Abstract
At the post-transcriptional and translational levels, microRNA (miRNA) represses protein-coding genes via seed pairing to the 3′ untranslated regions (UTRs) of mRNA. Although working models of miRNA-mediated gene silencing are successfully established using miRNA transfections and knockouts, the regulatory interaction between miRNA and long non-coding RNA (lncRNA) remain unknown. In particular, how the mRNA-resembling lncRNAs with 5′ cap, 3′ poly(A)-tail, or coding features, are regulated by miRNA is yet to be examined. We therefore investigated the functional interaction between miRNAs and lncRNAs with/without those features, in miRNA-transfected early zebrafish embryos. We observed that the greatest determinants of the miRNA-mediated silencing of lncRNAs were the 5′ cap and 3′ poly(A)-tails in lncRNAs, at both the post-transcriptional and translational levels. The lncRNAs confirmed to contain 5′ cap, 3′ poly(A)-tail, and the canonical miRNA target sites, were observed to be repressed in the level of both RNA and ribosome-protected fragment, while those with the miRNA target sites and without 5′ cap and 3′ poly(A)-tail, were not robustly repressed by miRNA introduction, thus suggesting a role as a miRNA-decoy.
Keywords: lncRNA, miRNA, sORF, 3′ poly(A)-tail, 5′ cap
INTRODUCTION
MicroRNA (miRNA), one of the conserved class of small non-coding RNAs of ~22 nucleotides (nt), participates in the control of protein-coding gene expression via base pairing to the 3′ untranslated region (3′ UTR) of messenger RNA (mRNA) (1). Since the discovery of their role in miRNA-mediated gene silencing (MGS), the regulatory modes and mechanisms of action involved have been studied, comparing between miRNA- and mock-transfected cells, or wild type and miRNA-knockout cells (2–6). The major determinants of efficient miRNA targeting include the conserved Watson-Crick base pairing (called seed pairing) between 3′ UTR of mRNA and the miRNA seed region; additional base pairing include position 8 of miRNA (7mer-m8, 7m8 site) and the presence of adenine opposing position 1 of miRNA (7mer-A1, 7A1 site), or both the additional base pairing at position 8 and the presence of adenine opposing position 1 of miRNA (8mer site) (1). With seed pairing, the global downregulation of mRNA targeted by miRNA was first evidenced by reverse transcription polymerase chain reaction (RT-PCR) experiments (7) and microarray data analysis (8); although, the first miRNA to be discovered (lin-4) was previously shown to control the expression of its target protein, LIN-14, at the translational level (9).
Multiple studies using miRNA-transfected cell lines (10–12), miRNA knockouts in zebrafish embryos (13) and mouse neutrophils (10, 11, 13), high-throughput RNA sequencing (RNA-seq), and ribosome-protected fragment (RPF) sequencing (Ribo-seq), have investigated the miRNA regulatory mechanisms, revealing that both post-transcriptional and translational regulation modes are involved in MGS. Debates over the relative contribution and order of the two regulatory modes have elucidated the dynamics of miRNA-mediated repression. In addition, miRNA-transfection in human cell lines and miRNA knockout experiments in mice revealed that the destabilization of target mRNAs, rather than translational repression, is most responsible for MGS (10, 11). Recent experiments using either a zygotic dicer mutant with significantly reduced levels of miR-430 (13) or miRNA-transfection (14), were conducted to study the early developmental stages of zebrafish embryos. These researches claim that the targets are translationally repressed early on (~4 hours after miRNA transfection), and post-transcriptionally downregulated later (~6 hours after miRNA transfection) (13), thus describing an early translational repression and a later dominant destabilization of the target mRNAs.
Although the majority of MGS-related studies mainly deal with the protein-coding genes, a handful of studies have examined the interaction between miRNA and long non-coding RNA (lncRNA), which is the other class of non-coding RNA longer than 200 nt (15–18). lncRNAs are k versatile, heterogeneous RNA molecules, involved in diverse biological processes, such as transcriptional, post-transcriptional, and translational regulation of gene expression (19). Although the concept of competing endogenous RNA (ceRNA) is not widely accepted, recent reports indicate the functional role of lncRNAs containing miRNA target sites as miRNA-decoys that quench the endogenous miRNAs to their binding sites (20, 21). Interestingly, some lncRNAs that quench the miRNAs via these sites get destabilized (22–24), whereas others are resistant to or not affected by the miRNA-mediated repression (25, 26). For example, a well-studied lncRNA, the metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) (MALAT1), is known to be repressed by miR-9 (22). Similarly, PTENP1, a pseudogene transcript of the tumor suppressor phosphatase and tensin homolog (PTEN), is repressed by endogenous miRNA targeting, and quenches the endogenous miRNAs (e.g., miR-20a, miR-19b, and miR-21), thus inhibiting the targeting of other mRNAs (23). Conversely, although the circular lncRNA (ciRS-7) that binds to miRNA-7 thorough its canonical target sites quenches miRNA, it is not repressed by MGS (25). So far, the factors that determine miRNA regulatory modes remain unknown.
lncRNAs lacking the open reading frames (ORFs) and protein-coding potential, share similar characteristics with protein-coding genes in terms of post-transcriptional processing (27). A substantial number, but not all of the lncRNAs, undergo similar post-transcriptional processing, such as 5′ capping, 3′ polyadenylation, and splicing (28). However, recent studies using Ribo-seq and mass spectrometry revealed that lncRNAs containing small ORFs (sORFs) translate small peptides (29–33). Moreover, 4 mouse and 12 human lncRNAs are now known to have sORFs, as predicted by Ribo-seq. Considering that lncRNAs containing 5′ cap, 3′ poly(A)-tail, and sORFs resemble protein-coding mRNAs in terms of having a similar structure, the lncRNAs with these three features may have regulatory functions (i.e., post-transcriptional and translational gene silencing) that are similar to those of protein-coding genes.
RESULTS
High-confidence set of lncRNAs
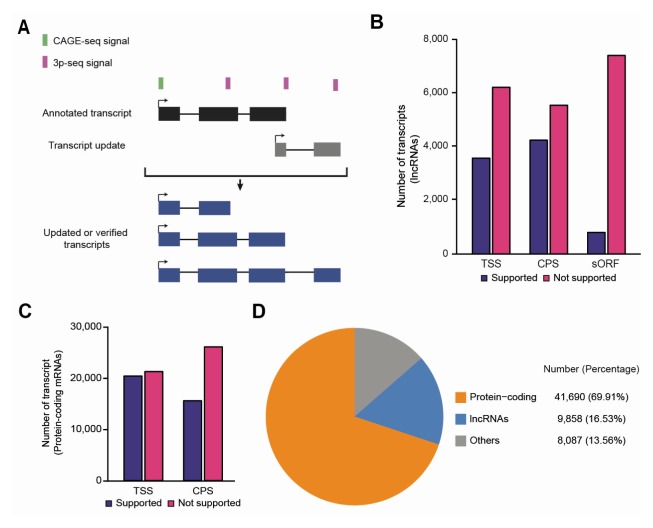
To acquire a high-confidence set of lncRNAs, the public lncRNA gene annotations (34–36) were re-examined, and a high-confidence set of mRNA-like lncRNAs embedding a 5′ cap, 3′ poly(A), sORF, or all three, were built (Supplementary Fig. S1). We first checked whether the lncRNAs are fragmented by examining the exon junctions between the lncRNAs and neighboring genes using publicly available RNA-seq data from the three early developmental stages in zebrafish (see Supplementary Material for more details). Both ends of the full-length lncRNAs were then updated with transcription start sites (TSSs) and cleavage and polya-denylation sites (CPSs), which denote the presence of 5′ cap and 3′ poly(A)-tail respectively; these were predicted from cap analysis gene expression sequencing (CAGE-seq) (37) and poly(A)-position profiling by sequencing (3P-seq) (38) data from the same developmental stages, respectively (Fig. 1A; see Supplementary Material for more details). If CPS was distant from the 3′ end of the transcript, only CPS supported by the newly assembled transcript was assigned to the corresponding transcript. This led to the identification of 4,276 lncRNAs that contain CPS, 3,598 that contain TSS, and 1,959 that contain both TSS and CPS (Fig. 1B). Next, the coding potential of lncRNAs was examined by searching for sORFs with 3 nt periodicity (Supplementary Fig. S2; see Supplementary Material for more details) in the publicly available Ribo-seq data from stage-matched samples, using Ribotaper (30). Consequently, 812 lncRNAs were predicted to be mRNA-like lncRNAs that have a putative sORF encoding less than 100 amino acids (Fig. 1B). In addition, 512 lncRNAs were predicted to have a putative ORF equal to or greater than 100 amino acids, and were analyzed separately from the lncRNAs discovered to have sORFs. Among the 8,534 lncRNAs predicted to have no ORFs, 1,083 were predicted as protein-coding mRNAs using the Coding Potential Calculator (CPC) (39), and were excluded from the lncRNA set without ORFs, thus yielding 7,451 lncRNAs without any ORF (Fig. 1B). For protein-coding mRNAs, the CPS and TSS updates were applied for better profiling of expression levels, as was done for lncRNAs. In total, 20,403 mRNAs contained TSS and 15,576 contained CPS (Fig. 1C), showing a higher proportion than that found for lncRNAs, which is consistent with observations in a previous study (40). For all the transcript annotations, ~70% (41,690) were protein-coding transcripts, ~17% (9,858) were lncRNA transcripts, and the remainder was considered as other ncRNA transcripts (Fig. 1D).
Fig. 1.
mRNA-like lncRNA annotations. (A) A schematic flow of TSS and CPS updates of transcripts. (B) Numbers of lncRNAs with TSSs, CPSs, or sORFs. (C) Numbers of protein-coding mRNAs with TSSs or CPSs. (D) Composition of zebrafish transcripts after the TSS and CPS updates.
The stability of lncRNAs is dependent on mRNA-like features
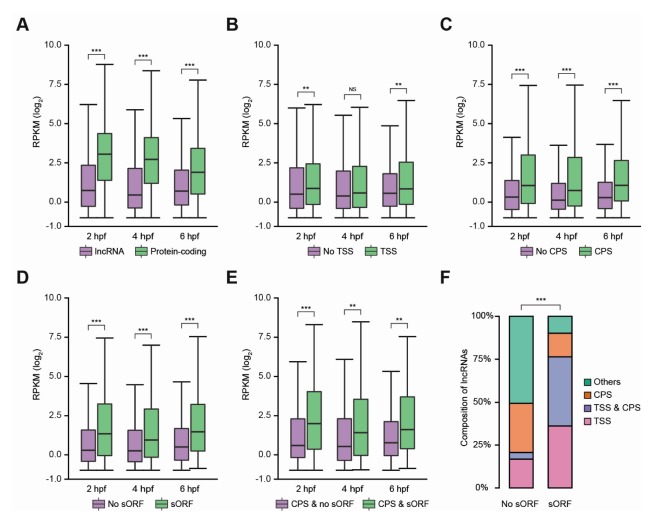
Since protein-coding mRNAs are more abundant and stable than lncRNA genes (41), we questioned whether the stability of lncRNAs is different based on mRNA-like features, such as the 5′ cap, 3′ poly(A)-tail or sORF. For miRNA targeting analysis, the expression levels of the updated protein-coding mRNAs and lncRNAs were calculated using the RNA-seq by expectation-maximization, RSEM (42) (Supplementary Fig. S1; see Supplementary Material for more details). We compared the expression levels of lncRNAs to those of protein-coding mRNAs, revealing consistently higher expression of protein-coding mRNAs at all stages (Fig. 2A). We then compared the lncRNAs with no mRNA-like features to those with the features. While the expression levels of lncRNAs containing TSSs was similar to that of lncRNAs with no mRNA-like features at the 4 hours post-fertilization (hpf) stage (Fig. 2B; P = 0.210), the lncRNAs containing CPSs were significantly more abundant and stable than those with no mRNA-like features at all stages (Fig. 2C; P = 1.709 × 10−9 for 2 hpf; P = 8.572 × 10−10 for 4 hpf; P = 1.215 × 10−8 for 6 hpf). This observation agrees with previous in vitro results, showing that the existence of 3′ poly(A)-tail, the product of a cleavage and polyadenylation event (43), is an important determinant of RNA stability, where the poly(A)-binding protein (PABP) binds to the 3′ poly(A)-tail and stabilizes the RNA, whereas the RNA without a 3′ poly(A)-tail is quickly degraded (44). Interestingly, the lncRNAs with sORFs were more abundant and stable than those with no sORFs (Fig. 2D; P = 9.302 × 10−8 for 2 hpf; P = 8.292 × 10−6 for 4 hpf; P = 6.951 × 10−8 for 6 hpf). As the lncRNAs with CPSs were observed to be more stable and highly expressed, we suspected that the higher expression levels of lncRNAs with sORFs could be explained by the presence of CPS. Therefore, lncRNAs with CPSs and sORFs were compared to those with CPSs but no sORFs, revealing greater abundance and stability for lncRNAs with CPSs and sORFs (Fig. 2E). Taken together, these results are evidence in support of a 5′ cap, 3′ poly (A)-tail and sORF being important determinants of lncRNA stability and expression levels. In fact, the lncRNAs with sORFs tended to have both TSS and CPS, indicating mRNA-like lncRNAs (P < 2.200 × 10−16; one-sided Fisher’s exact test, Fig. 2F).
Fig. 2.
Expression levels of lncRNAs with mRNA-like features at three developmental stages. (A–E) The expression levels of protein-coding mRNAs and lncRNAs are shown in box plots. The y-axis indicates the expression level, reads per kilobase of exons per million mapped reads (RPKM) in logarithm with base 2. The middle line in the boxes is the median level (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001; NS, not significant; two-sided Kolmogorov Smirnov test) (A) Box plots showing expression levels for all protein-coding mRNA and lncRNA. (B–D) Box plots showing expression levels for subsets of lncRNAs with mRNA-like features. (B) The expression levels of lncRNAs with or without TSS are shown in box plots. (C) The expression levels of lncRNAs with or without CPS in box plots. (D) The expression levels of lncRNAs with or without sORF in box plots. (E) The expression levels of polyadenylated lncRNAs with or without sORF in the box plots. (F) The composition of lncRNAs with TSSs, CPSs, or both (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001; one-sided Fisher’s exact test).
The lncRNAs are affected by miRNAs at an early stage of development
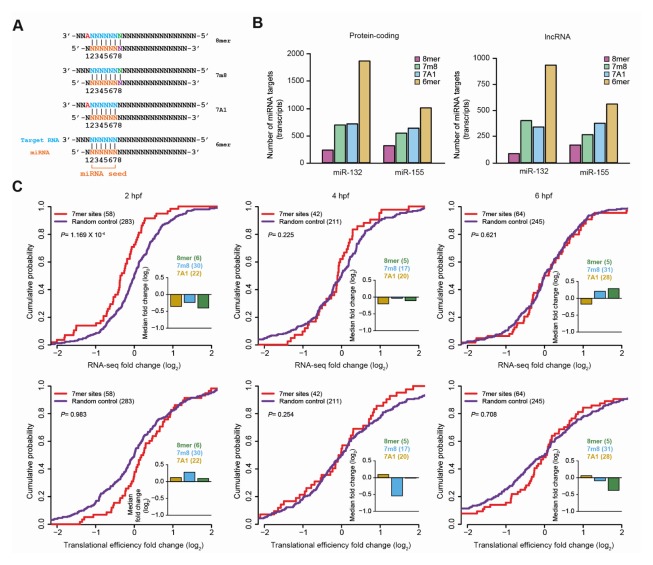
To investigate the impact of miRNA transfection on lncRNAs, we predicted canonical miRNA target sites, including 8mer, 7m8, 7A1, and 6mer sites in the whole exons and 3′ UTRs of lncRNAs, and protein-coding mRNAs, respectively (Fig. 3A and Supplementary Fig. S3). As expected, the proportion of miR-132 and miR-155 site types was very similar between protein-coding mRNAs and lncRNAs (Fig. 3B). Analyses of the changes in RNA and RPF levels between mock- and miR-132/155-injected zebrafish embryos revealed that protein-coding mRNAs displayed translational repression in early zebrafish embryo stages (pre-MZT), at 2 and 4 hpf, but not at the 6 hpf (Supplementary Fig. S4, bottom). Instead, the majority of the downregulation resulted from post-transcriptional repression at the 6 hpf stage (Supplementary Fig. S4, top). However, lncRNAs showed significant repression in RNA levels at an earlier stage of development (2 hpf) (Fig. 3C, top). Although the change of RPF levels at the subsequent time (4 hpf) was significant (Supplementary Fig. S5), the change of the RPF level was mainly caused by the change of the RNA level, resulting in no change of translation efficiency (Fig. 3C, bottom), which is the change of RPF levels normalized by those of RNA. These results demonstrate that a substantial portion of lncRNA population might interact with miRNA, and are mostly affected in RNA levels at an early stage of development, compared to protein-coding genes.
Fig. 3.
miRNA targeting on lncRNAs. (A) Canonical miRNA target sites. (B) The number of protein-coding mRNAs (left) and lncRNAs (right) embedding miR-132 or miR-155 target sites. In case of multiple sites, only the site with a greater impact was considered as the site corresponding to the transcript. (C) The cumulative distribution functions (CDFs) show the changes of RNA expression (top) and translational efficiency (bottom) for lncRNAs with miRNA (red) and random target sites (purple) (see Supplementary Material for more details), at each developmental stage in the zebrafish embryos. The number of miRNA and random target sites are shown in parenthesis in top left corner. P values were calculated using a one-tailed Wilcoxon rank-sum test and are shown at the top left corner. For each CDF, the median fold change (log2) and the number of 7mer sites of miR-132 and miR-155 (right), which are 8mer (green), 7m8 (sky blue) and 7A1 (dark orange), are depicted.
miRNA impacts mRNA-like lncRNAs
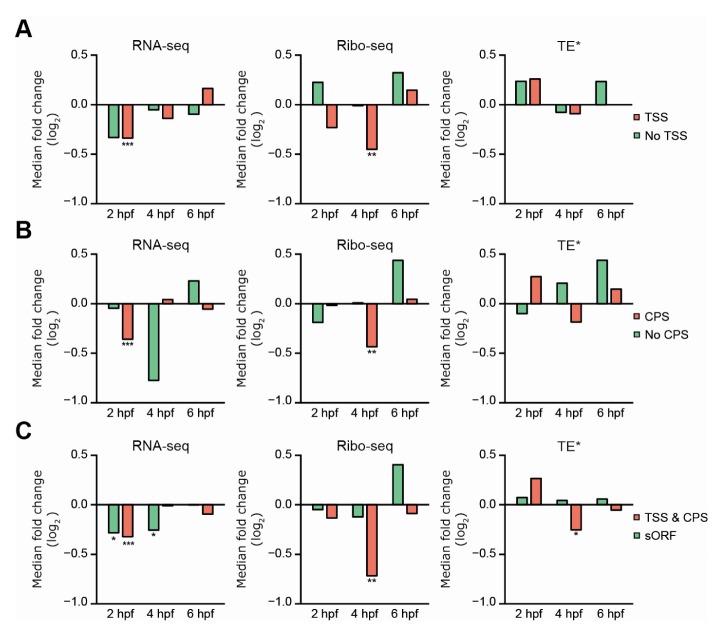
We hypothesized that lncRNAs with mRNA-like features can be regulated by miRNA transfection in a different scale or manner, compared to other lncRNAs. To test this hypothesis, the subset of lncRNAs containing mRNA-like features was compared to those without these features, with respect to the response to miRNA transfection (Fig. 4A–C). Since the number of lncRNAs is much less than the number of protein-coding mRNAs, log2 fold changes of miR-132 and -155 targets, and of non-targets, were combined in the downstream analysis, to achieve predictive statistical-power for the lncRNA subgroups. The lncRNAs with TSS or CPS, which are the evidence of 5′ cap and 3′ poly(A)-tail respectively, exhibited significant repression by miRNA transfection in the RNA and RPF levels at 2 and 4 hpf of zebrafish embryos, respectively, while lncRNA without TSS or CPS did not exhibit significant repression at any developmental stages of the zebrafish embryos (Fig. 4A, B). For further assessment of lncRNA with mRNA-like features, lncRNAs with both TSS and CPS, and with sORF, were respectively analyzed in the same manner (Fig. 4C). lncRNAs with both TSS and CPS showed significant repression of RNA and RPF levels at 2 hpf and 4 hpf of zebrafish embryos, respectively. While those with sORF did not exhibit significant translational repression mediated by miRNA at any developmental stages, lncRNAs with TSS and CPS exhibited significant reduction in translational efficiency at 4 hpf of zebrafish embryo (Fig. 4C). Taken together, these results support our hypothesis of miRNA-mediated repression on mRNA-like lncRNAs, as shown in exemplified lncRNAs containing miRNA target site with/without both TSS and CPS (Supplementary Fig. S6A, B). Note that ENSDART00000128177, a lncRNA classified as a processed transcript, supported by both TSS and CPS and predicted to contain miR-155 7m8 site, was downregulated in RNA and RPF levels at 2 hpf and 4 hpf of miR-155 injected zebrafish embryos (Supplementary Fig. S6A), while ENSDART00000152905, a lncRNA classified as an antisense transcript not supported by TSS and CPS, and predicted to contain miR-155 8mer target site, was not (Supplementary Fig. S6B).
Fig. 4.
miRNA-mediated repression of lncRNAs with 5′ cap and 3′ poly(A)-tail. (A–C) Median fold changes of RNA (left), RPF (middle) levels and translational efficiency (asterisk, *TE in right) of lncRNAs containing canonical miR-132 and -155 target sites with or without mRNA-like features, such as TSS (A), CPS (B), sORF or both TSS and CPS (C). The P values were calculated using one-tailed Wilcoxon rank sum test as in Fig. 3C, D. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
lncRNAs with putative ORFs are affected by miRNA as mRNA-like lncRNAs
In addition to lncRNAs with sORFs, the subsets of lncRNAs containing putative ORFs that possibly encode peptides longer than 100 amino acids, were separately analyzed (Supplementary Fig. S7). At all developmental stages in the zebrafish embryos, the expression levels of lncRNAs having putative ORFs were significantly higher than those lacking the evidence of any ORF (Supplementary Fig. S7A; P = 7.529 × 10−13 for 2 hpf; P = 2.913 × 10−11 for 4 hpf; P = 1.053 × 10−4 for 6 hpf). Although the expression levels for lncRNAs containing putative sORFs or ORFs were comparable to each other throughout the developmental stages (P = 0.062 for 2 hpf; P = 0.056 for 4 hpf; P = 0.621 for 6 hpf), their expression levels were much lower than that of protein-coding mRNAs at any stage (Supplementary Fig. S7A; P = 8.742 × 10−21 for 2 hpf; P = 1.069 × 10−22 for 4 hpf; P = 5.541 × 10−5 for 6 hpf). Next, the lncRNAs with putative ORFs were analyzed for similar mode of miRNA targeting as that for lncRNAs with sORFs. Akin to the lncRNAs with sORF, those with the putative ORFs exhibited a similar pattern and degree of repression of RNA levels at 2 hpf, but no repression of RPF levels and translational efficiency at any other stage (Supplementary Fig. S7B).
DISCUSSION
We used the RNA-seq and Ribo-seq data from multiple developmental stages of zebrafish embryos (14), to study the interaction between miRNAs and lncRNAs with mRNA-like features. While there is an established pattern of MGS observed in protein-coding genes — early translational repression and late, predominant post-transcriptional repression (10, 13, 14) — different pattern of MGS, the repression of RNA and RPF levels at 2 hpf and 4 hpf of zebrafish embryos, were observed in lncRNAs. A subsequent analysis of the lncRNA subsets containing TSS, CPS, or sORF signals revealed that the presence of 5′ cap and poly(A) tail might be required for efficient miRNA targeting and miRNA-mediated lncRNA repression at both the RNA and RPF levels. Post-transcriptional repression, observed on lncRNAs with both the TSS and CPS at an early stage of development (~2 hpf of zebrafish embryo), compared to those on protein-coding mRNAs, could be explained by the relatively shorter half-lives of lncRNAs (45). What was interesting was significant translational repression of lncRNA with mRNA like features, 5′ cap and 3′ poly(A)-tail mediated by miRNA. This result coincides with the observation from the previous study that circular RNA ciRS-7, which lack both 5′ cap and 3′ poly(A)-tail structure, were not affected by miRNAs and worked as a functional miRNA decoy, while polyadenylated, linear construct of ciRS-7 exhibited miRNA-mediated repression (25). Therefore, we propose that the presence of a 5′ cap and 3′ poly(A)-tail in lncRNAs would be one of important determinants of miRNA-mediated repression in lncRNAs (Supplementary Fig. S8). As many lncRNAs contain the 5′ cap and 3′ poly(A)-tail (28), this miRNA-mediated repression of lncRNAs may represent a new regulatory regime for non-coding RNA networks. Moreover, as lncRNAs without those features were not significantly affected by miRNAs, it is possible that the subset of lncRNAs without 5′ cap and 3′ poly(A)-tail might work as functional miRNA decoys.
MATERIALS AND METHODS
Detailed information is provided in the Supplementary Material.
Supplementary Information
ACKNOWLEDGEMENTS
We thank all BIG Lab members for helpful discussions. This work was supported by the Basic Science Research Program through the National Research Foundation of Korea, and funded by the Ministry of Science, ICT & Future Planning (NRF-2012M3A9D1054516) and supported by the Program for Agriculture Science & Technology Development of the Rural Development Administration, Republic of Korea (Project No. PJ01045303).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grimson A, Farh KK-H, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA Targeting Specificity in Mammals: Determinants beyond Seed Pairing. Molecular Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin C, Nam J-W, Farh KK-H, Chiang HR, Shkumatava A, Bartel DP. Expanding the MicroRNA Targeting Code: Functional Sites with Centered Pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bagga S, Bracht J, Hunter S, et al. Regulation by let-7 and lin-4 miRNAs Results in Target mRNA Degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 8.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 9.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 10.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichhorn SW, Guo H, McGeary SE, et al. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell. 2014;56:104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Huang J, Zhou N, et al. LncRNA loc285194 is a 53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cesana M, Cacchiarelli D, Legnini I, et al. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faghihi MA, Zhang M, Huang J, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 20.Denzler R, McGeary Sean E, Title Alexandra C, Agarwal V, Bartel David P, Stoffel M. Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression. Mol Cell. 2016;64:565–579. doi: 10.1016/j.molcel.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denzler R, Agarwal V, Stefano J, Bartel David P, Stoffel M. Assessing the ceRNA Hypothesis with Quantitative Measurements of miRNA and Target Abundance. Mol Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leucci E, Patella F, Waage J, et al. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep. 2013;3:2535. doi: 10.1038/srep02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon JH, Abdelmohsen K, Srikantan S, et al. LincRNA-21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 26.Franco-Zorrilla JM, Valli A, Todesco M, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 27.Nam JW, Bartel DP. Long non-coding RNAs in C. elegans. Genome Res. 2012;22:2529–2540. doi: 10.1101/gr.140475.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulitsky I, Bartel David P. lincRNAs: Genomics, Evolution, and Mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bazzini AA, Johnstone TG, Christiano R, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calviello L, Mukherjee N, Wyler E, et al. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods. 2016;13:165–170. doi: 10.1038/nmeth.3688. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM. Long non-coding RNAs as a source of new peptides. Elife. 2014;3:e03523. doi: 10.7554/eLife.03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith JE, Alvarez-Dominguez JR, Kline N, et al. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep. 2014;7:1858–1866. doi: 10.1016/j.celrep.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 34.Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11:1110–1122. doi: 10.1016/j.celrep.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pauli A, Valen E, Lin MF, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nepal C, Hadzhiev Y, Previti C, et al. Dynamic regulation of the transcription initiation landscape at single nucleotide resolution during vertebrate embryogenesis. Genome Res. 2013;23:1938–1950. doi: 10.1101/gr.153692.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong L, Zhang Y, Ye ZQ, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark MB, Johnston RL, Inostroza-Ponta M, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Kiledjian M. The Poly(A)-Binding Protein and an mRNA Stability Protein Jointly Regulate an Endoribonuclease Activity. Mol Cell Biol. 2000;20:6334–6341. doi: 10.1128/MCB.20.17.6334-6341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark MB, Johnston RL, Inostroza-Ponta M, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.