from outcompeting the PA wild type, such that the latter could drive BC to extinction. Taken together, our experiments identify within-species cheating as a novel mechanism, which can promote biodiversity in habitats with low spatial structure.
Materials and methods
Bacterial strains, media and growth conditions
We used the PA strain PAO1 (ATCC 15692) as the superior competitor and the BC strain H111 (LMG 23991), an isolate from a cystic fibrosis patient (Gotschlich et al., 2001), as the inferior competitor in our eco-system. The former produces pyoverdine and pyochelin, the primary and secondary siderophores of PA, respectively. H111 produces the primary siderophore ornibactin, but, as the PA wild type, it has also the ability to produce pyochelin as a secondary siderophore. Although it remains unknown whether this siderophore can be exchanged between species, it has been shown that under strong iron limitation, as imposed in our experiments, the production of pyochelin is negligible in both species (Dumas et al., 2013; Tyrrell et al., 2015). To rule out any pleiotropic effects associated with pyochelin production, we used the PA siderophore knock-out strain PAO1ΔpvdDΔpchEF as the PA cheat. This strain can neither produce pyoverdine nor pyochelin. While this strain possesses the receptor to take up pyoverdine, it cannot use ornibactin from BC. To be able to distinguish strains in mixed competitions, we used variants of the PA wild type and the PA cheat that constitutively expressed eGfp and mCherry, respectively (chromosomal insertions attTn7::Ptac-egfp and attTn7::Ptac-mcherry). H111 did not carry a fluorescent marker. This strain was easily distinguishable from PA based on the absence of a fluorescent signal and its small dot-like colony morphology.
Overnight cultures were grown in lysogeny broth (LB). We induced strongly iron-limiting growth conditions by using casamino acids (CAA) medium (per 1 l: 5 g casamino acids; 1.18 g K2HPO4∙3H2O; 0.25 g MgSO4∙7H2O) supplemented with 20 mm NaHCO3, 25 mm HEPES and 100 μg ml−1 of the natural iron chelator human apo-transferrin. For control experiments in iron-replete medium, we omitted transferrin from the above recipe to create conditions where siderophores are not required for growth. We further manipulated the spatial structure of the environment by growing bacteria either in: (a) liquid shaken (170 r.p.m.) medium reflecting a relatively unstructured environment; (b) liquid static medium representing an environment with intermediate spatial structure; or (c) viscous static medium containing 0.1% agar reflecting an environment with relatively high spatial structure. All experiments were conducted at 37 °C. All chemicals were purchased from Sigma-Aldrich, Buchs, Switzerland.
Growth of PA and BC in iron-limited CAA medium
To quantify the growth capacities of our community members in the iron-limited media used, we estimated their number of doublings in monocultures in 18 replicates each, across 24 h. Therefore, overnight cultures were washed twice with 0.8% NaCl solution and adjusted to OD600=1. Bacteria were then diluted and inoculated in 1.5 ml of CAA medium with supplements in 24-well plates to a starting OD600 of 1 × 10-4. Plates were subsequently incubated for 24 h. We measured cell density at the beginning and at the end of the experiment by serially diluting cultures in 0.8% NaCl and plating aliquots on LB agar plates supplemented with 20 μm FeCl3. These plates were incubated overnight and left at room temperature for another 24 h to allow bacterial colonies to fully mature. The colony-forming units (CFU) were then counted. The number of doublings was calculated as Doublings=(ln(xt/x0))/ln(2), where x0 and xt are the initial and the final CFUs, respectively (Harrison et al., 2008).
Competition experiments
To assess the competitive relationship between two strains, we washed overnight monocultures twice with 0.8% NaCl solution, adjusted them to OD600=1 and mixed the strains at specific ratios (see below for details). We inoculated mixes at a starting OD600 of 1 × 10-4 into 1.5 ml of iron-limited CAA medium on 24-well plates, and incubated the plates for 24 h. Before and after competition, we counted the CFUs of both competing strains by serially diluting cultures in 0.8% NaCl and plating competitions individually on LB agar plates supplemented with 20 μm FeCl3. The plates were incubated overnight and left at room temperature for another 24 h to allow fluorescent markers to fully mature. We used a fluorescent imaging device (Lumenera Infinity 3 camera connected to a dark chamber) to differentiate between fluorescently tagged and untagged colonies. We then calculated the relative fitness (v) of the focal strain as v=[at × (1−a0)]/[a0 × (1−a t)], where a0 and at are the initial and final frequency of the focal strain, respectively (Ross-Gillespie et al., 2007). We ln-transformed all fitness values in order to obtain normally distributed residuals. Values of ln(v)>0 or ln(v)<0 indicate whether the frequency of the focal strain increased or decreased relative to its competitor, respectively.
In a first set of competition assays, we checked for fitness effects of the fluorescent markers mCherry and eGfp. We competed the untagged PA wild type against both the mCherry and eGfp tagged PA wild type (eight replicates per competition). We found that the constitutive expression of egfp, but not mcherry, was associated with a slight but significant fitness cost (one-sample t-test testing whether the relative fitness differs from zero: for PAO1 egfp, ln(v)=−0.358±0.13 (mean±95% CI), t7=−6.58, P=0.0003; for PAO1 mcherry, ln(v)=−0.027±0.21, t7=−0.31, P=0.766). To ensure that this marker effect does not alter the competitive abilities of PA wild type and BC, we decided to tag the PA wild type (the strong competitor in our community) with eGfp, whereas BC remained untagged. Indeed, it turned out that even with the eGfp handicap, the PA wild type was able to displace BC (Figure 2a).
We then performed competitions between all pairwise combinations of our strains (PA wild type vs BC, PA cheat vs BC, and PA wild type vs PA cheat) using three different starting ratios (1:9, 1:1, 9:1). We varied the starting frequencies to examine the robustness of competitive outcomes across a range of mixing ratios. We further manipulated the spatial structure of the environment along a gradient from unstructured to highly structured according to the procedure described above. The minimum level of replication was 12 (for low and intermediate spatial structures) and 7 (for high spatial structure). Note that in competitions between the PA wild type and BC, CFU counts for BC often equalled zero (that is, BC frequency was below the detection limit after competitions). In order to estimate the relative fitness of BC, we substituted CFU=0 by CFU=0.1.
Finally, to verify that it was indeed iron limitation that caused the observed competitive interactions in our community, we performed a control experiment (in 9-fold replication), where we competed the strains in a relatively iron-rich environment (CAA without transferrin) where siderophores are less important for iron scavenging.
Evolutionary dynamics in experimental communities
We followed the evolutionary strain dynamics in our three-strain community (PA wild type vs PA cheat vs BC, 6 replicates) and in all combinations of 2-strain communities (four replicates each) over 6 days in iron-limited medium both under unstructured (shaken liquid) and structured (static viscous) conditions. As an additional control we also grew the three-strain community in unstructured iron-rich media where siderophores are unimportant for iron-uptake. In total, we had 42 evolving communities. We transferred 10% of each community to fresh medium daily, except for the community BC vs PA cheat, which grew poorly. For this combination, we transferred 25% of the culture to prevent over-dilution and subsequent population extinction. Following each daily transfer, we assessed the strain proportions in each community using the procedure described above.
Statistical analysis
We used linear models (LM) for statistical tests, and false discovery rate tests were applied for post hoc multiple pairwise comparisons (Benjamini and Hochberg, 1995). In cases where data distribution did not entirely meet the assumptions of linear models, we performed non-parametric Mann–Whitney tests and one-sample Wilcoxon signed rank tests to test the robustness of the parametric analyses. All statistical analysis was carried out in R 3.1.2 (R Development Core Team, 2015).
Results
The PA wild type outgrows BC and the PA cheat in monoculture
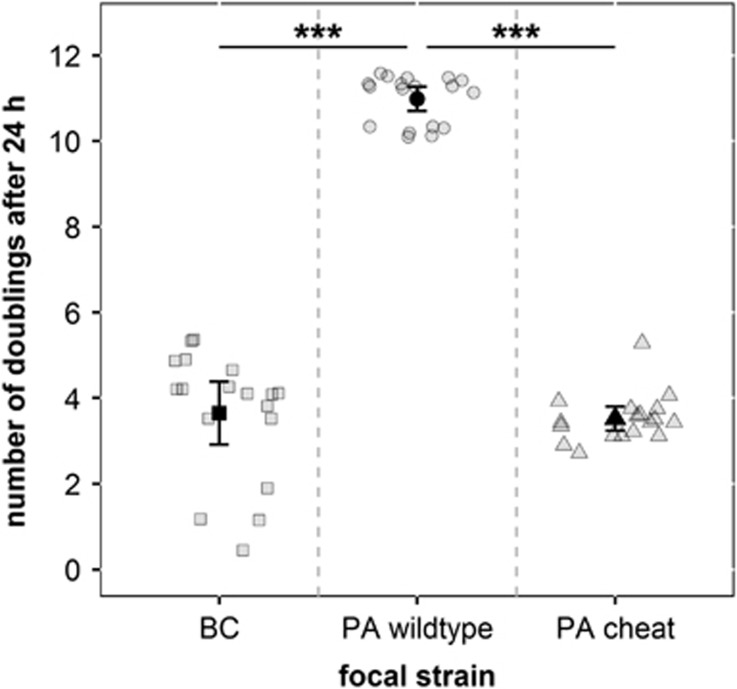
Our monoculture experiments show that the PA wild type divided significantly faster than BC under iron-limited conditions (LM: t51=22.72, P<0.0001). Based on the huge fitness difference observed, we expect the PA wild type to outcompete BC in direct competition. In contrast, BC and the PA cheat did not significantly differ in their number of doublings over 24 h (LM: t51=0.40, P=0.694; Figure 1). This suggests that cheats could have an equalizing fitness effect in mixed communities, as they drag down the overall growth potential of PA relative to BC. Moreover, because the PA wild type and the PA cheat only differ in their ability to produce siderophores, our data suggest that fitness differences between species are due to differences in siderophore efficiency: pyoverdine from PA is presumably the more efficient siderophore than ornibactin from BC.
Figure 1.
PA wild type grows significantly better than BC and the PA cheat in iron-limited medium (***P<0.001). This indicates that the PA cheats have an equalizing fitness effect in mixed communities, as they drag down the overall growth potential of PA relative to BC. As the PA wild type and the PA cheat are isogenic except for genes mediating siderophore production, these results indicate that pyoverdine is the most efficient iron-uptake system in this environment. Bacteria were grown as monocultures (18 replicates) in iron-limited medium for 24 h at 37 °C. Dark and faint symbols represent mean (±95% CI) and individual-replicate number of doublings, respectively.
PA wild type displaces BC in mixed cultures
As expected from the competitive exclusion principle (Hardin, 1960), our data show that co-existence between BC and the PA wild type is not possible in iron-limited medium. PA consistently outcompeted BC, regardless of the spatial structure of the medium (Figure 2a) and the starting frequency of BC (Supplementary Figure 1; one-sample t-test on pooled relative fitness values across all conditions: t98=−32.64, P<0.0001). In fact, BC was below the detection limit (mean detection limit=1.9%) in 75.8% of all competitions. BC persistence was typically confined to replicates where BC starting frequency was 90%, but even for those cases BC counts were low (ranging between 1 and 8 CFU, average frequency 2.9%, Supplementary Figure 2 and Supplementary Table 1). This shows that PA is a very strong competitor that can drastically suppress BC within 24 h, precluding co-existence of both species.
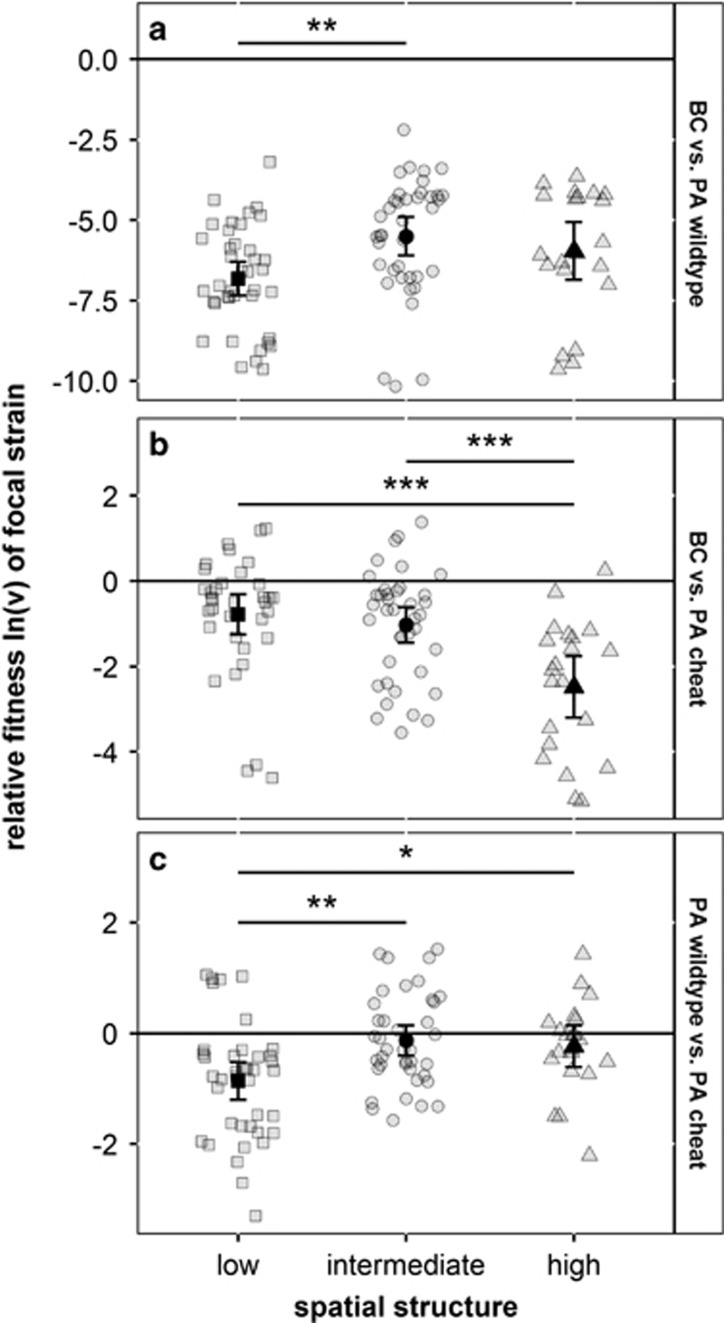
Figure 2.
Relative fitness ln(v) in pairwise competitions between BC, PA wild type and PA cheat in iron-limited CAA medium across three different spatial structures. BC is outcompeted by the PA wild type at all spatial structures (a). The PA cheat is a weaker competitor than the PA wild type, but still significantly outcompetes BC at all spatial structures (b). The PA wild type is outcompeted by the PA cheat only under low spatial structure (c). The relative fitness ln(v) of the focal strain (the first strain in the panel headers) is shown for three levels of spatial structure (square=low, circle=intermediate, triangle=high), and was calculated after 24 h of competition. The values of ln(v)<0, ln(v)>0, or ln(v)=0, indicate whether the focal strain respectively lost, won, or performed equally well in competition with its opponent. Gray symbols represent individual data points, whereas black symbols indicate means±95% CI. Asterisks indicate significant differences between treatments (*P<0.05, **P<0.01, ***P<0.001).
PA cheats change the competition dynamics
The PA cheat was significantly weaker in competition against BC than the PA wild type (LM: F1,189=447.9, P<0.0001; Figure 2a vs b), showing the importance of siderophores in the competition between PA and BC. Although the PA cheat significantly outcompeted BC under all spatial structures (Figure 2b; one-sample t-tests for low spatial structure: t95=−3.34, P=0.002; for intermediate spatial structure: t95=−5.02, P<0.0001; for high spatial structure: t95=−7.18, P<0.0001), there were significant differences between conditions. BC performed best in the medium with low spatial structure (low vs intermediate spatial structure, LM: t93=−3.86, P=0.0003; and low vs high spatial structure, LM: t93=−4.45, P<0.0001).
Spatial structure also significantly affected the relative fitness of the PA cheat in competition against the PA wild type (Figure 2c). In particular, the PA cheat was only able to significantly outgrow the PA wild type in low spatial structure (one-sample t-test: t38=−5.12, P<0.0001), whereas both strains were equally fit in intermediate (t38=−0.96, P=0.34) and high spatial structure (t20=−1.31, P=0.20). These results show that siderophore cheats can only invade a population of cooperating siderophore producers in the absence of any significant spatial structure (confirming previous findings, Kümmerli et al., 2009). This finding further demonstrates that the PA cheats’ capacity to increase intra-specific competition (that is, exerting a stabilizing effect (Chesson, 2000)) is highest with low spatial structure.
Iron limitation is required for PA cheats to alter competitive dynamics
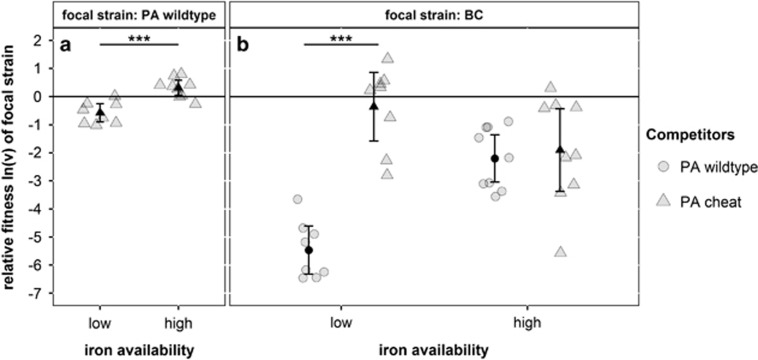
Control experiments revealed that it is indeed the ‘cheating effect’ – the PA cheat outcompetes the PA wild type, but is weaker in competition against BC – that affects the competitive dynamics in our iron-limited communities (Figure 3). When repeating all pairwise competitions in a medium where iron is more readily available and siderophores are no longer essential for iron scavenging, the cheating effect disappeared. Specifically, the relative fitness advantage the PA cheats enjoyed under low iron availability (one-sample t-test: t7=−4.19, P=0.0041) vanished under increased iron availability (Figure 3a). Moreover, our previous observation that BC experiences higher relative fitness in competition against the PA cheat than against the PA wild type was only apparent with low iron availability (LM: F1,14=65.43, P<0.0001), but not with high iron availability (LM: F1,16=0.162, P=0.693; Figure 3b). These findings show that both the equalizing and stabilizing fitness effects only prevail under low iron availability. The two effects disappear under increased iron availability because the PA wild type and the PA cheat become phenotypically similar (that is, reduced pyoverdine expression, Supplementary Figure 3) and therefore suppress BC to the same extent.
Figure 3.
Iron availability alters the competitive relationships between community members. The PA wild type (PA wt) is only outcompeted by the PA cheat in low iron availability, but not when iron is more readily available (a). Under low iron availability, BC performs significantly better against the PA cheat than against the PA wild type (b). This effect disappears in media with increased iron availability, where the PA wild type and the PA cheat both outcompeted BC to an intermediate but equal extent (b). The values of ln(v) indicate whether the focal strain lost (ln(v)<0), won (ln(v)>0), or performed equally well (ln(v)=0) in competition with its opponent. Gray symbols represent individual data points (high iron availability: 9 replicates; low iron availability: 8 replicates), whereas black symbols indicate means±95% CI. Asterisks indicate significant differences between treatments (***P<0.001).
Evolutionary dynamics in two-strain communities
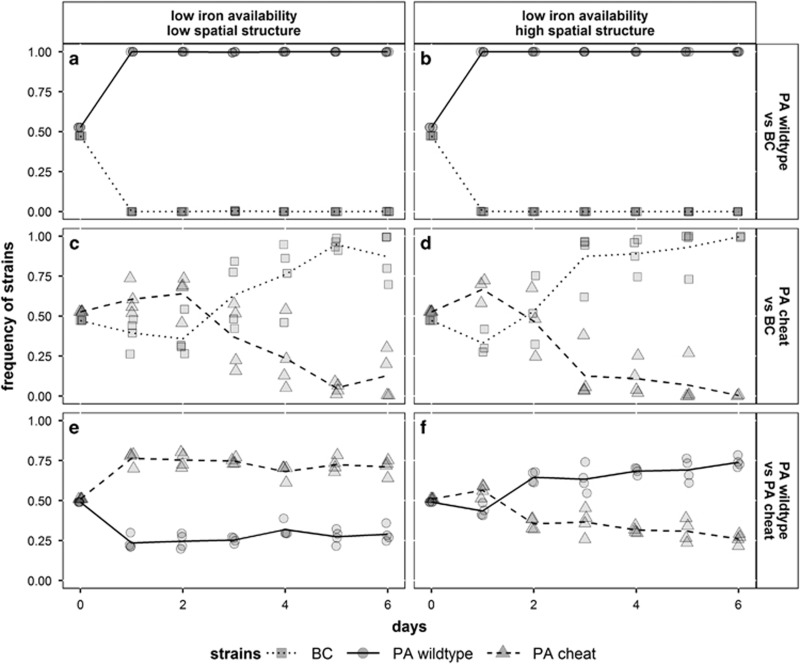
Our six-day evolution experiments confirmed that BC was drastically suppressed by the PA wild type regardless of the spatial structure (Figures 4a+b; mean detection limit of BC=0.27%, Supplementary Table 2). The dynamics significantly changed when replacing the PA wild type with the PA cheat. Here, the strain frequency fluctuated, under both low and high spatial structure, first in favor of the PA cheat and then, after the second day, in favor of BC (Figures 4c+d). The switch in the competitive strength between the two strains could be explained by phenotypic or genetic adaptations of BC to the presence of the PA cheat. Alternatively, it could be that PA cheats first benefited from internal iron stocks acquired during the pre-experimental growth in rich LB medium, which then became depleted during the course of the experiment. Finally, spatial structure strongly affected the long-term dynamics between the PA wild type and the PA cheat (Figures 4e+f). In line with our 24- h competition assays, we found that the PA cheat significantly outcompeted the PA wild type under low spatial structure, but was outcompeted itself by the PA wild type in the structured environment.
Figure 4.
Evolutionary dynamics in two-strain communities with BC (squares, dotted line), PA wild type (circles, solid line) and PA cheat (triangles, dashed line) in low and highly structured iron-limited CAA medium. Co-existence of BC and PA wild type is not possible, neither in unstructured (a) nor structured medium (b). BC becomes the dominant strain in competition with the PA cheat after 2 or 3 days regardless of spatial structure (c, d). The PA cheat outcompetes the PA wild type only in medium with low spatial structure (e) but not in highly structured medium (f). Symbols depict individual data points (from four replicates), whereas lines represent mean strain frequencies.
Evolutionary dynamics in three-strain communities
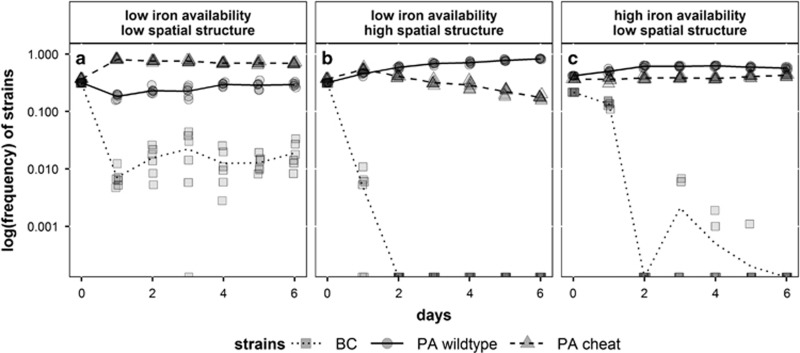
The two-strain evolutionary experiments above demonstrate that the PA cheat is a weak competitor against BC, but can itself significantly reduce the fitness of the PA wild type under low spatial structure and in iron-limited medium. Our experiments with three-strain communities demonstrate that this cheating effect was indeed sufficient to foster stable co-existence of BC, the PA wild type and the PA cheat over several days maintaining BC at low frequencies in the community (Figure 5a). Conversely, diversity could neither be sustained under high spatial structure (Figure 5b) nor under increased iron availability (Figure 5c; mean detection limit of BC on day 6 across all three conditions=0.19%, Supplementary Table 2).
Figure 5.
Evolutionary dynamics in three-strain communities consisting of BC (squares, dotted line), PA wild type (circles, solid line) and PA cheat (triangles, dashed line). BC was maintained at low frequency when iron availability and spatial structure were both low (a). In this scenario PA cheat could significantly outcompete the PA wild type and thereby buffer the competition between PA and BC. In contrast, species co-existence was neither possible when spatial structure was high (b) nor when iron availability was increased (c). In these scenarios, the PA cheat could not dominate the PA wild type, which led to the extinction of BC within 6 days. Symbols depict individual data points (from 6 replicates), whereas lines represent mean strain frequencies.
Discussion
Ecological theory predicts that species can locally co-exist if intra-specific competition exceeds inter-specific competition (Chesson, 2000; Lankau, 2009). We tested this prediction in bacterial communities consisting of the strong competitor (PA wild type) and its inferior counterpart BC – a species assembly where the PA wild type typically drives BC to extinction (Figure 4a). We introduced strong intra-specific competition through the addition of a PA cheat strain, which exploited the publically shared iron-scavenging pyoverdine secreted by the PA wild type. We demonstrate that this form of intra-specific competition indeed favors species co-existence under well-mixed culturing conditions, where the PA cheat efficiently exploits pyoverdine, thereby reducing both the growth of the PA wild type and the associated suppression of BC. The equalizing fitness effect exerted by the PA cheat was, however, reduced with increased spatial structuring of the environment. This is because structuring prevents successful pyoverdine exploitation, such that the PA wild type remained dominant in the population, driving BC rapidly to extinction. Since the sharing and exploitation of secreted compounds are common in the natural microbial world (West et al., 2007; Barrett et al., 2011; Cordero et al., 2012; Raymond et al., 2012; Andersen et al., 2015), we suggest that this form of intra-specific competition could take on an important role in maintaining biodiversity in habitats with relatively low spatial structure, such as marine ecosystems (Sogin et al., 2006; Rusch et al., 2007; Fuhrman et al., 2008).
Our finding that cheating reverses the relationship between spatial structure and biodiversity contradicts a common paradigm in ecology (Tilman, 1994; Kerr et al., 2002; Reichenbach et al., 2007; Hibbing et al., 2010; Celiker and Gore, 2012; Vos et al., 2013; Coyte et al., 2015). With regard to bacteria, our results particularly differ from those reported for toxin-mediated interference competition, where co-existence between toxin producers, susceptible and resistant strains has been shown to explicitly rely on spatial structure (Kerr et al., 2002; Kirkup and Riley, 2004; Narisawa et al., 2008; Biernaskie et al., 2013; Pérez-Gutiérrez et al., 2013; Abrudan et al., 2015). The reason for this difference resides in the different types of social interactions involved. In the case of strong interference competition via toxins, spatial structure has a stabilizing fitness effect. It physically separates competing species, and thereby indirectly increases intra-specific competition. In contrast, public goods cheating has both a stabilizing (that is, increased intra-specific competition) and an equalizing (fitness of competing strains converge the more cheats there are) component. These two components are maximized with low spatial structure where public goods exploitation by cheats is most efficient (Figure 2c; Kümmerli et al., 2009; Yang et al., 2010; Mitri et al., 2011). It is conceivable that the importance of the two social interaction types varies across habitat type: toxin-mediated interactions might be more relevant for biodiversity in well-structured soil- and surface-attached communities, whereas public goods cheating might be an important driver of biodiversity in relatively unstructured aquatic planktonic communities.
There are two recent studies, which present different mechanisms that can also maintain microbial biodiversity in the absence of significant spatial structure (Kelsic et al., 2015; Inglis et al., 2016). Kelsic et al. (2015) extended the classic models of toxin production (Czaran et al., 2002; Kerr et al., 2002) to include the cooperative degradation of toxins outside the cell, which offers immunity to resistant and susceptible strains alike. Their experimental work on agar plates showed that biodiversity can be stabilized by cooperative toxin degradation with high spatial structure. Their theoretical models suggest that the same mechanism could also stabilize biodiversity in the absence of spatial structure. Inglis et al. (2016) on the other hand, established the loner effect, where cooperators and cheats can co-exist in the presence of a loner strain, which has an independent lifestyle and does not partake in social interactions with the other community members. In their system, loners beat cheats, cheats beat cooperators and cooperators beat loners, leading to cyclical rock-paper-scissors dynamics and strain co-existence. In principle, BC could act as a loner in our community fostering co-existence between cooperators and cheats. However, this is unlikely to be the case because the cheat-cooperator dynamics were unaffected by the presence of BC (Figures 4e and f and Figures 5a and b), and there was no evidence for cyclical succession of strains as observed in Inglis et al. (2016). Rather BC stayed at a constant low frequency in the population. This suggests that the cheating effect described here is a novel mechanism promoting species co-existence under low spatial structure.
In our scenario, community stability clearly depended on the relative success of cheats. Our results show that if the PA cheat cannot sufficiently supress the PA wild type then the weak competitor BC goes extinct (Figure 5). On the other hand, if the PA cheat were too efficient in exploiting the PA wild type then community stability would also not be possible, because the PA cheat would be expected to first displace the PA wild type, to then be displaced by BC. Thus, for cheating to sustain biodiversity in the long-term, an intermediate level of cheat success is required. There are three mutually non-exclusive factors that may contribute to an intermediate cheat success. First, the relative success of public good cheats often declines when they become more common, which can maintain cooperators at relatively low frequency in the population (Ross-Gillespie et al., 2007; Gore et al., 2009; Zhou et al., 2014). Second, even in relatively well-mixed habitats, such as the ocean, there is some level of spatial structuring, which might prevent cheats from completely outcompeting cooperators (Cordero et al., 2012; Kümmerli et al., 2014). Finally, experimental evolution studies have shown that cooperators can rapidly adapt to the presence of cheats, thereby preventing cheats from fixing in the population (Zhang et al., 2009; Kümmerli et al., 2015).
While our results highlight that intra-specific competition can counterbalance inter-specific competition and foster species co-existence, we found that BC was typically maintained at rather low frequencies (that is, below 5%) in the community (Figure 5a). This finding is reminiscent of species abundance patterns in natural communities, where a few abundant species often occur together with an enormous diversity of rare species, the so-called rare biosphere (Sogin et al., 2006; Rusch et al., 2007). Although hypothetical at this stage, it would be interesting to test whether the cheating mechanism described in this paper can explain the occurrence and maintenance of the rare biosphere in natural habitats.
In conclusion, our study establishes cheating as a mechanism that can maintain biodiversity in habitats with low spatial structure. For this mechanism to work, cheating must be prevalent in dominant species but rare in subordinate species of the community. Although it remains to be seen whether this assumption indeed holds for natural multi-species communities, it seems a realistic assumption, as populations of dominant species are necessarily larger, which should favor the evolution of de novo cheats more readily due to higher mutation supply rates. Taken together, we show that cheating, which is typically considered as a destructive force in biology, undermining group-beneficial traits, can take on an important constructive role in shaping species co-existence at the level of the community.
Acknowledgments
We thank Martin Ackermann and Andy Gardner for comments, Leo Eberl for comments and providing strains, and the Swiss National Science Foundation for funding (grant no. PP00P3-139164).
Author contributions
AL and RK designed the research. AL conducted the experiments. AL, RFI and RK analyzed the data. AL, RFI and RK wrote the manuscript.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Abrudan MI, Smakman F, Grimbergen AJ, Westhoff S, Miller EL, van Wezel GP et al. (2015). Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc Natl Acad Sci USA 112: 11054–11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SB, Marvig RL, Molin S, Johansen HK, Griffin AS. (2015). Long-term social dynamics drive loss of function in pathogenic bacteria. Proc Natl Acad Sci USA 112: 10756–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabás G, Michalska-Smith MJ, Allesina S. (2016). The effect of intra- and interspecific competition on coexistence in multispecies communities. Am Nat 188: E1–E12. [DOI] [PubMed] [Google Scholar]
- Barrett LG, Bell T, Dwyer G, Bergelson J. (2011). Cheating, trade-offs and the evolution of aggressiveness in a natural pathogen population. Ecol Lett 14: 1149–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Eisenhauer N, Scheu S, Jousset A. (2012). Increasing antagonistic interactions cause bacterial communities to collapse at high diversity. Ecol Lett 15: 468–474. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate: a pratical and powerful approach to multiple testing. J Roy Stat Soc Ser A 57: 289–300. [Google Scholar]
- Biernaskie JM, Gardner A, West SA. (2013). Multicoloured greenbeards, bacteriocin diversity and the rock-paper-scissors game. J Evol Biol 26: 2081–2094. [DOI] [PubMed] [Google Scholar]
- Bragonzi A, Farulla I, Paroni M, Twomey KB, Pirone L, Lore NI et al. (2012). Modelling co-infection of the cystic fibrosis lung by Pseudomonas aeruginosa and Burkholderia cenocepacia reveals influences on biofilm formation and host response. PLoS One 7: e52330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst MA, Hochberg ME, Bell T, Buckling A. (2006). Character displacement promotes cooperation in bacterial biofilms. Curr Biol 16: 2030–2034. [DOI] [PubMed] [Google Scholar]
- Celiker H, Gore J. (2012). Competition between species can stabilize public-goods cooperation within a species. Mol Syst Biol 8: 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesson P. (2000). Mechanisms of maintenance of species diversity. Annu Rev Ecol Evol Syst 31: 343–366. [Google Scholar]
- Cordero OX, Ventouras L-A, DeLong EF, Polz MF. (2012). Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci USA 109: 20059–20064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello A, Reen FJ, O'Gara F, Callaghan M, McClean S. (2014). Inhibition of co-colonizing cystic fibrosis-associated pathogens by Pseudomonas aeruginosa and Burkholderia multivorans. Microbiology 160: 1474–1487. [DOI] [PubMed] [Google Scholar]
- Coyte KZ, Schluter J, Foster KR. (2015). The ecology of the microbiome: Networks, competition, and stability. Science 350: 663–666. [DOI] [PubMed] [Google Scholar]
- Curtis TP, Sloan WT, Scannell JW. (2002). Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA 99: 10494–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaran T, Hoekstra RF, Pagie L. (2002). Chemical warfare between microbes promotes biodiversity. Proc Natl Acad Sci USA 99: 786–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas Z, Kümmerli R. (2012). Cost of cooperation rules selection for cheats in bacterial metapopulations. J Evol Biol 25: 473–484. [DOI] [PubMed] [Google Scholar]
- Dumas Z, Ross-Gillespie A, Kümmerli R. (2013). Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proc Biol Sci 280: 20131055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KR, Bell T. (2012). Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol 22: 1845–1850. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL et al. (2008). A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA 105: 7774–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore J, Youk H, van Oudenaarden A. (2009). Snowdrift game dynamics and facultative cheating in yeast. Nature 456: 253–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotschlich A, Huber B, Geisenberger O, Togl A, Steidle A, Riedel K et al. (2001). Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst Appl Microbiol 24: 1–14. [DOI] [PubMed] [Google Scholar]
- Griffin A, West SA, Buckling A. (2004). Cooperation and competition in pathogenic bacteria. Nature 430: 1024–1027. [DOI] [PubMed] [Google Scholar]
- Hardin G. (1960). The competitive exclusion principle. Science 131: 1292–1297. [DOI] [PubMed] [Google Scholar]
- Harrison F, Paul J, Massey RC, Buckling A. (2008). Interspecific competition and siderophore-mediated cooperation in Pseudomonas aeruginosa. ISME J 2: 49–55. [DOI] [PubMed] [Google Scholar]
- Hersman LE, Huang A, Maurice PA, Forsythe JE. (2000). Siderophore production and iron reduction by Pseudomonas mendocina in response to iron deprivation. Geomicrobiol J 17: 261–273. [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB. (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RD, Grover J, Tilman D. (1994). Simple rules for interspecific dominance in systems with exploitative and apparent competition. Am Nat 144: 741–771. [Google Scholar]
- Inglis RF, Biernaskie JM, Gardner A, Kümmerli R. (2016). Presence of a loner strain maintains cooperation and diversity in well-mixed bacterial communities. Proc Biol Sci 283: 20152682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsic ED, Zhao J, Vetsigian K, Kishony R. (2015). Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature 521: 516–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B, Riley MA, Feldman MW, Bohannan BJM. (2002). Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418: 171–174. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. (2008). Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci USA 105: 18188–18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkup BC, Riley MA. (2004). Antibiotic-mediated antagonism leads to a bacterial game of rock–paper–scissors in vivo. Nature 428: 412–414. [DOI] [PubMed] [Google Scholar]
- Kümmerli R, Griffin AS, West SA, Buckling A, Harrison F. (2009). Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc Biol Sci 276: 3531–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmerli R, Schiessl KT, Waldvogel T, McNeill K, Ackermann M. (2014). Habitat structure and the evolution of diffusible siderophores in bacteria. Ecol Lett 17: 1536–1544. [DOI] [PubMed] [Google Scholar]
- Kümmerli R, Santorelli L, Granato E, Dumas Z, Dobay A, Griffin AS et al. (2015). Co-evolutionary dynamics between public good producers and cheats in the bacterium Pseudomonas aeruginosa. J Evol Biol 28: 2264–2274. [DOI] [PubMed] [Google Scholar]
- Lambiase A, Raia V, Del Pezzo M, Sepe A, Carnovale V, Rossano F. (2006). Microbiology of airway disease in a cohort of patients with cystic fibrosis. BMC Infect Dis 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankau RA. (2009). Genetic variation promotes long-term coexistence of Brassica nigra and its competitors. Am Nat 174: E40–E53. [DOI] [PubMed] [Google Scholar]
- Miethke M, Marahiel MA. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71: 413–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitri S, Xavier JB, Foster KR. (2011). Social evolution in multispecies biofilms. Proc Natl Acad Sci USA 108: 10839–10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narisawa N, Haruta S, Arai H, Ishii M, Igarashi Y. (2008). Coexistence of antibiotic-producing and antibiotic-sensitive bacteria in biofilms is mediated by resistant bacteria. Appl Environ Microbiol 74: 3887–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Gutiérrez RA, López-Ramírez V, Islas Á, Alcaraz LD, Hernández-González I, Olivera BCL et al. (2013). Antagonism influences assembly of a Bacillus guild in a local community and is depicted as a food-chain network. ISME J 7: 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. (2015) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Rainey PB, Travisano M. (1998). Adaptive radiation in a heterogeneous environment. Nature 394: 69–72. [DOI] [PubMed] [Google Scholar]
- Rainey PB, Rainey K. (2003). Evolution of cooperation and conflict in experimental bacterial poplations. Nature 425: 72–74. [DOI] [PubMed] [Google Scholar]
- Raymond B, West SA, Griffin AS, Bonsall MB. (2012). The dynamics of cooperative bacterial virulence in the field. Science 337: 85–88. [DOI] [PubMed] [Google Scholar]
- Reichenbach T, Mobilia M, Frey E. (2007). Mobility promotes and jeopardizes biodiversity in rock-paper-scissors games. Nature 448: 1046–1049. [DOI] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Rosenzweig RF, Sharp RR, Treves DS, Adams J. (1994). Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics 137: 903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Gillespie A, Gardner A, West SA, Griffin AS. (2007). Frequency dependence and cooperation: theory and a test with bacteria. Am Nat 170: 331–342. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored ‘‘rare biosphere’’. Proc Natl Acad Sci USA 103: 12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G et al. (2015). Structure and function of the global ocean microbiome. Science 348: 1261359. [DOI] [PubMed] [Google Scholar]
- The Human Microbiome Project. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D. (1994). Competition and biodiversity in spatially structured habitats. Ecology 75: 2–16. [Google Scholar]
- Tyrrell J, Callaghan M. (2015). Iron acquisition in the Cystic Fibrosis lung and potential for novel therapeutic strategies. Microbiology 162: 191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell J, Whelan N, Wright C, Sa-Correia I, McClean S, Thomas M et al. (2015). Investigation of the multifaceted iron acquisition strategies of Burkholderia cenocepacia. Biometals 28: 367–380. [DOI] [PubMed] [Google Scholar]
- Vos M, Wolf AB, Jennings SJ, Kowalchuk GA. (2013). Micro-scale determinants of bacterial diversity in soil. FEMS Microbiol Rev 37: 936–954. [DOI] [PubMed] [Google Scholar]
- Weaver BW, Kolter R. (2004). Burkholderia spp. alter Pseudomonas aeruginosa physiology through iron sequestration. J Bacteriol 186: 2376–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. (2007). The social lives of microbes. Annu Rev Ecol, Evol Syst 38: 53–77. [Google Scholar]
- Yang J, Evans BA, Rozen DE. (2010). Signal diffusion and the mitigation of social exploitation in pneumococcal competence signalling. Proc Biol Sci 277: 2991–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q-G, Buckling A, Ellis RJ, Godfray HCJ. (2009). Coevolution between cooperators and cheats in a microbial system. Evolution 63: 2248–2256. [DOI] [PubMed] [Google Scholar]
- Zhou L, Slamti L, Nielsen-LeRoux C, Lereclus D, Raymond B. (2014). The social biology of quorum sensing in a naturalistic host pathogen system. Curr Biol 24: 2417–2422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.