Abstract
miR-23~27~24 was recently implicated in restricting Th2 immunity as well as the differentiation and function of other effector T cell lineages. Interestingly, miR-24, unlike other family members, actually promotes Th1 and Th17 responses. Here, we show that miR-24 drives the production of IFNγ and IL-17 in T cells at least in part through targeting TCF1, a transcription factor known for its role in limiting Th1 and Th17 immunity. Surprisingly, while TCF1 was previously shown to promote Th2 responses through inducing GATA3, enforced TCF1 expression in miR-24-overexpresing T cells led to further downregulation of IL-4 and GATA3 expression, suggesting miR-24-mediated inhibition of Th2 immunity cannot be attributed to TCF1 repression by miR-24. Together, our data demonstrates a novel miR-24-TCF1 pathway in controlling effector cytokine production by T cells and further suggests miR-24 could function as a key upstream molecule regulating TCF1-mediated immune responses.
Introduction
During immune responses, naive T cells become activated and differentiate into distinct populations of helper T (Th) cells. In the process of T cell differentiation, each Th cell subset acquires lineage-defining “master regulators” which orchestrate unique transcriptional programs allowing the different Th cell populations to exert their respective immune functions through secreting corresponding effector cytokines (1). In addition to transcriptional regulation, the establishment and maintenance of each functional Th cell subset also involve the expression of epigenetic regulators such as microRNAs (miRNAs) that could either promote or inhibit the differentiation and function of a given Th cell lineage (1).
In the past decade, many miRNAs have been identified to be crucial in regulating Th cell fate decisions and controlling their effector functions (2). For example, members of the miR-29 family have been shown to inhibit Th1 immunity not only by repressing the Th1 master regulators T-bet and Eomes, but also through directly targeting the effector cytokine IFNγ (3–5). Recently, we and others have demonstrated that miR-24 and miR-27 could potently control Th2 immunity through coordinately repressing Gata3, IL-4 and many other Th2-associated molecules (6, 7). Interestingly, while the whole miR-23~27~24 family also negatively regulates the differentiation and function of other Th cell lineages, miR-24, unlike miR-23 or miR-27, actually promotes Th1 and Th17 responses, suggesting that under certain circumstances individual members can antagonize rather than cooperate with each other to fine tune a given biological effect of the entire miRNA family (6).
In this study, we showed that miR-24 drives the production of IFNγ and IL-17 by conventional T (Tconv) cells in a cell-intrinsic manner and enforced expression of miR-24 in Tconv cells lead to dysregulated Th1 and Th17 responses even in the presence of wildtype (WT) Treg cells. Mechanistically, through analyzing whole transcriptome sequencing data combined with high-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation (HITS-CLIP) results (6, 8), we identified TCF1, a transcription factor known for its role in restricting Th1 and Th17 responses (9, 10), as a bona-fide miR-24 target. Retrovirally overexpressing TCF1, with or without the β-catenin-interacting domain, in miR-24-overexpressing T cells reduced their production of effector cytokines under Th1 and Th17 polarizing conditions. On the other hand, partially ablating TCF1 in T cells so that it is expressed to a level similar to that detected in T cells with excessive miR-24 expression led to elevated Th1 and Th17 responses. Interestingly, although TCF1 was previously shown to promote Th2 responses through inducing GATA3 expression (9), miR-24 does not seem to limit Th2 immunity though targeting TCF1, as enforcing TCF1 expression in miR-24-overexpressing T cells led to further downregulation of IL-4 and GATA3 expression. Collectively, our results identify a novel miR-24-TCF1 axis in modulating effector T (Teff) cell cytokine production and further suggest that miR-24 could serve as a key regulator in fine tuning the amount of TCF1 to control a wide range of immune responses.
Materials and Methods
Mice
R24Tg (6) and TCF1fl mice (11) were described previously. All mice were maintained and handled in accordance with the Institutional Animal Care and Use Guidelines of UCSD and National Institutes of Health Guidelines for the care and Use of Laboratory Animals and the ARRIVE Guidelines.
Flow cytometry and antibodies
Single-cell suspension of different lymphoid organs as well other tissues including lung and lamina propria were prepared as described previously (6). For FACS analysis, cells were stained with Ghost Dye Red 780 (Tonbo Biosciences) or Fixable Viability Dye eflour 450 (eBiosciences) followed by surface staining with antibodies against CD4, CD8, CD62L, CD44, Ly5.1. Intracellular staining was performed with antibodies against Foxp3, T-bet, GATA3, RORγT (eBiosciences), TCF1 (Cell signaling) and GFP (eBioscience). To assess IFNγ, IL-17, IL-4 production, cells were incubated with PMA (50ng/ml), ionomycine (0.5μg/ml) and Brefeldin A (1μg/ml) for 4 h at 37 °C, followed by standard staining as described above. BD LSRFortessa or BD LSRFortessa 20x cell analyzer (BD Biosciences) was used for data collection while Flowjo software (Tree Star) was used for data analysis.
Gene expression profiling analysis
RNA-sequencing results (GSE75909) of CD4+CD25−CD62Lhi T cells isolated from WT or R24Tg mice were analyzed as described previously (6). Briefly, gene expression values were generated for RefSeq annotated transcripts using HOMER (12). For the cumulative distribution function (CDF) plots, target sites were restricted to perfect seed complementarity between positions 2 and 7 of the corresponding miRNA with positive Argonaute binding peaks in the HITS-CLIP database (8). Empirical cumulative distributions were computed using Matlab (R2014b) to display the log2(miRNA Tg/WT) against the cumulative frequency of Th1- or Th17- associated or all genes (13).
Generation of mixed bone marrow chimeras
BM cells were isolated from WT, R24Tg or Ly5.1+ B6 mice followed by T cell depletion using mouse Pan T (90.2) kit (Dynabead). WT or R24Tg BM cells were mixed with Ly5.1+ B6 BM cells at 1:1 prior transferring into lethally irradiated (950 rads) Rag1−/− recipients intravenously. 8 weeks after BM reconstitution, lymphocytes from different tissues were isolated and assessed using FACS analysis.
Quantitative PCR analysis
CD4+CD25−CD62Lhi T cells in spleen were sorted by FACSAria flow cytometry (BD Biosciences). Total mRNA were isolated by using miRNeasy kit (Qiagen). cDNA were generated by using iScript cDNA synthesis kit (Bio-Rad) followed by real-time PCR reactions using SYBR green PCR kits (Applied Biosystems). Primer sequences are as follows: TCF1 Forward 5′-TCAATCTGCTCATGCCCTAC-3′, TCF1 Reverse 5′-TGGA CTGCTGAAATGTTCGT-3′, GAPDH Forward 5′-CGTCCCGTAGACAAAATGGT-3′, GAPDH reverse 5′-TCAA TGAAGGGGTCGTTGAT-3′
Retroviral transduction
Long TCF1 isoform (L-TCF1) was PCR amplified from mouse CD4+ T cell mRNA and short TCF isoform (S-TCF1) was PCR amplified by using pCMV6-TCF7 (Origene) as a template. L-TCF1 and S-TCF1 were subcloned into retroviral vector, pMIG-RI, respectively. Retrovirus was generated as described previously (6). FACS sorted CD4+CD25−CD62Lhi T cells were stimulated in αCD3 (2μg/ml) and αCD28 (2μg/ml) coated wells for 24 h under indicated Th cell polarizing conditions as described previously (6), followed by spin-infection for 90 min at 2000 rpm in the presence of 8 μg/ml of polybrene (Millipore). After 4 d of retrovirus transduction, cells were harvested and stained with selected sets of antibodies.
Immunoblotting
Cells were subjected to lysis in RIPA buffer (Cell signaling) supplemented with 1mM PMSF for 20 min. Cell lysates were separated by SDS-PAGE and transferred onto PVDF membrane (Bio-Rad). Antibodies against TCF1 (Cell signaling) and β-Actin (Sigma) were used to detect corresponding proteins. The proteins were quantified with Image J (National Institutes of Health).
Luciferase reporter assay
3′UTR of TCF1 was amplified from mouse genomic DNA and cloned into psiCheck2 vector (Promega). Site-direct mutagenesis (Agilent) was performed to obtain TCF1 3′UTR mutants. One day before transfection, HEK293 T cells were plated in 24 well. psiCheck2 bearing WT or mutant 3′UTR were transfected into HEK293T cells with miR-24-expressing plasmid or control empty vector. Luciferase activities were assessed using Dual luciferase reporter assay system (Promega) according to the manufacturer’s instruction at 24 h after transfection.
Statistical Methods
Statistical tests were performed using Prism 5.0c (GraphPad). Significance was determined by unpaired Student’s t-test with a 95% confidence interval. Error bars indicate SD. *p<0.05, **p<0.01, ***p<0.001.
Results
Enforced expression of miR-24 in T cells results in elevated IFNγ production before the onset of spontaneous lympho-hyperactivation diseases
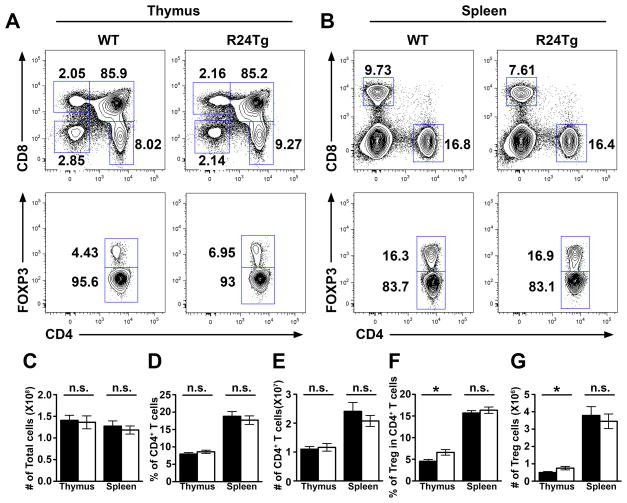
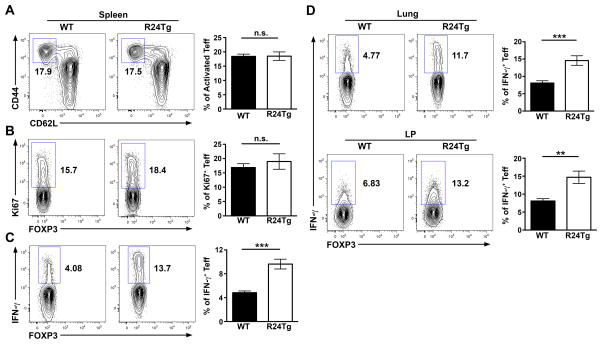
Considering that miR-24 can either cooperate with or antagonize the other members in a given miR-23~27~24-dependent biology (6), we sought to further examine mice harboring T cells with elevated miR-24 expression (R24Tg) to determine the precise role of miR-24 in controlling T cell immunity. As shown in Fig. 1, like mice with T cell-specific overexpression of the miR-23~27~23 cluster (R23CTg), R24Tg mice did not display any abnormality in total thymic or splenic cellularities nor in any peripheral T cell compartment compared to their WT littermates. However, in contrast to the reduced thymic Treg cell frequencies detected in R23CTg mice as well as mice with T cell-specific overexpression of miR-27 (R27Tg) (6, 14), enforced expression of miR-24 in T cells alone resulted in appreciably increased thymic Treg cell numbers (Fig. 1F and G), consistent with the previous notion that miR-24 can antagonize some functions of other members in the miR-23~27~24 family (6). On the other hand, our recent study has demonstrated that overexpression of miR-24 alone in T cells could result in the development of lympho-hyperactivation phenotypes similar to what was observed in R23CTg and R27Tg mice (6). Further analysis of R24Tg mice of young ages revealed that even before T cell hyperactivation could be detected, those mice already harbored elevated numbers of IFNγ-producing T cells both in secondary lymphoid organs and other tissues (Fig. 2), supporting the reported role of miR-24 in promoting Th1 differentiation and IFNγ production (6).
Figure 1. Increased thymic Treg cell numbers in mice with T cell-specific miR-24 overexpression.
FACS analysis of (A) thymus and (B) spleen of 6–8 wks old R24Tg mice or WT littermates. (C) Cellularity of thymus and spleen as welll as (D–G) frequencies and absolute numbers of thymic and splenic total CD4+ T cells and CD4+Foxp3+ Treg cells in R24Tg mice and WT littermates are shown. Data are representative of four independent experiments (n = 10–11). *p<0.05.
Figure 2. R24Tg mice exhibited heightened IFNγ responses prior to the development of lympho-hyperactivation phenotypes.
FACS analysis and frequencies of (A) activated CD44hiCD62Llow, (B) Ki67+ subsets and (C) IFNγ+ cells in Foxp3−CD4+ Tconv cells in spleen from 6–8 wks old R24Tg mice or their WT littermates. (D) FACS analysis and frequencies of IFNγ+ cells in Foxp3−CD4+ Tconv cells in lung or lamina propria (LP) of small intestine are shown. Data are representative of 4 independent experiments (n = 10–11). **p<0.01, ***p<0.001.
Tconv cells with excessive miR-24 expression from healthy R24Tg mice already exhibit Th1 and Th17 gene signatures
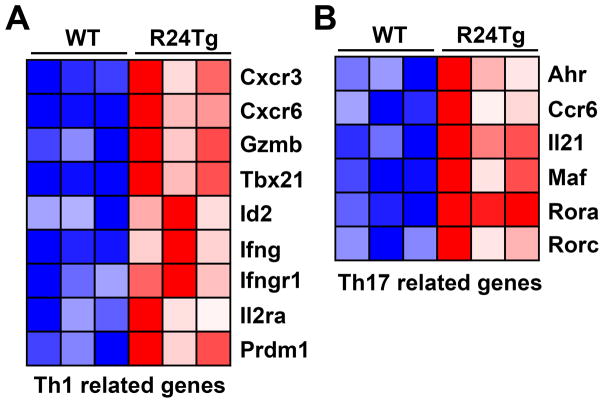
While we did not detect significant changes in IL-17 production by T cells in R24Tg mice before the onset of the autoimmune diseases, in addition to enhanced Th1 responses, increased Th17 polarization was also observed in T cells with excessive miR-24 expression (6). To further explore the molecular impacts of miR-24 on T cell immunity, we re-analyzed the transcriptom profiling results of CD4+CD25−CD62Lhi Tconv cells isolated from R24Tg mice of 6~8 weeks of age (6). While overexpression of miR-24 was reported to negatively regulate the Th2-related gene network (6, 7), R24Tg T cells isolated from mice prior to the onset of T cell hyperactivation already exhibited elevated expression of many Th1- and Th17-signature genes (Fig. 3). These results together with the previous findings obtained from the in vitro polarization studies strongly suggested that miR-24 directly promotes Th1 and Th17 responses in the Tconv cell compartment that would subsequently lead to the development of the aforementioned lympho-hyperactivation phenotypes (6).
Figure 3. Tconv cells with miR-24 overexpression exhibited Th1 and Th17 gene signatures.
Heat map of representative gene associated with (A) Th1 and (B) Th17 cell differentiation and function differentially expressed in CD4+CD25−CD62Lhi Tconv cells in the presence or absence of excessive miR-24 expression.
miR-24 drives dyrsregulated IFNγ and IL-17 responses despite the presence of WT Treg cells
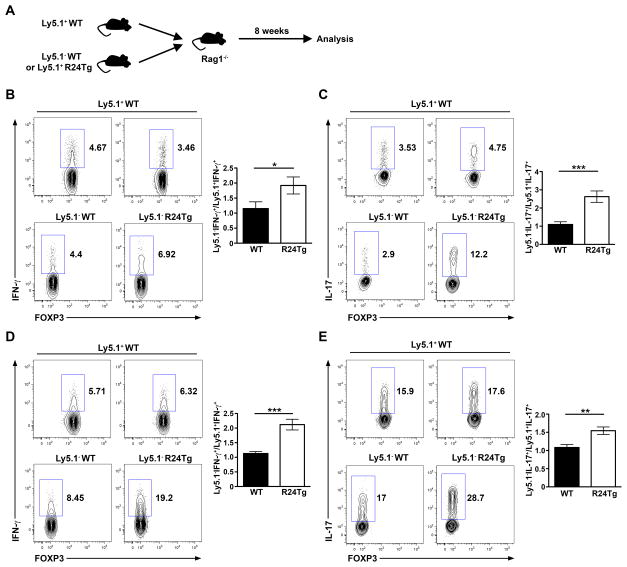
Our recent findings in mice harboring T cells with miR-27 overexpression suggested that the observed autoimmunity was not driven by miR-27 in Tconv cells in a cell-autonomous fashion but rather resulted from a perturbed Treg cell compartment (14). While it is likely miR-24 can directly enhance the production of effector cytokines by Tconv cells, it remains probable that such Tconv cell phenotypes could only be detected in mice lacking a functional Treg cell population as a result of exaggerated miR-24-mediated gene regulation. Despite the fact that miR-24-overexpressing Treg cells exhibited normal suppressor function in vitro (Fig. S1), to directly examine this possibility, we performed bone marrow (BM) chimeras studies by transferring BM cells from R24Tg mice or WT littermates mixed with BM cells from Ly5.1+ B6 mice at a 1:1 ratio into irradiated Rag1-deficient recipients as described previously (15) (Fig. 4A). 8 weeks after the initial BM transfer, R24Tg/Ly5.1+ B6 chimeric mice remained healthy and showed no sign of T cell hyperactivation, similar to our observation in young R24Tg mice. On the other hand, despite the presence of WT Treg cells from the Ly5.1+ compartment, noticeably increased frequencies of IFNγ- and IL-17-producing Tconv cells could be detected in T cells with excessive miR-24 expression but not in the WT counterparts in the same chimeric mice (Fig. 4B–E). These results confirmed the previous notion that miR-24 promotes IFNγ and IL-17 production by Tconv cells in a cell-intrinsic manner and further demonstrated that such Tconv cell phenotypes would occur independently of any potential impact of miR-24 on Treg cell-mediated regulation.
Figure 4. Cell-intrinsic role of miR-24 in driving the production of IFNγ and IL-17 by Tconv cells.
(A) Schematic of generation of mixed BM chimeras. FACS analysis and ratios of frequencies of (B) Ly5.1−IFNγ+ and Ly5.1+IFNγ+; (C) Ly5.1−IL-17+ and Ly5.1+IL-17+ Tconv cells in lung and (D) Ly5.1−IFNγ+ and Ly5.1+IFNγ+; (E) Ly5.1−IL-17+ and Ly5.1+IL-17+ Tconv cells in LP of small intestine. Data are representative of 3 independent experiments (n = 16–19). *p<0.05, **p<0.01, ***p<0.001.
miR-24 promotes the generation of IFNγ- and IL-17-producing T cells through targeting TCF1
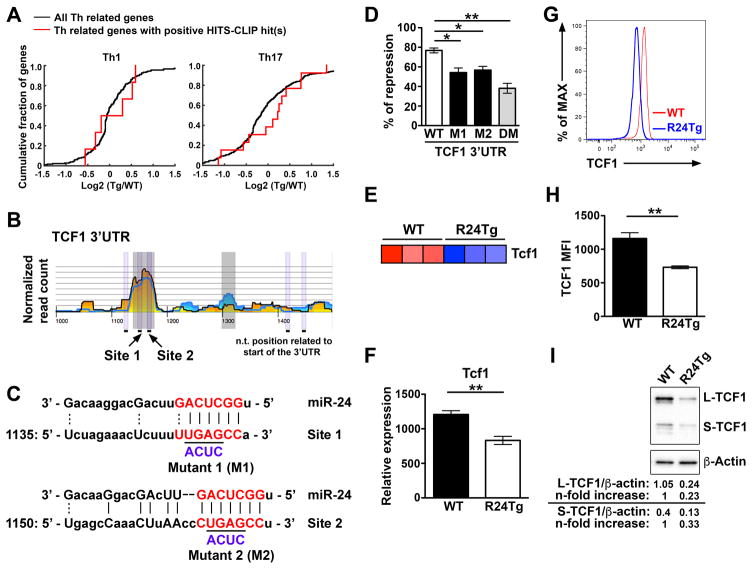
To identify the molecular mechanisms underlying miR-24-mediated regulation of effector cytokine production by Tconv cells, we examined the aforementioned whole transcriptome sequencing results obtained from naive miR-24-overexpressing T cells in conjunction with the analysis of the previously reported HITS-CLIP database (6, 8). While miR-24 does not generally repress Th1- or Th17-related genes at the mRNA level similar to what was reported in the regulation of Th2-related genes (Fig. 5A)(6), TCF1, a high mobility group (HMG) box transcription factor which has been previously shown to inhibit the expression of IFNγ and IL-17 in T cells (9, 10), was identified as a potential miR-24 target responsible for the elevated IFNγ and IL-17 responses observed in R24Tg mice (Fig. 5B and C). Luciferase reporter analysis confirmed that TCF1 is indeed a direct target of miR-24 as co-transfection of miR-24 with the TCF1 3′UTR resulted in appreciable repression of reporter activity, whereas such repression was markedly abolished when the two putative miR-24-binding sites were mutated (Fig. 5D). Consistently, diminished TCF1 mRNA was detected in T cells overexpressing miR-24 in the aforementioned transcriptome profiling study and by qPCR analysis (Fig. 5E and F). Moreover, R24Tg T cells exhibited 40~50% reduction in TCF1 protein expression (Fig. 5G and H). Finally, as TCF1 exists in two major isoforms: a long isoform (L-TCF1) that can interact with β-catenin to activate the Wnt signaling pathway and a short one (S-TCF1) that cannot (16), we next examined the effect of miR-24-mediated repression on different TCF1 isoforms. Immunoblotting results revealed that both TCF1 isoforms were repressed in T cells overexpressing miR-24 (Fig. 5I), suggesting that miR-24 can modulate both β-catenin-dependent and -independent TCF1-mediated biology.
Figure 5. TCF1 is a direct target of miR-24.
(A) Cumulative distribution frequency (CDF) plots depicting the effect of overexpression of miR-24 on mRNA expression of Th1 and Th17 cell-associated genes. Levels of mRNAs of Th1 or Th17 cell-associated genes bearing HITS-CLIP identified miR-24 sites (red line) were compared with mRNAs of all Th1 or Th17 cell-associated genes (black line). (B) HITS-CLIP analysis of putative miR-24 binding sites in the 3′UTR of TCF1. Reads from the 12 replicates have been stacked. Coordinates along the x-axis indicate nucleotide position relative to the beginning of the 3′UTR. The y axis indicates normalized read counts, which are square root transformed after individual library normalization (8). (C) Sequence alignment of putative miR-24 binding sites in the 3′UTR of TCF1. Mutations of the two corresponding miR-24 target sites are shown below. (D) Ratios of repressed luciferase activity of cells in the presence of WT or different mutated TCF1 3′ UTRs (M1: mutant 1, M2: mutant 2 or DM: double mutants) transfected with miR-24 compared with cells transfected with empty vector. (E) TCF1 mRNA expression results from the naive CD4+ T cell RNA-sequencing study was presented as a heat map. (F) qPCR of TCF1 mRNA levels, (G) FACS analysis and (H) mean fluorescence intensity (MFI) of TCF1 protein amounts in naïve CD4+ T cells from R24Tg mice or WT littermates were shown. (I) Expression of different TCF1 isoforms in naïve CD4+ T cells isolated from R24Tg or control mice were assessed by immunoblotting. Densitometric values of different TCF1 isoform were normalized to β-actin expression values and n-fold increase on the basis of each corresponding WT. All data are representative of at least 3 independent experiments (n = 6–7). *p<0.05, **p<0.01.
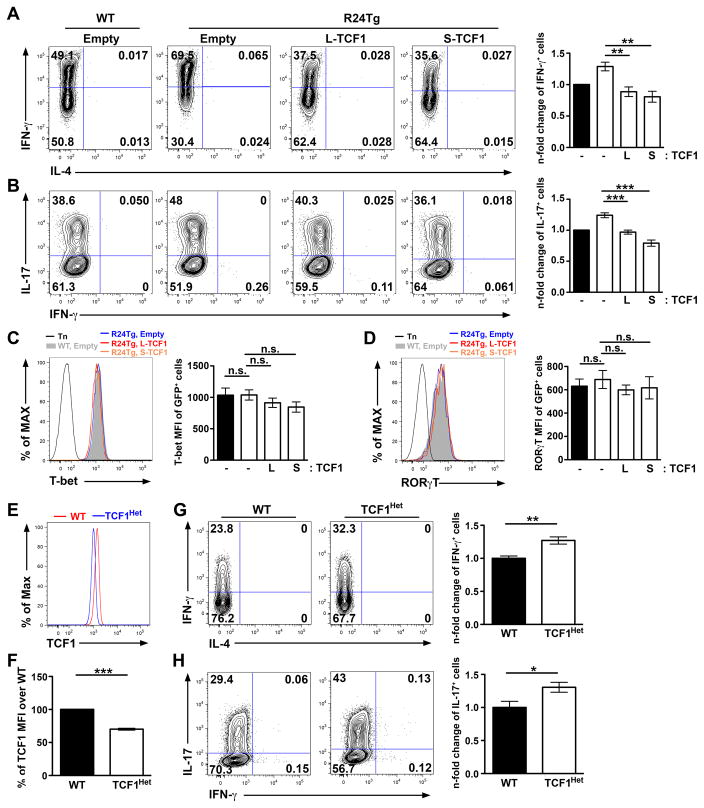
Next, we sought to determine whether the augmented miR-24-mediated TCF1 repression could account for the aforementioned elevated IFNγ and IL-17 responses observed in R24Tg T cells. To this end, we first retrovirally overexpressed TCF1 in R24Tg T cells and assessed their abilities to differentiate into IFNγ- and IL-17-secreting T cells, respectively. Consistent with previous findings where TCF1 was shown to negatively regulates IFNγ and IL-17 production in a β-catenin-independent manner (9, 10), both long and short TCF1 isoforms were equally effective in reducing IFNγ and IL-17 responses in T cells overexpressing miR-24 (Fig. 6A and B). On the other hand, on a per-cell basis we did not detect any alteration in the levels of T-bet or RORγT, in the IFNγ-producing T cell and IL-17-secreting T cells, respectively (Fig. 6C and D), also supporting a previous notion that TCF1 does not directly inhibit the lineage-specific transcription regulators in differentiated Th1 or Th17 cells (9, 10). Nevertheless, the level of TCF1 overexpression resulted from retroviral transduction is much higher than that could be normally detected in WT T cells (Fig. S2A). Consequently, WT T cells with “supra-optimal” TCF1 expression also exhibited a similar reduction in IFNγ and IL-17 production (Fig. S2B), thus precluding the possibility to formally determine whether miR-24-mediated TCF1 repression is indeed responsible for controlling IFNγ and IL-17 responses in T cells via this approach. To examine the miR-24 effect on TCF1 at a more physiologically relevant level, we then generated CD4-cre TCF1fl/+ mice in which all T cells express reduced TCF1 levels similar to what we could observed in our R24Tg T cells (Fig.6E and F). As shown in Fig. 6G and H, a ~30–40% reduction in TCF1 expression was sufficient to elevate IFNγ and IL-17 secretion in T cells upon Th1 and Th17 differentiation, respectively. Together, these results not only support our hypothesized role of a miR-24-TCF1 axis in regulating Th1 and Th17 immunity but further suggest that the expression of TCF1 needs to be tightly regulated as small changes in TCF1 amounts could have significant impacts on lymphocyte development and responses.
Figure 6. Small changes in TCF1 amounts is sufficient to impact effector T cell responses.
FACS analysis of the production of (A) IFNγ or (B) IL-17 and the expression of (C) T-bet or (D) RORγT in GFP+ R24Tg CD4+ T cells transduced with control vector or vectors expressing either long (L-TCF1) or short TCF1 (S-TCF1) isoforms with a GFP reporter under Th1- or Th17- polarizing conditions. n-fold changes (on the basis of corresponding WT controls) of IFNγ+ or IL-17+ cell frequencies or T-bet or RORγT MFI in GFP+ R24Tg CD4+ T cells were shown on the right panel. (E) FACS analysis and (F) % of TCF1 MFI in naïve CD4+ T cells from CD4-cre TCF1fl/+ mice (TCF1Het) over WT littermates were shown. FACS analysis of the production of (G) IFNγ or (H) IL-17 in WT or TCF1Het CD4+ T cells under Th1- or Th17- polarizing conditions. n-fold changes (on the basis of corresponding WT controls) of IFNγ+ or IL-17+ cell frequencies were shown on the right panel. All data are representative of at least three independent experiments (n = 3–6). *p<0.05, **p<0.01, ***p<0.001.
Repression of TCF1 by miR-24 does not contribute to miR-24-dependent impairment in Th2 differentiation
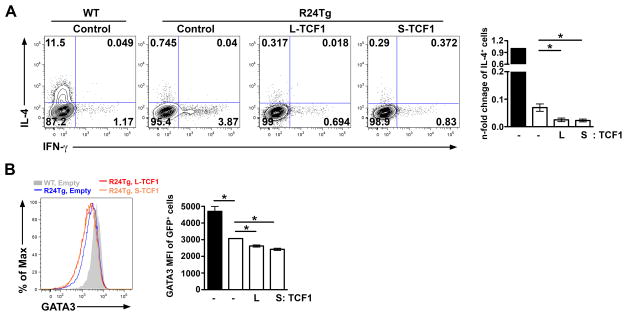
Recently, we have demonstrated that miR-24 controls Th2 immunity in part through targeting IL-4 directly (6). The fact that miR-24 represses TCF1 and TCF1 was shown to promote Th2 responses and IL-4 production through inducing GATA3 expression raised a possibility that miR-24-mediated TCF1 repression could also contribute to the impaired Th2 responses observed in T cells with excessive miR-24 expression (9). It should be noted, however, that while TCF1 inhibits IFNγ and IL-17 in a β-catenin-independent manner, it was shown that interaction between TCF1 and β-catenin is critical to promote GATA3 expression and the resultant IL-4 production (9). In the absence of the β-catenin-interacting domain, the short TCF1 isoform does not function as a transcriptional activator but rather a repressor that inhibits IL-4 expression (17). Since both TCF1 isoforms with opposite effects on IL-4 production were repressed by miR-24, we sought to examine the exact role of miR-24-mediated TCF1 repression in regulating Th2 immunity. To our surprise, while overexpression of miR-24 resulted in a drastic reduction in IL-4-producing T cells consistent with our previous findings (6), the introduction of neither the long nor the short TCF1 isoform restored the ability of miR-24-overexpressing T cells to differentiate into IL-4-secreting cells; instead, both isoforms further decreased IL-4 production as well as GATA3 expression (Fig. 7A and B). These results clearly demonstrated that miR-24 does not limit IL-4 production and Th2 differentiation through targeting TCF1.
Figure 7. Enforced expression of TCF1 regardless of its capacity to interact with β-catenin in R24Tg T cells led to further decrease in IL-4 production and GATA3 expression.
FACS analysis of (A) IL-4 production and (B) GATA3 expression in GFP+ R24Tg CD4+ T cells transduced with control vector or vectors expressing either L-TCF1 or S-TCF1 with a GFP reporter under Th2- polarizing conditions. n-fold changes (on the basis of corresponding WT controls) of IL-4+ cell frequencies or GATA3 MFI in GFP+ R24Tg CD4+ T cells were shown on the right panels. Data are representative of at least four independent experiments (n = 4–6). *p<0.05.
Discussion
While it is generally believed that miRNA clusters are evolutionarily conserved to ensure their biological impact through having different cluster members collaboratively target the same gene or different components of a common biological process (18), recent studies have revealed that individual members could also antagonize each other’s function to fine tune a given biological effect of the entire miRNA family (6, 19, 20). Specifically, we have recently reported that enforced miR-24 expression resulted in enhanced Th1 and Th17 differentiation whereas overexpression of the whole miR-23~27~24 cluster or other family members inhibited those Th lineages (6). In this study, by further examining mice with T cell-specific miR-24 overexpression as well as employing the mixed BM chimera approach, we comprehensively demonstrated a Tconv cell-intrinsic role of miR-24 in promoting IFNγ and IL-17 responses. Moreover, by analyzing whole transcriptome sequencing data together with the HITS-CLIP database, we further identified a novel miR-24-TCF1 axis in regulating the effector function of multiple Th cell subsets.
Despite the fact that enforced expression of either miR-24 or miR-27 in T cells can both lead to spontaneous lympho-hyperactivition (6), the effector mechanisms underlying the observed autoimmunity were very different. Unlike miR-27, the overexpression of which impaired Teff activation and function but promoted disease development through impeding Treg cell-mediated immunoregulation (14), exaggerated miR-24-mediated regulation directly augmented the generation of effector cytokine producing Tconv cells preceding the onset of the autoimmune disorder. It is also noteworthy that R24Tg mice harbored increased thymic Treg cells as opposed to the reduced number observed in R27Tg mice (14). A recent report has suggested a regulatory role of TCF1 in limiting thymic Treg cell development (21). It is thus plausible that miR-24 could also promote thymic Treg cell generation through targeting TCF1 and further suggests that miR-24 might act more as a positive player rather than a negative regulator like miR-27 in controlling Treg cell biology. While we could not exclude the possibility that excessive miR-24 expression might still hamper some aspects of Treg cell function thus contributing to the autoimmune phenotypes detected in R24Tg mice, our studies have unequivocally demonstrated that excessive miR-24 expression in Tconv cells alone is sufficient to cause dysregulated IFNγ and IL-17 responses and would lead to the subsequent autoimmunity.
TCF1 functions as a key downstream effector of the Wnt signaling pathway to control T cell development in the thymus as well as mature T cell differentiation in the periphery (22–24). Specifically, it was previously shown that TCF1 inhibits IFNγ and IL-17 responses without directly impacting the expression of T-bet and RORγT (9, 10). Interestingly, while our analysis of differentiated Th1 and Th17 cells regarding the effect of TCF1 on lineage-specific transcription factors was in agreement with the aforementioned reports, our transcriptome profiling results revealed that enhanced expression of T-bet and RORγT as well as other Th1- and Th17-associated genes could already be observed in CD4+CD25−CD62Lhi T cells isolated from healthy R24Tg mice. These results suggested that TCF1 might likely play a role in regulating T-bet and RORγT expression during the early phase of T cell differentiation as T cells with reduced TCF1 amount are better poised to become effector T cell subsets. However, once cells are fully differentiated, while TCF1 could still modulate the production of effector cytokines, its impact on the expression of lineage-specific transcription factors diminishes as those molecules would be stabilized by many transcriptional and epigenetic regulations (25).
While our work has clearly shown that miR-24 could drive the generation of IFNγ- and IL-17-producing T cells through repressing TCF1, it seemed to argue against the reported role of TCF1 in promoting Th2 responses (9). Restoring the expression of the long TCF1 isoform, the one with the capacity to induce GATA3 expression through interacting with β-catenin, in miR-24-overexpressing cells resulted in a further decrease in GATA3 expression and IL-4 production. Despite the lack of immediate experimental evidence to reconcile the discrepancy between our study and the previous work, it is conceivable that miR-24-mediated regulation of other molecules could play a key role in influencing the outcome of TCF1-mediated GATA3 induction. Finally, while it seems that the miR-24-TCF1 axis reported in this study generally promotes effector function of all T cell subsets, expression of TCF1 has been recently shown to be crucial for establishing the transcriptional program of the follicular T help (Tfh) cell population (11, 26). As such, excessive miR-24 expression is likely to be detrimental to Tfh responses and miR-24 might function as a central molecular regulator in controlling the reciprocal development of Tfh cells vs. other Teff cell lineages through targeting TCF1.
Our study has revealed a novel miR-24-TCF1 axis in regulating effector cytokine production and further demonstrated that the expression of TCF1 needs to be tightly regulated as small changes in TCF1 amounts could have significant impact on lymphocyte development and responses. Although there is always a concern over the physiological relevance of using a mouse model with selectively enforced expression of only one member of the whole miRNA family, it should be noted that despite the fact that the expression of different miR-23~27~24 cluster members are usually regulated in a similar fashion, they could exhibit very distinct expression profiles under certain conditions (27). To this end, miR-27 expression was found to be highly upregulated in T cells isolated from multiple sclerosis patients and was suggested to play a pivotal role in promoting autoimmune inflammation (28). Likewise, elevated miR-24 expression was also detected in patients with ulcerative colitis and rheumatoid arthritis (29, 30). While the cellular sources of miR-24 under those settings remain to be further investigated, considering the proinflammatory role of IFNγ and IL-17 in driving the aforementioned autoimmune disease progression, the miR-24-TCF1 axis could serve as a molecular basis to develop new therapeutics for autoimmunity with elevated miR-24 expression. Alternatively, targeting this axis could also provide potential translational benefits in disease settings when a strong Teff cytokine response is desired.
Supplementary Material
Acknowledgments
We thank B. Nizamova and K. Mccunney for superb technical assistance, and all members of our laboratory for discussions. The authors declare no competing financial interests.
This work was supported by grants from the National Institutes of Health grants AI089935, AI103646, AI108651 and AI123782 (to L.-F. Lu).
References
- 1.Nakayamada S, Takahashi H, Kanno Y, O’Shea JJ. Helper T cell diversity and plasticity. Current opinion in immunology. 2012;24:297–302. doi: 10.1016/j.coi.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nature reviews. Immunology. 2013;13:666–678. doi: 10.1038/nri3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nature immunology. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 4.Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, Matloubian M, Blelloch R, Ansel KM. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith KM, Guerau-de-Arellano M, Costinean S, Williams JL, Bottoni A, Mavrikis Cox G, Satoskar AR, Croce CM, Racke MK, Lovett-Racke AE, Whitacre CC. miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. Journal of immunology. 2012;189:1567–1576. doi: 10.4049/jimmunol.1103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho S, Wu CJ, Yasuda T, Cruz LO, Khan AA, Lin LL, Nguyen DT, Miller M, Lee HM, Kuo ML, Broide DH, Rajewsky K, Rudensky AY, Lu LF. miR-23 approximately 27 approximately 24 clusters control effector T cell differentiation and function. The Journal of experimental medicine. 2016;213:235–249. doi: 10.1084/jem.20150990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pua HH, Steiner DF, Patel S, Gonzalez JR, Ortiz-Carpena JF, Kageyama R, Chiou NT, Gallman A, de Kouchkovsky D, Jeker LT, McManus MT, Erle DJ, Ansel KM. MicroRNAs 24 and 27 Suppress Allergic Inflammation and Target a Network of Regulators of T Helper 2 Cell-Associated Cytokine Production. Immunity. 2016;44:821–832. doi: 10.1016/j.immuni.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Molecular cell. 2012;48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Q, Sharma A, Oh SY, Moon HG, Hossain MZ, Salay TM, Leeds KE, Du H, Wu B, Waterman ML, Zhu Z, Sen JM. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nature immunology. 2009;10:992–999. doi: 10.1038/ni.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu Q, Sharma A, Ghosh A, Sen JM. T cell factor-1 negatively regulates expression of IL-17 family of cytokines and protects mice from experimental autoimmune encephalomyelitis. Journal of immunology. 2011;186:3946–3952. doi: 10.4049/jimmunol.1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, Love PE, Peng W, Xue HH, Crotty S. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nature immunology. 2015;16:980–990. doi: 10.1038/ni.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stubbington MJ, Mahata B, Svensson V, Deonarine A, Nissen JK, Betz AG, Teichmann SA. An atlas of mouse CD4(+) T cell transcriptomes. Biology direct. 2015;10:14. doi: 10.1186/s13062-015-0045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruz LO, Hashemifar SS, Wu CJ, Cho S, Nguyen DT, Lin LL, Khan AA, Lu LF. Excessive expression of miR-27 impairs Treg-mediated immunological tolerance. The Journal of clinical investigation. 2017;127:530–542. doi: 10.1172/JCI88415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ioannidis V, Beermann F, Clevers H, Held W. The beta-catenin--TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nature immunology. 2001;2:691–697. doi: 10.1038/90623. [DOI] [PubMed] [Google Scholar]
- 17.Maier E, Hebenstreit D, Posselt G, Hammerl P, Duschl A, Horejs-Hoeck J. Inhibition of suppressive T cell factor 1 (TCF-1) isoforms in naive CD4+ T cells is mediated by IL-4/STAT6 signaling. The Journal of biological chemistry. 2011;286:919–928. doi: 10.1074/jbc.M110.144949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang T, Yu J, Liu C, Guo L. An exploration of evolution, maturation, expression and function relationships in mir-23 approximately 27 approximately 24 cluster. PloS one. 2014;9:e106223. doi: 10.1371/journal.pone.0106223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olive V, Sabio E, Bennett MJ, De Jong CS, Biton A, McGann JC, Greaney SK, Sodir NM, Zhou AY, Balakrishnan A, Foth M, Luftig MA, Goga A, Speed TP, Xuan Z, Evan GI, Wan Y, Minella AC, He L. A component of the mir-17-92 polycistronic oncomir promotes oncogene-dependent apoptosis. eLife. 2013;2:e00822. doi: 10.7554/eLife.00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuziwara CS, Kimura ET. High iodine blocks a Notch/miR-19 loop activated by the BRAF(V600E) oncoprotein and restores the response to TGFbeta in thyroid follicular cells. Thyroid: official journal of the American Thyroid Association. 2014;24:453–462. doi: 10.1089/thy.2013.0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barra MM, Richards DM, Hansson J, Hofer AC, Delacher M, Hettinger J, Krijgsveld J, Feuerer M. Transcription Factor 7 Limits Regulatory T Cell Generation in the Thymus. Journal of immunology. 2015;195:3058–3070. doi: 10.4049/jimmunol.1500821. [DOI] [PubMed] [Google Scholar]
- 22.Staal FJ, Clevers HC. Wnt signaling in the thymus. Current opinion in immunology. 2003;15:204–208. doi: 10.1016/s0952-7915(03)00003-7. [DOI] [PubMed] [Google Scholar]
- 23.Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinke FC, Xue HH. From inception to output, Tcf1 and Lef1 safeguard development of T cells and innate immune cells. Immunologic research. 2014;59:45–55. doi: 10.1007/s12026-014-8545-9. [DOI] [PubMed] [Google Scholar]
- 25.Kanno Y, Vahedi G, Hirahara K, Singleton K, O’Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annual review of immunology. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu L, Cao Y, Xie Z, Huang Q, Bai Q, Yang X, He R, Hao Y, Wang H, Zhao T, Fan Z, Qin A, Ye J, Zhou X, Ye L, Wu Y. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nature immunology. 2015;16:991–999. doi: 10.1038/ni.3229. [DOI] [PubMed] [Google Scholar]
- 27.Sun F, Wang J, Pan Q, Yu Y, Zhang Y, Wan Y, Li X, Hong A. Characterization of function and regulation of miR-24-1 and miR-31. Biochemical and biophysical research communications. 2009;380:660–665. doi: 10.1016/j.bbrc.2009.01.161. [DOI] [PubMed] [Google Scholar]
- 28.Guerau-de-Arellano M, Smith KM, Godlewski J, Liu Y, Winger R, Lawler SE, Whitacre CC, Racke MK, Lovett-Racke AE. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain: a journal of neurology. 2011;134:3578–3589. doi: 10.1093/brain/awr262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635 e1624. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 30.Murata K, Furu M, Yoshitomi H, Ishikawa M, Shibuya H, Hashimoto M, Imura Y, Fujii T, Ito H, Mimori T, Matsuda S. Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PloS one. 2013;8:e69118. doi: 10.1371/journal.pone.0069118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.