Abstract
Proper maintenance of biological components is crucial for longevity and healthy aging. Although the role of homeostatic maintenance systems for DNA and protein in longevity is established, it remains largely unknown for RNA. In our recent work, we show that nonsense-mediated mRNA decay (NMD) promotes longevity in the roundworm C. elegans by enhancing RNA quality control. We find that the activity of NMD decreases during aging, raising the possibility that RNA quality declines in old animals. We then show that key components of NMD complex are required for prolonged lifespan in C. elegans. In addition, animals with reduced insulin/insulin-like growth factor-1 (IGF-1) signaling (IIS), a representative longevity model, display increased NMD activity. Thus, up-regulation of NMD appears to play crucial roles in longevity conferred by reduced IIS via enhancing mRNA quality control. As both IIS and NMD pathways are evolutionarily conserved, mammals including humans may be equipped with similar RNA quality control systems to achieve longevity.
Keywords: Aging, Caenorhabditis elegans, daf-2/insulin/IGF-1 signaling, Nonsense-mediated mRNA decay, smg-2
DNA and protein quality control systems deteriorate with aging, and this causes diverse aging-related diseases such as Werner syndrome and Alzheimer’s disease. However, whether RNA quality control is affected by aging or itself regulates aging remains largely unexplored. For the last several years, our group in collaboration with Dr. Hong Gil Nam’s group has focused on this issue by using the roundworm C. elegans, an excellent model for aging research.
We have focused on RNA helicases that are involved in diverse aspects of RNA biology ranging from transcription, splicing, translation, to RNA degradation. We first performed a large-scale RNA interference-based lifespan screen upon knocking down each of 78 genes that encode proteins that contain RNA helicase domains in C. elegans (Seo M et al (2015) Proc Nat Acad Sci U S A 112, E4246–4255). We identified 11 RNA helicases, which are crucial for longevity conferred by reduced insulin/insulin-like growth factor-1 (IGF-1) signaling (IIS), an evolutionarily conserved aging-regulatory pathway. We reported the functional characterization of two RNA helicases, HEL-1 (DEAD-box helicase 39A/UAP56/BAT1) (Seo M et al (2015) Proc Nat Acad Sci U S A 112, E4246–4255) and SACY-1 (DEAD-box helicase 41 (Seo M et al (2016) Cell Cycle (Georgetown, Tex) 15, 1821–1829). Herein, we introduce our most recent paper showing the life-extending roles of SMG-2 (UPF1 RNA helicase), a core component of nonsense-mediated mRNA decay (NMD) responsible for mRNA quality control.
NMD degrades aberrant RNAs such as premature termination codon (PTC)-containing mRNAs. As PTC-containing mRNAs are translated into potentially toxic truncated proteins, NMD is critical for maintaining normal cellular physiology. NMD is also known to degrade other endogenous mRNAs, such as upstream open reading frame (uORF)- and long 3′ untranslated region (UTR)-containing transcripts. We show that NMD activity decreases during C. elegans aging by using a green fluorescent protein (GFP)-fused PTC-containing NMD reporter. We then find that the levels of various NMD targets, including PTC-, uORF- and long 3′ UTR-containing transcripts, are decreased in long-lived daf-2/insulin IGF-1 receptor mutant animals, which display reduced IIS and delayed aging phenotypes. Furthermore, we show that reduced IIS increases the degradation rate of NMD target transcripts. Thus, age- dependent decreases in NMD activity appear to play a causative role in C. elegans longevity.
Different tissues have distinct roles in the aging of a whole organism, and therefore we investigated in which tissues NMD contributes to longevity in C. elegans. Interestingly, NMD in neurons plays the most important role in long lifespan. Through the analysis of neuron-specific RNA-seq. data (Kaletsky R et al (2016) Nature 529, 92–96), we find that the levels of NMD targets are down-regulated in the neurons of animals with reduced IIS. In addition, we show that RNA quality in neurons is better maintained than that in other tissues during aging. These data indicate the importance of the neuronal NMD for C. elegans longevity. Interestingly, most neurons in the mammalian central nervous system (CNS) are post-mitotic and may need to maintain cellular components longer than those in other tissues. Therefore, we speculate that NMD in neurons of the mammalian CNS may also be enhanced, as compared to that in other tissues.
We next aimed to identify key NMD target transcripts for longevity. We show that the level of a yars-2 (tyrosyl-tRNA synthetase 2) splice variant, a canonical NMD target, is decreased by reduced IIS. Importantly, down-regulation of yars-2 is responsible for long lifespan in animals with reduced IIS. These data imply that reduced IIS enhances NMD, which decreases the levels of the yars-2 transcript, and this in turn promotes longevity. As tRNA synthetases are essential components for mRNA translation, these results raise the possibility that enhanced NMD may extend lifespan by regulating both mRNA decay and translation. Our data are also in agreement with previous reports showing that translation rates are down-regulated in C. elegans with reduced IIS (Stout GJ et al (2013) Mol Syst Biol 9, 679) and that genetic inhibition of several tRNA synthetases increases lifespan (Lee SS et al (2003) Nat Genet 33, 40–48; Kim Y and Sun H (2007) Aging Cell 6, 489–503).
In conclusion, our study demonstrates that proper removal of mRNAs via NMD-mediated RNA quality control, particularly in neurons, is pivotal for organismal longevity (Fig. 1). Interestingly, we and others recently reported that factors regulating mRNA splicing are also important for C. elegans longevity (Seo M et al (2016) Cell Cycle (Georgetown, Tex) 15, 1821–1829; Heintz et al (2017) Nature 541, 102–106). Therefore, precise regulation of mRNA quality at multiple steps, including splicing and degradation, appears to regulate organismal lifespan. As these RNA-processing mechanisms are evolutionarily conserved, it will be of interest to test whether similar RNA quality control systems play roles in longevity in mammals, including humans.
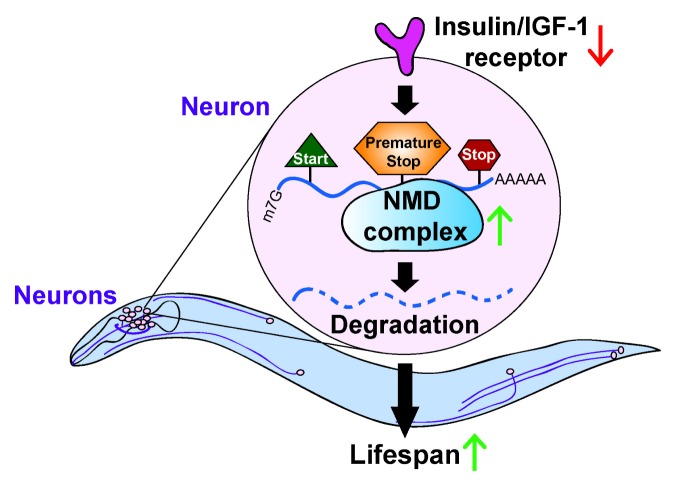
Fig. 1.
A model for the roles of nonsense-mediated mRNA decay (NMD) in C. elegans longevity. When insulin/IGF-1 signaling is reduced, for example by genetic inhibition of the insulin/IGF-1 receptor, NMD is enhanced. The NMD complex then degrades aberrant mRNAs such as premature termination (stop) codon (PTC)-containing transcripts, and contributes to lifespan extension. Among several tissues, NMD in neurons appears to be the most crucial for longevity.
ACKNOWLEDGEMENTS
This work was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health and Welfare (HI14C2337) to S.-J.V.L. We thank Lee laboratory members for helpful comments on this manuscript.
Abbreviations
- C. elegans
Caenorhabditis elegans
- IIS
insulin/IGF-1 signaling
- NMD
nonsense-mediated mRNA decay
- PTC
premature termination codon
- uORF
upstream open reading frame
- 3′ UTR
3′ untranslated region
- GFP
green fluorescent protein
Footnotes
Perspective to: Son HG, et al. RNA surveillance via nonsense- mediated mRNA decay is crucial for longevity in daf-2/insulin/ IGF-1 mutant C. elegans. Nature Communications 8, 14749 (2017) DOI: 10.1038/ncomms14749