Abstract
MicroRNAs (miRNAs) regulate gene expression by guiding the Argonaute (Ago)-containing RNA-induced silencing complex (RISC) to specific target mRNA molecules. It is well established that miRNAs are stabilized by Ago proteins, but the molecular features that trigger miRNA destabilization from Ago proteins remain largely unknown. To explore the molecular mechanisms of how targets affect the stability of miRNAs in human Ago (hAgo) proteins, we employed an in vitro system that consisted of a minimal hAgo2-RISC in HEK293T cell lysates. Surprisingly, we found that miRNAs are drastically destabilized by binding to seedless, non-canonical targets. We showed that miRNAs are destabilized at their 3′ ends during this process, which is largely attributed to the conformational flexibility of the L1-PAZ domain. Based on these results, we propose that non-canonical targets may play an important regulatory role in controlling the stability of miRNAs, instead of being regulated by miRNAs.
Keywords: microRNA, Argonaute, Non-canonical target, Target-directed microRNA destabilization
MicroRNAs (miRNAs) are ~22-long non-coding RNA molecules that are a key regulatory component in eukaryotes. Bioinformatics combined with functional analysis have established that Watson-Crick base-pairing between the 5′ seed region (positions 2–8) of mature miRNAs and their target mRNAs is the key determinant for target recognition. This is further supported by structural analysis of Argonaute (Ago) proteins showing that seed sequences are favorably positioned for efficient target recognition. However, several studies also suggested that the presence of a seed site alone does not guarantee the functional interactions (Didiano et al (2006) Nat Struct Mol Biol 13, 849–851). These observations led to the development of methods to directly identify the targets of miRNAs. These methods are usually accompanied by biochemical isolation of the miRNA-target RNA complex via cross-linking and immunoprecipitation, followed by high-throughput sequencing (CLIP-seq) (Chi et al (2009) Nature 460, 479–486). More recently, an improved CLIP-seq method, called CLASH (cross linking, ligation, and sequencing of a miRNA-target chimera), included an additional step by which miRNAs are ligated to their target RNAs within the Ago-complex (Helwak et al (2013) Cell 153, 654–665). These miRNA-target chimeric reads are likely to contain sequence features at high confidence that are directly relevant to miRNA target recognition. Taking advantages of these powerful methods, it has been turned out that a substantial amount of miRNA target sites were non-canonical with imperfect or non-seed interactions. However, these non-canonical sites did not confer efficient target silencing, raising an intriguing question regarding their functional roles (Agarwal et al (2015) eLife 4, e05005).
It is well established that mature miRNAs are stabilized by Ago loading, and thereby can guide RISCs to complementary target RNAs for silencing purposes. By contrast, it has been recently suggested that a highly complementary target binding may have the opposite effect that leads to destabilization of miRNAs from Ago proteins (Ameres et al (2010) Science 328, 1534–1539). How these processes are controlled and which molecular players are important to remain largely elusive. We hypothesized that non-canonical targets are not subjected to miRNA-mediated silencing or at least not efficient ones, but instead, they might have regulatory roles in miRNA stability. To test this hypothesis, we first established an in vitro system that consisted of a minimal human Ago-RISC prepared from human embryonic kidney 293T cells and aimed to understand how the stabilities of miRNAs in human Ago proteins were affected by non-canonical interactions. Because seedless targets were expected to have low binding affinities to RISC, the in vitro system was particularly useful to create crowded environments with an excess amount of target RNAs. We found that the seedless, non-canonical targets negatively regulated the stabilities of miRNAs in human Ago proteins. In other words, a complete seed pairing dramatically inhibited miRNA destabilization, and thus, the most effective sites for stimulating destabilization did not only have extensive pairing to the 3′ region of the miRNAs but also had mismatches in the seed region.
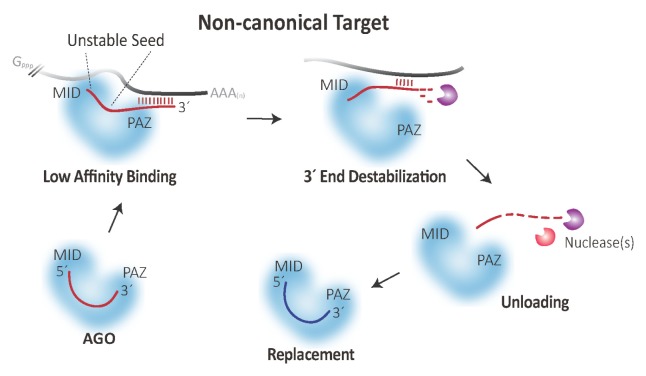
In addition, we observed two different mechanisms of non-canonical target-mediated miRNA destabilization (Fig. 1): (i) miRNAs were destabilized at their 3′ ends, while they were still anchored in Ago proteins (i.e., 3′ end destabilization within Ago), and (ii) miRNAs were completely released from the Ago protein (i.e., unloading) (De et al (2013) Mol Cell 50, 344–355). How can these mechanisms be explained? Structural studies have shown that 3′ end of miRNAs are initially anchored to the PAZ domain and released when the base-pairing between miRNAs and their targets is propagated toward the 3′ end. We reasoned that miRNAs are initially destabilized at their 3′ end during these conformational changes upon binding to non-canonical targets. To gain insights into features of Ago proteins that promote destabilization, we extended our analysis with mutated Ago proteins whose residues are expected to be involved in these processes. From this, we found that residues in the PAZ domain and its hinge L1 are required for efficient miRNA destabilization. The complete release of miRNAs from Ago proteins is expected to occur after the 3′ end destabilization, possibly due to the instability of the 5′ seed region (Fig. 1).
Fig. 1.
Proposed reaction mechanisms of non-canonical targets in the regulation of miRNA stability in human Ago proteins. miRNAs bind to non-canonical targets with low affinities because of mismatches in the seed region. Upon an extensive 3′ pairing between miRNAs and their targets, the 3′ ends of miRNAs are released from the PAZ domain, and this, in turn, leads to destabilization of miRNAs from Ago proteins.
What is the biological significance of these findings? The Ago protein is the main limiting factor for efficient RNA silencing pathway in cells. Therefore, Ago proteins should be reused for other miRNA molecules, and the proposed mechanisms may be efficient strategies for Ago proteins for the removal of unnecessary or less important miRNAs and the replacement with more important ones (Fig. 1). Our results indicate that Ago proteins, at least for humans, are able to efficiently distinguish between the natural (canonical) and non-natural (non-canonical) targets during the lateral diffusion of target search. These mechanisms can be especially useful to prevent the fitness costs of overactive miRNA-mediated silencing and to reduce high levels of transcription/translation of Ago genes that incur large metabolic costs.
What would be the endogenous correlation between miRNAs and their non-canonical targets, in terms of their expression levels? There are two possible perspectives for this question. It is tempting to speculate that a negative correlation would be beneficial for miRNAs that do not have targets with intact seed matches (natural targets). Such miRNAs should be removed quickly to avoid a possible saturation of Ago proteins. In contrast, we also postulate that the abundance of miRNAs and their non-canonical targets can be positively correlated with one another. Highly expressed miRNAs are more likely to have important functions, and thus, their turnover and 3′ end heterogeneities should be kept under strict control.
The term “non-canonical” is used as a broad term to describe the target sites that contain imperfect complementarity to the 5′ seed region of the miRNAs. From our results, it is of importance to note that not all non-canonical targets can effectively destabilize miRNAs in human Ago proteins unless such targets contain extensive complementarity to the 3′ ends of miRNAs. Because a large proportion of non-canonical sites identified from CLIP-seq do not appear to have a contiguous pairing to the 3′ end of miRNAs, the degree of regulation mediated by non-canonical targets can be limited in cells. Although CLIP-seq experiments have shown that non-canonical sites are more prevalent than previously appreciated, the results from our in vitro binding assay demonstrated that miRNAs bind to non-canonical targets with approximately 10-fold less affinity to RISC. Based on these observations, we postulate that, while canonical seed interactions constitute a general pathway of miRNA function, the non-canonical interactions may provide a means to fine-tune, refine and diversify miRNAs in cells. Further analyses, including bioinformatics and statistics, are required to unravel to what extent such mechanisms contribute to the dynamically controlled regulations of miRNAs and their target transcripts under steady-state conditions.
ACKNOWLEDGEMENTS
This work was supported by the Next-Generation BioGreen 21 Program (No. PJ01115601), Rural Development Administration, Republic of Korea.
Abbreviations
- Ago
Argonaute
- CLASH
Crosslinking, ligation, and sequencing of hybrids
- CLIP
Cross-linking and immunoprecipitation
- HEK293T
human embryonic kidney 293T
- miRNA
microRNA
- RISC
RNA-induced silencing complex
Footnotes
Perspective to: Park et al. (2017), Non-canonical targets destabilize microRNAs in human Argonautes, Nucleic Acids Research, 45 (4): 1569–1583, doi: 10.1093/nar/gkx029