Abstract
In mammals, cap-dependent translation of mRNAs is initiated by two distinct mechanisms: cap-binding complex (CBC; a heterodimer of CBP80 and 20)-dependent translation (CT) and eIF4E-dependent translation (ET). Both translation initiation mechanisms share common features in driving cap-dependent translation; nevertheless, they can be distinguished from each other based on their molecular features and biological roles. CT is largely associated with mRNA surveillance such as nonsense-mediated mRNA decay (NMD), whereas ET is predominantly involved in the bulk of protein synthesis. However, several recent studies have demonstrated that CT and ET have similar roles in protein synthesis and mRNA surveillance. In a subset of mRNAs, CT preferentially drives the cap-dependent translation, as ET does, and ET is responsible for mRNA surveillance, as CT does. In this review, we summarize and compare the molecular features of CT and ET with a focus on the emerging roles of CT in translation.
Keywords: CBC, eIF4E, NMD, Translation
INTRODUCTION
Eukaryotic transcripts synthesized by RNA polymerase II are sequentially and coordinately processed into mature messenger RNAs (mRNAs) through various nuclear events such as transcription, 5′-end capping, splicing, and 3′-end processing/polyadenylation (1). Early in transcription, the 5′-end of the primary transcript is covalently modified by the addition of a 7-methylguanosine cap, which co-transcriptionally occurs via nuclear capping enzyme (2, 3). The 5′-cap structure is recognized by the nuclear cap-binding complex (CBC), composed of a heterodimer of cap-binding protein 80 (CBP80) and cap-binding protein 20 (CBP20) [also known as nuclear cap-binding protein 1 (NCBP1) and nuclear cap-binding protein 2 (NCBP2), respectively]. The CBC-bound pre-mRNA is further processed by a splicing machinery. As a consequence of splicing, a specialized protein complex named the exon junction complex (EJC), which consists of four core components (eIF4A3, MAGOH, Y14, and MLN51) and a number of auxiliary factors, is deposited approximately 20–24 nucleotides upstream of each exon-exon junction (4, 5). In addition, the 3′-end of transcript is cleaved and poly-adenylated. The 3′-poly(A) tail is recognized by nuclear poly(A)-binding protein 1 (PABPN1; also known as PABP2) (6). Mature mRNAs are exported from the nucleus to the cytoplasm only after the completion of proper mRNA processing in the nucleus, and they contain a 5′-cap bound by the CBC, 3′-poly(A) tail bound by PABPN1, and exon-exon junctions associated with EJCs. The mRNAs either being exported from the nucleus or completely exported via the nuclear pore complex serve as a template for translation in the cytoplasm.
In the cytoplasm, the 5′-cap directs cap-dependent translation via two distinct pathways: CBC-dependent translation (CT) and eukaryotic translation initiation factor 4E (eIF4E)-dependent translation (ET) (7). CT is believed to precede ET because CBC-bound mRNA is a precursor of eIF4E-bound mRNA (8). With respect to their functional roles, it is generally considered that CT is largely involved in mRNA quality control, whereas ET is in charge of the bulk of protein synthesis (7). However, recent studies have made it harder to define the functional boundary between CT and ET. Therefore, in this review, we describe the recent findings on CT and compare the molecular features of CT and ET with a focus on the emerging roles of CT in eukaryotic gene expression.
MOLECULAR FEATURES OF CT: SIMILARITIES AND DIFFERENCES BETWEEN CT AND ET
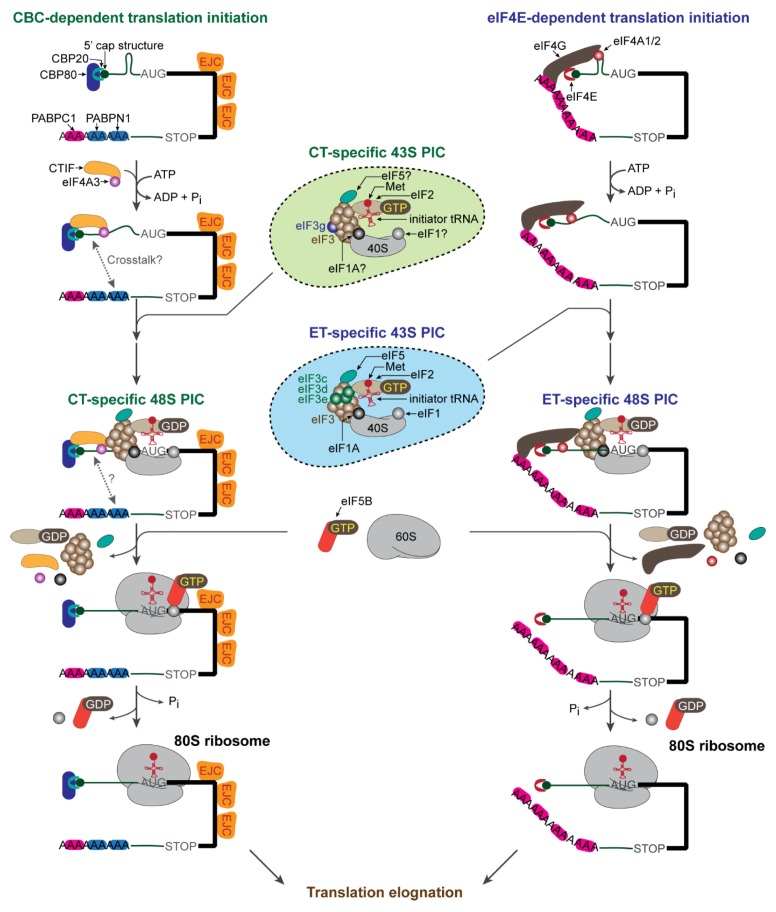
Eukaryotic translation initiation occurs through two distinct steps (Fig. 1); the first is the formation of the 48S preinitiation complex (PIC) with codon-anticodon base pairing between the initiator tRNA and a start codon in the P site of a 40S ribosome (a small subunit of ribosome), and the second is the joining of a 60S ribosome (a large subunit of ribosome) to the PIC to form an elongation-competent 80S ribosome (9, 10). To this end, the 40S ribosome should be physically attached to the cap-binding protein bound to the 5′-cap structure for efficient ribosome scanning through the 5′ untranslated region (5′UTR) to locate the start codon in the proper context. The physical association is mediated by a scaffold protein, which directly binds to the cap-binding protein and eIF3 complex (11). Notably, the eIF3 complex physically associates with the 40S ribosome and various translation initiation factors such as (i) eIF1 and eIF1A, which confer the fidelity of start codon selection (12, 13), (ii) the ternary complex consisting of the eIF2 complex (composed of eIF2α, eIF2β, and eIF2γ), GTP, and methionine-charged initiator tRNAi (14), and (iii) eIF5, which stimulates the GTPase activity of eIF2γ and consequently leads to the conversion of GTP-bound eIF2 to GDP-bound eIF2 (9, 10). The resulting multi-factor complex is called the 43S PIC (or the scanning complex) (9).
Fig. 1.
Mechanistic comparison of CBC-dependent translation (CT) and eIF4E-dependent translation (ET). The details are described in the text.
For ET, the 40S ribosome is attached to the 5′-cap structure via eIF4G, which directly binds to eIF4E (9, 10) and the eIF3 complex (15, 16). In the case of CT, a majority of mRNA-bound CBC recruits the 40S ribosome through physical interactions of a CT-specific factor, CBC-dependent translation initiation factor (CTIF) with CBP80 (17) and the eIF3 complex (18). However, an ET-specific factor, eIF4G also has an ability to bridge the CBC and the eIF3 complex, although there are some controversial reports (17–21). Intriguingly, eIF4G and CTIF serve as a binding platform for the eIF3 complex because they share an evolutionarily conserved domain called the middle domain of eukaryotic translation initiation factor 4G (MIF4G domain) with approximately 46% similarity (17, 22, 23). However, they directly bind to different subunits of the eIF3 complex to recruit the 40S ribosome to mRNA (eIF3c, eIF3d, and eIF3e for eIF4G and eIF3g for CTIF). Initial study of in vitro reconstitution experiments using purified eIF3 complex revealed that eIF3g is dispensable in eIF3 complex formation (24). However, a recent mass spectrometric analysis showed that eIF3g is located in close proximity to eIF3j (25), which is positioned at the decoding center of the 40S ribosome (26). In addition, a recent cryoelectron microscopic analysis of the mammalian 48S PIC revealed that eIF3g is relocated from the peripheral region of the 40S ribosome to the P site at its intersubunit face to promote efficient subunit joining (27). Therefore, eIF3g is thought to serve as a platform for the CT complex to easily access the start codon and consequently to assemble an elongation-competent 80S ribosome.
After attachment of the 40S ribosome to the cap structure, a scanning complex scans for the start codon in the 5′ to 3′ direction along the 5′UTR. Efficient ribosome scanning requires an ATP-dependent DEAD-box RNA helicase for unwinding RNA secondary or tertiary structures in the 5′UTR (28). Interestingly, CT and ET exhibit different factor requirements for unwinding the secondary or tertiary structures. For ET, eIF4A1 or eIF4A2, which directly interacts with eIF4G, facilitates efficient unwinding (29–31). In contrast, the CT complex engages eIF4A3 through its interaction with CTIF (32). Accordingly, the downregulation of eIF4A3 but not eIF4A1 and eIF4A2 reduces the translational efficiency of CBP80-bound mRNAs harboring moderate secondary structures and blocks CT-associated mRNA quality control (32).
It should be noted that eIF4A itself has weak helicase activity (33, 34) and thus requires a cofactor to efficiently unwind RNA secondary or tertiary structures in the 5′UTR. For ET, the binding of eIF4G to eIF4A1/2 converts eIF4A1/2 from a closed to an open conformation, promoting its helicase activity (31). In addition, the helicase activity of eIF4A1/2 is stimulated by eIF4B and eIF4H (28, 35). However, it is not yet known what cellular factor(s) can modulate the helicase activity of eIF4A3 during CT. One plausible candidate is MLN51, a component of the EJC, as MLN51 has been found to enhance the helicase activity of eIF4A3 (36, 37). As eIF4G does (28, 31), it is also likely that CTIF could facilitate conformational change of eIF4A3 to promote the helicase activity.
After the scanning complex finally reaches the start codon, the 48S PIC undergoes drastic remodeling. eIF5 in association with the eIF3 complex stimulates the GTPase activity of eIF2γ of the eIF2 complex, converting eIF2-GTP to eIF2-GDP (14). Then, eIF5B (a ribosome-dependent GTPase) triggers the dissociation of eIF1, eIF1A, eIF2-GDP, eIF3, and eIF5 from the 40S ribosome and facilitates the joining of the 60S ribosome (38–40). Subsequently, the GTPase activity of eIF5B is stimulated, releasing eIF5B from the ribosome. A majority of studies on the mechanism of this step have focused on ET, particularly by adopting in vitro reconstitution approaches. Considering that the overall initiation process of CT is similar to that of ET, it is possible that CT may follow a comparable route for 48S PIC assembly and subsequent 60S joining using canonical initiation factors as well as CT-specific factors such as CTIF and eIF4A3.
In addition to the 5′-cap structure, the poly(A) tail is critical for efficient cap-dependent translation (41). During ET, cytoplasmic poly(A)-binding protein 1 (PABPC1; also known as PABP1) associates with the poly(A) tail of mRNAs and interacts with eIF4G via direct binding to the PAM1 motif in the N-terminus of eIF4G (42). This interaction enhances the efficiency of ET either by augmenting the cap-binding ability of eIF4E (43, 44), which may lead to stable mRNA circularization (45), or by directly or indirectly facilitating the recruitment of the 60S ribosome to the 48S PIC (46). In addition, PABPC1 binds to eukaryotic polypeptide chain release factor 3 (eRF3), one of translation release factors that promotes translation termination (47, 48). In contrast, there is no known CT-specific poly(A)-binding protein, and whether the poly(A) tail of CBC-bound mRNAs is involved in CT has not been confirmed. Notably, there is no characterized binding motif or domain within CBP80 and CTIF for binding to poly(A)-binding proteins (either PABPN1 or PABPC1). However, considering that PABPN1 forms a complex with the CBC but not with eIF4E (20) and that it is detected in actively translating polysomes (49), the possibility that PABPN1 may participate in CT cannot be completely ruled out.
CT VS. ET IN NONSENSE-MEDIATED mRNA DECAY
During mRNA processing in eukaryotic cells, premature termination codons (PTCs) can arise on mRNAs as a consequence of mutations in somatic cells, inaccurate transcription or splicing, or alternative splicing that leads to intron retention or inclusion of a PTC-containing exon (50). These aberrant mRNAs would express truncated polypeptides, the accumulation of which is potentially deleterious to the cells. Fortunately, eukaryotic cells have evolved several mRNA quality control pathways, which are typified by nonsense-mediated mRNA decay (NMD). The aberrant mRNAs harboring PTCs would be selectively recognized and rapidly degraded by the NMD pathway before truncated polypeptides are accumulated within the cells (7, 51–53).
Efficient NMD requires at least a single round of translation to recognize the PTCs on mRNAs. NMD has long been considered to occur during CT because (i) CBC-bound mRNAs are precursors of eIF4E-bound mRNAs, (ii) the CBC can drive cap-dependent translation, and (iii) the majority of NMD occurs on CBC-bound mRNAs (7, 8, 20, 49, 54, 55). Therefore, the traditional model of CT-coupled NMD can be described as follows: newly synthesized CBC-bound and EJC-deposited mRNAs are subject to the first round of translation during or after their export from the nucleus to the cytoplasm. The first round of translation would be predominantly mediated by the CBC rather than eIF4E because newly synthesized mRNAs have a 5′-end cap bound by the CBC. During the first round of CBC-mediated translation, an elongating 80S ribosome removes EJCs deposited 20–24 nucleotides upstream of exon-exon junctions after splicing. When the elongating 80S ribosome reaches a termination codon, eRF1 and eRF3 are recruited to the terminating ribosome along with UPF1 (a key NMD factor) and SMG1 kinase, forming the SMG1-UPF1-eRF1/3 (SURF) complex. The stability of the SURF complex is dependent on the presence or absence of EJCs downstream of the termination codon. The SURF complex would be unstable if there is no EJC downstream of the termination codon; consequently, the termination codon would be recognized as “normal”, and the mRNA would be stable. On the other hand, if EJC is present downstream of the termination codon, it interacts with the SURF complex and induces complex remodeling; consequently, the termination codon would be recognized as a “PTC”, and the mRNA would be rapidly degraded via exoribonucleolytic or endoribonucleolytic cleavage.
Recently, two separate studies claimed that NMD can occur on eIF4E-bound mRNAs as well as CBC-bound mRNAs (56, 57). To verify their hypothesis, they examined the efficiency of NMD under conditions where ET is compromised. The studies found that treatment of cells with 4EGI-1, a specific translational inhibitor, which disrupts the interaction between eIF4E and eIF4G (58), and overexpression of eIF4E-BP1, which competes with eIF4G for binding to eIF4E (59), suppressed the NMD of PTC-containing mRNAs (e.g., β-globin and GPx1). Furthermore, the half-life of PTC-containing mRNAs (e.g., β-globin, immunoglobulin μ, and TCRβ) was also decreased in eIF4E-bound messenger ribonucleoproteins (mRNPs) as well as in CBP80-bound mRNPs. In addition, eIF4E was found to be associated with UPF1 in an RNA-dependent manner. Based on these observations, the authors proposed that both CBC-bound and eIF4E-bound mRNAs are subject to NMD.
Although the data are compelling, several key issues should be carefully considered. First, considering that CBC-bound mRNA is a precursor of eIF4E-bound mRNA, it is likely that a majority of the first round of translation may be predominantly mediated by the CBC rather than eIF4E. Therefore, it should be clearly determined whether the observed reduction in the level and half-life of eIF4E-bound NMD substrates simply reflects the reduction in the level and half-life of CBC-bound NMD substrates. Second, although 4EGI-1 was originally reported as a specific compound that inhibits ET (58), several subsequent studies have reported additional 4EGI-1 targets besides the interaction between eIF4E and eIF4G (60, 61). Notably, it has been reported that treatment of cells with 4EGI-1 in relatively low concentrations strongly inhibited translation without significant changes in eIF4F levels; on the other hand, 4EGI-1 treatment increased the association of the 5′-cap with ribosomal complexes containing the phosphorylated (inactive) form of eIF2 (60). This effect would inhibit the efficiency of both CT and ET. Third, CBC-bound mRNPs but not eIF4E-containing mRNPs contain detectable amounts of EJCs, which are critical for stimulating the NMD of PTC-containing mRNAs (8, 17, 20, 32, 49, 62). Therefore, as mentioned earlier, the rapid degradation of NMD substrates in eIF4E-containing mRNPs would reflect the rapid degradation of NMD substrates in CBC-containing mRNPs. However, another equally plausible explanation is that CBC-bound NMD substrates may be predominantly subject to EJC-dependent NMD, whereas eIF4E-bound NMD substrates may be subject to EJC-independent NMD. Fourth, the replacement of the CBC by eIF4E occurs in a translation-independent manner (55). Therefore, even before the complete termination of CT, CBC would be replaced by eIF4E, suggesting the possibility of a transient interaction between eIF4E and UPF1.
Recently, two independent studies revealed that mammalian target of rapamycin complex 1 (mTORC1), which specifically controls ET efficiency by phosphorylating eIF4E-BP (63), can modulate the efficiency of NMD (64, 65). One study demonstrated that insulin treatment increased the interaction between UPF1 and eIF4E-bound PTC-containing mRNAs and augmented eIF4E-associated NMD, which was blocked by treatment with rapamycin (a selective inhibitor of mTORC1) (65). On the other hand, another study reported that rapamycin treatment promoted the NMD of a subset of mRNAs, probably by blocking the replacement of CBC by eIF4E (64). Therefore, it would be interesting to determine whether there are CT-specific or ET-specific NMD substrates depending on cellular conditions.
REPLICATION-DEPENDENT HISTONE mRNAs ARE LARGELY TRANSLATED VIA CT
Histone genes are essential as structural and functional units for packaging genomic DNA into chromatin to sustain genomic integrity and for regulating transcriptional activity, respectively (66, 67). In particular, the expression of replication-dependent histone (RDH) genes is tightly controlled during the cell cycle. Upon entry into the S phase, the levels of RDH mRNAs increase by approximately 35-fold at the transcriptional level (68). Newly synthesized RDH transcripts are processed into mature RDH mRNAs with the aid of (i) U7 small nuclear ribonucleoprotein (U7 snRNP), which interacts with histone downstream element (HDE), (ii) stem-loop binding protein (SLBP), which directly binds to the histone stem-loop (HSL) located in the 3′UTR, and (iii) a 3′-end cleavage complex containing cleavage and polyadenylation specificity factor subunit 73 (CPSF73), CPSF100, and symplekin (69, 70). The site between the HSL (recognized by SLBP) and the HDE (base-paired with U7 snRNA) is cleaved by CPSF73 and further trimmed by ERI1, a 3′-to-5′ exonuclease. The resulting RDH mRNAs, which have a HSL instead of a poly(A) tail, are exported from the nucleus to the cytoplasm for translation. On the other hand, at the end of S phase or under genotoxic stresses, actively translating RDH mRNAs are rapidly degraded via hyperphosphorylation of UPF1, which recruits various mRNA degradation factors to RDH mRNAs (70–72).
RDH mRNAs are unique in their structure because they lack a poly(A) tail but instead have a conserved HSL at their 3′UTR. In the case of canonical mRNAs, the interaction between PABPC1 and eIF4G triggers mRNA circularization for efficient translation. Similarly, the circularization of RDH mRNAs has been proposed to be mediated by an indirect interaction between SLBP and eIF4G via SLBP-interacting protein 1 (SLIP1), suggesting a possible role of ET in the translation of RDH mRNAs (73). However, this view was recently challenged by two reports demonstrating that the degradation of RDH mRNAs largely occurs on CBC-bound RDH mRNAs (32, 74). Considering that (i) RDH mRNA degradation requires translation (75, 76) and (ii) SLIP1-mediated RDH mRNA circularization increases the ET of RDH mRNAs (73), the eIF4E-bound RDH mRNPs are expected to be subject to rapid decay. In contrast, the authors found that a majority of RDH mRNA degradation occurs on CBC-bound RDH mRNAs depending on the direct association between CTIF and SLBP (74). Downregulation of a CT-specific factor, either CTIF or eIF4A3, was found to increase the abundance of RDH mRNAs. Furthermore, overexpression of eIF4E-BP1 or eIF4E-T did not affect the levels of RDH mRNAs. These observations support that the majority of RDH mRNAs are translated via CT instead of ET. Although the two studies described above have conflicting results, the findings indicated that at least a subset of mRNAs (e.g. RDH mRNAs) may preferentially use CT instead of ET for protein synthesis and mRNA degradation.
DIFFERENTIAL REGULATION OF CT AND ET UNDER STRESS CONDITIONS
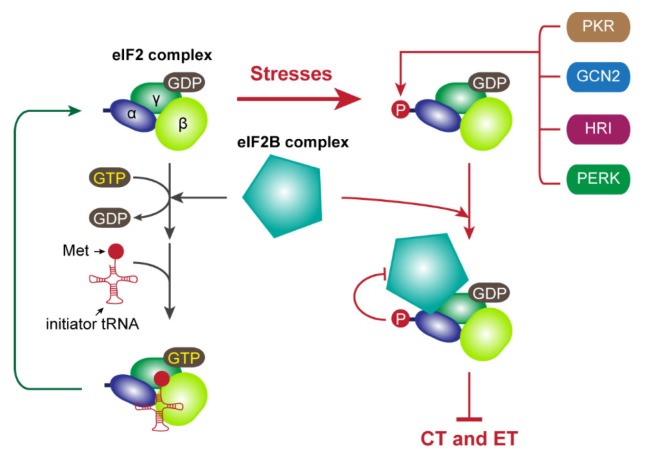
Eukaryotic cap-dependent translation is regulated at various steps under certain conditions such as oxidative stress, hypoxia, viral infection, apoptosis, nutrient starvation, differentiation, and development (77, 78). Under stress conditions, two main factors involved in translation initiation are post-translationally regulated: phosphorylation of eIF2α (Fig. 2) and eIF4E-BP (Fig. 3). GTP-bound eIF2 associates with the initiator tRNA (14). After establishment of the 48S PIC onto the start codon, eIF5 stimulates the conversion of eIF2-GTP to eIF2-GDP, which is released from the ribosome after the joining of the 60S ribosome (38–40). The released eIF2-GDP binds to eIF2B, which is an eIF2-specific guanosine exchange factor (GEF), and is converted to eIF2-GTP for recycling (14). However, under stress conditions, several eIF2-specific kinases are activated and phosphorylate Ser51 in eIF2α, which include PERK (ER stress), PKR (double-stranded RNA upon viral infection), HRI (heme deficiency), and GCN2 (amino acid starvation) (14, 77–79). The phosphorylated eIF2 complex sequesters eIF2B, leading to a decrease in the amount of available eIF2-GTP and shutdown of translation. Therefore, both CT and ET can be blocked by eIF2α phosphorylation as both types of translation require a functionally active eIF2 complex (Fig. 2).
Fig. 2.
Translational control mediated by eIF2α phosphorylation under stress conditions. The details are described in the text.
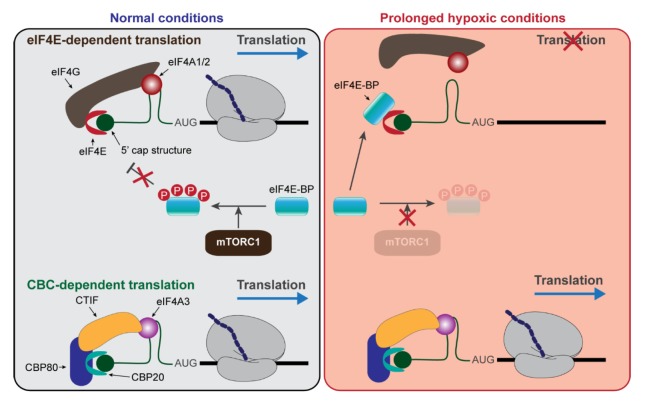
Fig. 3.
Translational control under prolonged hypoxic conditions. The details are described in the text.
In addition to eIF2α, the phosphorylation status of eIF4E-BP is changed in response to intrinsic or extrinsic stresses. eIF4E-BP competes with eIF4G for binding to eIF4E (59). Under normal or actively growing conditions, active mTORC1 phosphorylates eIF4E-BP, leading to the decreased binding of eIF4E-BP to eIF4E and consequently resulting in an increase in ET efficiency (59, 63). However, under stress conditions, eIF4E-BP becomes hypophosphorylated and strongly binds to eIF4E, resulting in the inhibition of ET. Therefore, eIF4E-BP is an ET-specific translation regulator, and CT is insensitive to eIF4E-BP activity (Fig. 3).
In comparison with ET, CT is relatively insensitive to certain environmental stresses, and this can be demonstrated by translational regulation during hypoxia, a condition in which cells or tissues have low oxygen levels. Global translation is regulated during hypoxic stresses at two independent stages: (i) eIF2α phosphorylation at the early stage (acute/moderate hypoxia) and (ii) eIF4E-BP hypophosphorylation and eIF4E sequestration into the nucleus or the cytoplasmic processing bodies at the late stage (prolonged hypoxia) (80–83). The former induces the global shutdown of protein synthesis by targeting both CT and ET, whereas the latter blocks ET but not CT. Accordingly, acute hypoxia inhibits NMD (84), whereas prolonged hypoxia does not affect NMD even though ET is markedly compromised (85). The relative resistance of CT to environmental stresses is further demonstrated by translational regulation under serum starved conditions, in which mTORC1 is inactivated and the amount of hypophosphorylated eIF4E-BP is elevated. Under these conditions, ET efficiency is drastically reduced. However, CT-coupled NMD remains efficient (86). Taken together, CT could support the translation of a subset of mRNAs under stress conditions when ET is compromised.
CONCLUSION REMARKS
CT and ET have long been considered to be largely involved in mRNA quality control and the bulk of protein synthesis, respectively. However, recent studies have reassessed their roles in translation initiation, revealing that both CT and ET are functionally interchangeable. Although definitive conclusions are difficult given the contradictory data, the following general conclusions can be made:
NMD occurs during CT or ET; however, NMD is predominantly coupled to CT.
CT instead of ET is preferred in the protein synthesis of a subset of mRNAs. In other words, CT can drive multiple rounds of translation of a subset of mRNAs, similar to ET (17, 87–89).
In comparison with ET, CT is relatively resistant to intrinsic or extrinsic stresses.
In this context, future studies should elucidate the molecular features of CT, identify mRNAs that are specific to CT or ET, and evaluate CT- or ET-specific NMD.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation (NRF) of Korea grant funded by the Korean government (Ministry of Science, ICT, and Future Planning; NRF-2015R1A3A203 3665) and by a Korea University grant. I.R. was in part supported by NRF (NRF-2016R1D1A1B03933894).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Singh G, Pratt G, Yeo GW, Moore MJ. The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. Ann Rev Biochem. 2015;84:325–354. doi: 10.1146/annurev-biochem-080111-092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shatkin AJ, Manley JL. The ends of the affair: capping and polyadenylation. Nat Struct Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 3.Cowling VH. Regulation of mRNA cap methylation. Biochem J. 2010;425:295–302. doi: 10.1042/BJ20091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Hir H, Sauliere J, Wang Z. The exon junction complex as a node of post-transcriptional networks. Nat Rev Mol Cell Biol. 2016;17:41–54. doi: 10.1038/nrm.2015.7. [DOI] [PubMed] [Google Scholar]
- 5.Woodward LA, Mabin JW, Gangras P, Singh G. The exon junction complex: a lifelong guardian of mRNA fate. Wiley Interdiscip Rev RNA. 2016:e1411. doi: 10.1002/wrna.1411. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee A, Apponi LH, Pavlath GK, Corbett AH. PABPN1: molecular function and muscle disease. FEBS J. 2013;280:4230–4250. doi: 10.1111/febs.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Ann Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 11.Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Majumdar R, Bandyopadhyay A, Maitra U. Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40 S preinitiation complex. J Biol Chem. 2003;278:6580–6587. doi: 10.1074/jbc.M210357200. [DOI] [PubMed] [Google Scholar]
- 13.Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 14.Proud CG. eIF2 and the control of cell physiology. Semin Cell Dev Biol. 2005;16:3–12. doi: 10.1016/j.semcdb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Villa N, Do A, Hershey JW, Fraser CS. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J Biol Chem. 2013;288:32932–32940. doi: 10.1074/jbc.M113.517011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeFebvre AK, Korneeva NL, Trutschl M, et al. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem. 2006;281:22917–22932. doi: 10.1074/jbc.M605418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim KM, Cho H, Choi K, et al. A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20-dependent translation. Genes Dev. 2009;23:2033–2045. doi: 10.1101/gad.1823409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choe J, Oh N, Park S, et al. Translation initiation on mRNAs bound by nuclear cap-binding protein complex CBP80/20 requires interaction between CBP80/20-dependent translation initiation factor and eukaryotic translation initiation factor 3g. J Biol Chem. 2012;287:18500–18509. doi: 10.1074/jbc.M111.327528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lejeune F, Ranganathan AC, Maquat LE. eIF4G is required for the pioneer round of translation in mammalian cells. Nat Struct Mol Biol. 2004;11:992–1000. doi: 10.1038/nsmb824. [DOI] [PubMed] [Google Scholar]
- 20.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/S0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 21.McKendrick L, Thompson E, Ferreira J, Morley SJ, Lewis JD. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol Cell Biol. 2001;21:3632–3641. doi: 10.1128/MCB.21.11.3632-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Moeller H, Lerner R, Ricciardi A, Basquin C, Marzluff WF, Conti E. Structural and biochemical studies of SLIP1-SLBP identify DBP5 and eIF3g as SLIP1-binding proteins. Nucleic Acids Res. 2013;41:7960–7971. doi: 10.1093/nar/gkt558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 24.Masutani M, Sonenberg N, Yokoyama S, Imataka H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26:3373–3383. doi: 10.1038/sj.emboj.7601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou M, Sandercock AM, Fraser CS, et al. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci U S A. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser CS, Berry KE, Hershey JW, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 27.Simonetti A, Brito Querido J, Myasnikov AG, et al. eIF3 Peripheral Subunits Rearrangement after mRNA Binding and Start-Codon Recognition. Mol Cell. 2016;63:206–217. doi: 10.1016/j.molcel.2016.05.033. [DOI] [PubMed] [Google Scholar]
- 28.Parsyan A, Svitkin Y, Shahbazian D, et al. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 29.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marintchev A, Edmonds KA, Marintcheva B, et al. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oberer M, Marintchev A, Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choe J, Ryu I, Park OH, et al. eIF4AIII enhances translation of nuclear cap-binding complex-bound mRNAs by promoting disruption of secondary structures in 5′UTR. Proc Natl Acad Sci U S A. 2014;111:E4577–4586. doi: 10.1073/pnas.1409695111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers GW, Jr, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 34.Li Q, Imataka H, Morino S, et al. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol Cell Biol. 1999;19:7336–7346. doi: 10.1128/MCB.19.11.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu WT, Wilczynska A, Smith E, Bushell M. The diverse roles of the eIF4A family: you are the company you keep. Biochem Soc T. 2014;42:166–172. doi: 10.1042/BST20130161. [DOI] [PubMed] [Google Scholar]
- 36.Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 37.Noble CG, Song H. MLN51 stimulates the RNA-helicase activity of eIF4AIII. PLoS One. 2007;2:e303. doi: 10.1371/journal.pone.0000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JH, Pestova TV, Shin BS, Cao C, Choi SK, Dever TE. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc Natl Acad Sci U S A. 2002;99:16689–16694. doi: 10.1073/pnas.262569399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 41.Weill L, Belloc E, Bava FA, Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol. 2012;19:577–585. doi: 10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- 42.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Der Haar T, Ball PD, McCarthy JE. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-Cap by domains of eIF4G. J Biol Chem. 2000;275:30551–30555. doi: 10.1074/jbc.M004565200. [DOI] [PubMed] [Google Scholar]
- 44.Borman AM, Michel YM, Kean KM. Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5′-end. Nucleic Acids Res. 2000;28:4068–4075. doi: 10.1093/nar/28.21.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/S1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 46.Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- 48.Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Str. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 49.Chiu SY, Lejeune F, Ranganathan AC, Maquat LE. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He F, Jacobson A. Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story. Ann Rev Gene. 2015;49:339–366. doi: 10.1146/annurev-genet-112414-054639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fatscher T, Boehm V, Gehring NH. Mechanism, factors, and physiological role of nonsense-mediated mRNA decay. Cell Mol Life Sci. 2015;72:4523–4544. doi: 10.1007/s00018-015-2017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karousis ED, Nasif S, Muhlemann O. Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdiscip Rev RNA. 2016;7:661–682. doi: 10.1002/wrna.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hwang J, Kim YK. When a ribosome encounters a premature termination codon. BMB Rep. 2013;46:9–16. doi: 10.5483/BMBRep.2013.46.1.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurosaki T, Maquat LE. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci. 2016;129:461–467. doi: 10.1242/jcs.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato H, Maquat LE. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin beta. Genes Dev. 2009;23:2537–2550. doi: 10.1101/gad.1817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durand S, Lykke-Andersen J. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat Struct Mol Biol. 2013;20:702–709. doi: 10.1038/nsmb.2575. [DOI] [PubMed] [Google Scholar]
- 57.Rufener SC, Muhlemann O. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2013;20:710–717. doi: 10.1038/nsmb.2576. [DOI] [PubMed] [Google Scholar]
- 58.Moerke NJ, Aktas H, Chen H, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 59.Qin X, Jiang B, Zhang Y. 4E-BP1, a multifactor regulated multifunctional protein. Cell cycle. 2016;15:781–786. doi: 10.1080/15384101.2016.1151581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McMahon R, Zaborowska I, Walsh D. Noncy-totoxic inhibition of viral infection through eIF4F-independent suppression of translation by 4EGi-1. J Virol. 2011;85:853–864. doi: 10.1128/JVI.01873-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mokas S, Mills JR, Garreau C, et al. Uncoupling stress granule assembly and translation initiation inhibition. Mol Biol Cell. 2009;20:2673–2683. doi: 10.1091/mbc.E08-10-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kashima I, Yamashita A, Izumi N, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 64.Martinez-Nunez RT, Wallace A, Coyne D, et al. Modulation of nonsense mediated decay by rapamycin. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw1109. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park J, Ahn S, Jayabalan AK, Ohn T, Koh HC, Hwang J. Insulin Signaling Augments eIF4E-Dependent Nonsense-Mediated mRNA Decay in Mammalian Cells. Biochim Biophys Acta. 20161859:896–905. doi: 10.1016/j.bbagrm.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 66.Rattray AM, Muller B. The control of histone gene expression. Biochem Soc T. 2012;40:880–885. doi: 10.1042/BST20120065. [DOI] [PubMed] [Google Scholar]
- 67.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harris ME, Bohni R, Schneiderman MH, Ramamurthy L, Schumperli D, Marzluff WF. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/MCB.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romeo V, Schumperli D. Cycling in the nucleus: regulation of RNA 3′ processing and nuclear organization of replication-dependent histone genes. Curr Opin Cell Biol. 2016;40:23–31. doi: 10.1016/j.ceb.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 70.Hoefig KP, Heissmeyer V. Degradation of oligouridylated histone mRNAs: see UUUUU and goodbye. Wiley Interdiscip Rev RNA. 2014;5:577–589. doi: 10.1002/wrna.1232. [DOI] [PubMed] [Google Scholar]
- 71.Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 72.Choe J, Ahn SH, Kim YK. The mRNP remodeling mediated by UPF1 promotes rapid degradation of replication-dependent histone mRNA. Nucleic Acids Res. 2014;42:9334–9349. doi: 10.1093/nar/gku610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cakmakci NG, Lerner RS, Wagner EJ, Zheng L, Marzluff WF. SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol Cell Biol. 2008;28:1182–1194. doi: 10.1128/MCB.01500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choe J, Kim KM, Park S, et al. Rapid degradation of replication-dependent histone mRNAs largely occurs on mRNAs bound by nuclear cap-binding proteins 80 and 20. Nucleic Acids Res. 2013;41:1307–1318. doi: 10.1093/nar/gks1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stimac E, Groppi VE, Jr, Coffino P. Inhibition of protein synthesis stabilizes histone mRNA. Mol Cell Biol. 1984;4:2082–2090. doi: 10.1128/MCB.4.10.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Graves RA, Pandey NB, Chodchoy N, Marzluff WF. Translation is required for regulation of histone mRNA degradation. Cell. 1987;48:615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- 77.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 78.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamasaki S, Anderson P. Reprogramming mRNA translation during stress. Curr Opin Cell Biol. 2008;20:222–226. doi: 10.1016/j.ceb.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koritzinsky M, Magagnin MG, van den Beucken T, et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 2006;25:1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koumenis C, Naczki C, Koritzinsky M, et al. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol. 2002;22:7405–7416. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arsham AM, Howell JJ, Simon MC. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J Biol Chem. 2003;278:29655–29660. doi: 10.1074/jbc.M212770200. [DOI] [PubMed] [Google Scholar]
- 83.Dostie J, Ferraiuolo M, Pause A, Adam SA, Sonenberg N. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 2000;19:3142–3156. doi: 10.1093/emboj/19.12.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol. 2008;28:3729–3741. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oh N, Kim KM, Choe J, Kim YK. Pioneer round of translation mediated by nuclear cap-binding proteins CBP80/20 occurs during prolonged hypoxia. FEBS Lett. 2007;581:5158–5164. doi: 10.1016/j.febslet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 86.Oh N, Kim KM, Cho H, Choe J, Kim YK. Pioneer round of translation occurs during serum starvation. Biochem Bioph Res Co. 2007;362:145–151. doi: 10.1016/j.bbrc.2007.07.169. [DOI] [PubMed] [Google Scholar]
- 87.Apcher S, Daskalogianni C, Lejeune F, et al. Major source of antigenic peptides for the MHC class I pathway is produced during the pioneer round of mRNA translation. Proc Natl Acad Sci U S A. 2011;108:11572–11577. doi: 10.1073/pnas.1104104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sharma A, Yilmaz A, Marsh K, Cochrane A, Boris-Lawrie K. Thriving under Stress: Selective Translation of HIV-1 Structural Protein mRNA during Vpr-Mediated Impairment of eIF4E Translation Activity. PLoS Pathog. 2012;8:e1002612. doi: 10.1371/journal.ppat.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]