Abstract
O-GlcNAc hydrolase, OGA, removes O-linked N-acetylglucosamine (O-GlcNAc) from myriad nucleocytoplasmic proteins. Through co-expression and assembly of OGA fragments we determined the 3-D structure of human OGA, revealing an unusual helix exchanged dimer that lays a structural foundation for an improved understanding of substrate recognition and regulation of OGA. Structures of OGA in complex with a series of inhibitors define a precise blueprint for the design of inhibitors having clinical value.
The dynamic O-GlcNAc modification of hundreds of nuclear and cytoplasmic proteins plays diverse roles in a range of cellular processes including, for example, transcriptional regulation and stress response (reviewed in Ref1,2). Dysregulation of O-GlcNAcylation has been implicated in diseases including cancer3, obesity4, and neurodegenerative diseases.5,6 Notably, therapeutic agents targeting the O-GlcNAc modification have entered phase I clinical trials, stimulating interest in the molecular and chemical basis of O-GlcNAcylation and its manipulation with small molecules.7
Within mammals, this modification of serine and threonine residues is installed by O-GlcNAc transferase, OGT, for which extensive structural data are available.8,9 The O-GlcNAc modification is removed by O-GlcNAc hydrolase, OGA.10 Structures of bacterial homologs of OGA from CAZY family GH8411 (originally, and most notably Bacteroides thetaiotaomicron (BtGH84)12 and Clostridium perfringens (CpNGA )13), having a conserved active site with OGA, have aided glycomimetic inhibitor design. Indeed, compounds based on the neighboring-group catalytic mechanism12,14 have been applied in cellular and animal studies. The absence of structural data for mammalian OGA, however, has limited efforts toward inhibitor design and curtailed insight into peptide substrate binding and association with binding partners. Accordingly, as part of our long-standing effort to understand O-GlcNAcase, we set out to dissect human OGA (hOGA) and study its structure as well as its binding to different inhibitor classes.
We first sought to establish a functional construct of human OGA for structural analyses. hOGA is a complex multi-domain protein, produced as two splice variants; long-form OGA-L and a less-active short form, OGA-S (reviewed in ref 15). OGA-L consists of an N-terminal catalytic domain (GH84), a helical domain, extensive regions predicted as disordered, and a C-terminal domain of unknown function having similarity to histone acetyl transferase (HAT) domains (Supplementary Results, Supplementary Fig. 1a). We generated various truncated constructs and screened possible domain boundaries15, none of these (Supplementary Table 1), however, yielded protein amenable to structural analysis.
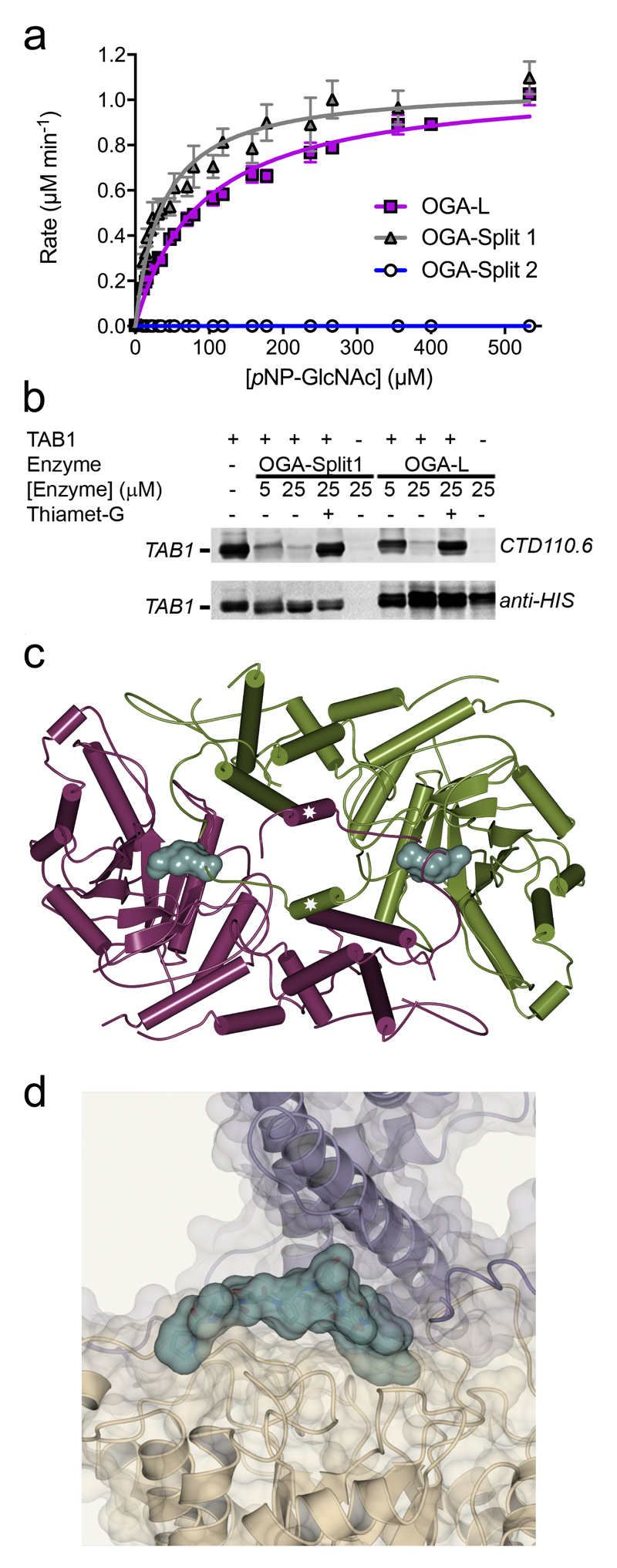
Central to our successful strategy was that hOGA can be cleaved by caspase-3 into two fragments, which remain tightly associated and active in solution.16 Accordingly, co-expression of two hOGA fragments should permit their systematic truncation and removal of putative disordered regions, yet allow their assembly into active hOGA in a form suitable for crystallization. We co-expressed an extensive series of N- and C-terminal constructs in which the putative disordered regions were systematically truncated (Supplementary Table 1). Constructs were screened for formation of stable complexes and screened for crystallization. Ultimately, a construct comprising amino acids 11-396 (N-terminal fragment) and 535-715 (C-terminal fragment) (Supplementary Fig. 1b) yielded crystals suitable for structure determination. Furthermore, this construct, which we term “Split1”, has essentially wild-type catalytic activity toward synthetic substrates (Fig. 1a) and processes O-GlcNAcylated proteins (Fig. 1b, Supplementary Fig. 2, 3).
Figure 1. Function and structure of hOGA.
a) Michaelis-Menten kinetics of crystallized construct Split 1, full-length hOGA (OGA-L) and Split 2. The kinetic parameters for the OGA variants are, OGA-L: Vmax = 1.10 ± 0.02μM min-1; KM = 92.47 ± 4.86µM, and OGA-Split 1: Vmax = 1.11 ± 0.03μM min-1; KM = 40.93 ± 3.30µM. Therefore, the catalytic efficiency (kcat/KM) of OGA-L is roughly similar (2.3 fold lower) to OGA-Split 1 when using pNP-GlcNAc as the substrate. Data represent average of quadruplicate rate measurements ± s. d. b) Immunoblot analysis showing that Split1 can digest recombinant O-GlcNAcylated TAB1, evaluated using anti-O-GlcNAc antibody CTD110.6 anti-Histidine antibody as a loading control. Thiamet-G inhibits digestion. c) Ribbon diagram of the hOGA dimer, colored by chain. The helix (Q676-P694) swapped between the two helical bundles is marked with a star. The position of the active site (-1 sugar binding site) is indicated by the Van der Waals' surface of the inhibitor Thiamet-G in bluegreen. d) Binding of the C-terminal peptide (shown with its Van der Waals surface in bluegreen) of the helical bundle fragment to the hOGA peptide-binding groove. Full blots are provided in the Supplementary Fig. 15.
We solved the structure of Split1 (Supplementary Table 2) by molecular replacement using a sculpted model of BtGH84 having 31% identity in the catalytic domain. This initial model was greatly improved by extensive rebuilding, guided by the positions of sulfur atoms from methionine and cysteine residues, observed using a long wavelength dataset, (Supplementary Fig. 4). The final model of Split1 (11-396 / 535-715; in which residues 11-58, 341-370, 535-536, 596-598, 674-675 and 696-706 are disordered) reveals a two-domain structure, with an N-terminal catalytic domain followed by a C-terminal helical bundle (Fig. 1c). hOGA forms a dimer with a contact interface of 4390 Å2, calculated using PISA.17 We confirmed that the dimer also forms in solution by size exclusion chromatography with multi-angle light scattering (SEC-MALS) (Supplementary Fig. 5a). Notably, a dimer “swap” of the C-terminal helix from each of the helical domains (Q676-P694) completes the open 3-helix bundle to yield a closed 4-helix coiled coil bundle (Fig. 1c). This swap is essential for stable interactions between both the two domains and the two monomers. Deletion of this helix compromised dimer formation as assessed by SEC-MALS, and led to disassembly into its peptide components to yield a near inactive construct (Fig. 1a, Supplementary Fig. 5b, 6). SEC-MALS also showed near full-length hOGA (His(6)9-916) is primarily a dimer (Supplementary Fig. 5c). Previous evidence on the native multimerization status of hOGA was conflicting and depended on the method used.10,18,19 When superposed with the bacterial homolog structures, BtGH8412, CpNGA13 and Oceanicola granulosus OGA (OgOGA)20 there is structural conservation of the catalytic domain and in particular the “−1” sugar binding subsite that we define as the active site pocket (Supplementary Fig. 7). The majority of active site residues comprising this subsite are conserved and mutations of these residues in these enzymes BtGH8412,13,20 and in hOGA21, coupled with detailed mechanistic studies, 14,21,22 provide clear support for a catalytic mechanism involving substrate-assisted catalysis from the substrate acetamido group. The helical regions that contribute to the putative peptide-binding cleft (see below) are, however, markedly different; both in organization and residue identity (Supplementary Fig. 7-9). The most similar helical domain is seen for OgOGA20, though that model was assigned a monomeric organization lacking the helix exchange observed in hOGA.
The active site pocket of hOGA is located at the base of a V-shaped cleft (~22 x 25 Å with an angle of ~70°) formed between the catalytic domain of monomer 1 and the C-terminal helical bundle of the other monomer of the dimer (Fig. 1c, Supplementary Fig. 10). The unusual dimer topology thus plays an unforeseen functional –as well as structural – role by contributing to formation of this groove, which likely binds the peptide component of substrates. The helical bundle contributes a rigid structure (Supplementary Fig. 10), but also a flexible loop connecting the swapped helix, which can adopt different conformations as discussed below. This mobility may confer plasticity to this groove, perhaps allowing it to accommodate different substrates and inhibitors. Notably, our analysis revealed unexpected density in this groove (Fig. 1d, Supplementary Fig. 11a) that we assigned as the C-terminal end (P707-Y715) of the helical bundle with Y715 adopting a position that is consistent with the position of an O-GlcNAcylated serine/threonine residue (Supplementary Fig. 12). Notably, this crystal-packing derived peptide runs in the opposite direction to that observed in bacterial structures20 and thus is only indicative of the peptide binding surface; further study of peptide complexes will be needed to clarify binding of protein substrates.
Key to our analysis of hOGA is how it binds inhibitors. Extensive soaking of diverse chemical inhibitors was performed to displace the C-terminal peptide. Structures of complexes with the mechanism-derived inhibitor Thiamet-G (Ki=0.9 nM),23 the “PUGNAc-imidazole” hybrid (Ki=3.9 μM)24 and a potent derivative (VV347) of recently described pyrrolidine inhibitors (Ki=8 nM),25 were determined (Fig. 2, Supplementary Fig. 11b, 13, Supplementary Note). The sugar-like moieties of these inhibitors occupy the −1 GlcNAc binding site, making hydrogen bonds with, for example G67, K98, N280, D285 and N313. The alkyl amino-group of Thiamet-G fills the pocket, formed by C215, Y219 and W278. In all three cases the catalytic residues D174 and D175 are engaged with the inhibitors, representing a “closed” conformation. D174 interacts with both nitrogens of the N-aminothiazoline moiety as expected for this transition state mimic. The acetamido groups of VV347, and PUGNAc-imidazole, point into the same pocket with the acetamido nitrogen interacting with D174 (Fig. 2, Supplementary Fig. 14), consistent with its role as polarizing residue. D175 points towards the anomeric carbon as expected for its role as general acid/base in the catalytic cycle.22 Interactions of the sugar-like moiety of these inhibitors within the −1 sugar binding subsite are conserved between hOGA and their bacterial counterparts, explaining the success of inhibitors targeting this site. Not conserved, however, are residues outside the -1 sugar binding subsite. These residues participate in the recognition of the aglycon component of inhibitors that project out of the active site pocket and can be observed here for the first time (Fig. 2, Supplementary Fig. 11b).
Figure 2. Ligand binding to hOGA.
a) Binding of Thiamet-G (left), the PUGNAc-imidazole” hybrid (middle) and the pyrrolidine derivative “VV347” in the active site of hOGA. The corresponding electron density is shown at 2.0 σ r.m.s.d. (0.28 e/Å3) for Thiamet-G. For the “PUGNAc-imidazole” hybrid (middle) at 1.5 σ r.m.s.d. (0.17 e/Å3), and the pyrrolidine derivative “VV347” at 1.5 σ r.m.s.d. (0.21 e/Å3). The catalytic residues as well the acetamido pocket forming residues are shown as sticks. c) Van der Waals' surface of the inhibitor binding sites showing that the -1-subsite is deeply buried with the aglycon units extending outwards interacting with both domains of hOGA.
The aglycon of the glucoimidazole inhibitor mainly interacts with residues of the catalytic domain that are outside the -1 binding site pocket, notably F223 and V254. The phenyl group is within 4.5 Å of the loop comprising residues 677-683 of the helical bundle of monomer 2 (680-loop) suggesting a direct contribution of the helical bundle domain to the binding of the inhibitor and potential substrates. The complex with VV347 (Fig. 2) reveals interactions with multiple residues of the helical bundle domain. The helix comprising residues 633-662 and the 680-loop, are core structural elements of the peptide-binding groove. The trifluoromethyl-phenyl group of VV347 binds in a pocket formed by the side chain of W645, part of helix α4 and W679, part of the 680-loop, which undergoes a major reorientation (Supplementary Fig. 15). Previous mutagenesis studies suggest the 680-loop interacts with protein substrates20 though further study will illuminate the precise roles of this feature.
In summary, we report a functional construct of human OGA obtained by exploiting the ability of the N- and C-terminal fragments of hOGA to associate stably. The structure revealed an unusual obligate dimer with intertwined helical bundle domains that leads to residues from both domains contributing to formation of the substrate-binding site. Structures of OGA in complex with rationally-designed high affinity inhibitors define both the active site pocket and, crucially, the surrounding peptide-binding and aglycon regions in a manner that is unique to mammalian OGA. Strikingly, part of this peptide binding site is a flexible loop connecting the swapped helix of the helical bundle domain with the opposite peptide binding groove, which may open-up possible communication between active sites in response to peptide binding. Exploitation of this peptide binding groove will offer new opportunities for the design of OGA inhibitors as research tools and for potential clinical use in treating O-GlcNAc related diseases.
Online Methods
Cloning, expression and purification
The gene of the longest isoform of hOGA (hOGA-L (Uniprot Accession number: O60502)) was synthesized in a codon optimized form for recombinant expression in Escherichia coli. The successful expression construct encoded the N-terminal region of hOGA, comprising amino acids 11-396, and the C-terminal region, comprising amino acids 535-715. The N-terminal construct was cloned in the vector pACYC-Duet (Millipore) using the sequence and ligation independent cloning method 26 in frame with an N-terminal His6-Tag. The C-terminal construct was cloned into the vector pET-YSBLIC3C 27 with an N-terminal His6-Tag followed by a 3C-protease cleavage site using the same method. The nucleotide sequences of all made constructs were confirmed by sequencing. Both vectors were simultaneously transformed into E. coli Bl21(DE3)-Gold (Agilent) for subsequent protein expression. Cells were grown in 2L TB-medium to an OD600 of ~1.0 and protein synthesis was then induced by adding IPTG to a final concentration of 0.1 mM. Protein expression was carried out at 16 °C with an induction time of 20 h. The cells were harvested by centrifugation at 4500xg for 20 min, flash frozen and stored at -20°C until required.
For purification of the hOGA complex, cells were resuspended in 50 mM HEPES pH 7.0, 750 mM NaCl, 20 mM imidazole, and 0.5 mM DTT (resuspension buffer). Cells were lysed using a French Press at 25 kPsi. The lysate was cleared by centrifugation at 50,000 g for 1h and the supernatant was passed through a 10 mL HisTrap FF column (GE Healthcare) pre-equilibrated with resuspension buffer. The bound hOGA was purified by gradient elution over 10 column volumes using 0 to 50% of elution buffer (50 mM HEPES pH 7.0, 750 mM NaCl, 500 mM imidazole and 0.5 mM DTT). hOGA containing fractions were combined, concentrated by ultrafiltration using Vivaspin columns (Sartorius) with a molecular weight cut-off (MWCO) of 30 kDa, and applied to a Superdex S200 column (GE Healthcare) pre-equilibrated with size-exclusion buffer (10 mM HEPES pH 7.0, 250 mM NaCl, 1 mM DTT). Fractions corresponding to the dimeric form of hOGA were combined and concentrated to 20 mg/ml by ultrafiltration with a Vivaspin (MWCO: 30 kDa) column, flash frozen using liquid nitrogen, and stored at -80°C until required.
Crystallization and data collection
Initial crystallization conditions were identified using commercially available screens from Hampton and Molecular Dimension in a 96 well sitting drop screening format. Further optimization in a 48 well sitting drop format provided suitable conditions for reliable crystallization (crystallization solution: 0.1-0.2 M (NH4)3-citrate pH 6.5-7.5; 16-24 % PEG 3350). Optimal crystals were reliably obtained by micro seeding with previously obtained crystals. For data collection, protein crystals were transferred into crystallization solution containing 25% PEG3350 (cryoprotectant solution), which enabled cryoprotection of the crystals. Crystals were recovered using a Nylon microfibre loop (Hampton) and flash frozen in liquid nitrogen. For soaking experiments the inhibitors were dissolved in 10 % (v/v) DMSO to a concentration of 100 mM and added to a drop containing the cryoprotectant solution to a final inhibitor concentration of 10 mM. Crystals were soaked with inhibitors for times ranging from 48 hours to 1 week. The resulting crystals were handled as described above. Data were collected at the Diamond light source beamlines I02, I03 or I04 using a Pilatus 6M detector (Dectris) at a wavelength of 0.979 Å. Data were collected over 180° with an oscillation angle of 0.1°. Data were integrated with XDS 28, integrated in the XIA2 pipeline 29 and scaled using AIMLESS 30.
Structure solution and refinement
The structure was solved by molecular replacement using Phaser31 in conjunction with a sculpted model of BtGH84 (PDB-ID 2CHO)12. The initial model was rebuilt and refined using Buccaneer 32. The final model was obtained by alternating rounds of manual model building in COOT33, followed by reciprocal refinement with Refmac34 or Phenix35. For inhibitor complexes the apo-structure was refined against data from a crystal soaked with the respective inhibitor. If clear density for a bound inhibitor could be identified a model of the ligand was built using Acedrg, part of the CCP4- software package36 and incorporated in the apo model which was then subsequently refined. The quality of the final models were judged using MolProbity 37. The number of outliers is between 0.2 to 0.4 %. Figures of the structural models were prepared using CCP4MG 38.
Enzyme kinetics
Initial rate experiments of OGA-L, OGA-Split 1, and OGA-Split 2 catalyzed pNP-GlcNAc hydrolysis were carried out in PBS buffer (pH 7.4) and monitored continuously at 25°C at a wavelength of 405 nm using a SpectraMax i3x multi-mode plate reader from Molecular Devices. Reactions were performed in a 384 well clear assay plate from Corning (Product #3702) in a final reaction volume of 45 µl. Steady state kinetic values were attained from substrate dose response curves using 50 nM of the OGA-L, OGA-Split 1, and OGA-Split 2 variants unless otherwise stated and varying concentrations of pNP-GlcNAc. Reaction velocities were determined by linear regression of the progress curves over a 15 min period. The amount of product formed was assessed by creating a pNP standard curve in PBS buffer. The substrate dose-response curves were then fit to the Michaelis-Menten equation using the GraphPad Prism5 software package.
Inhibition assays were performed using a final concentration of 20 nM OGA-L and 200 μM of 4-methylumbelliferyl-2-acetamido-2-deoxy-β-D-glucopyranoside with various concentrations of VV-347 and Thiamet-G. The assay was performed at 37°C in a final volume of 45µl in inhibition buffer: PBS pH7.4, 0.0025% BSA, 0.0025mM DTT. Reactions were initiated with the addition of enzyme, and reaction progress was monitored continuously (excitation and emission wavelengths: 350, and 445nm) over a 10-minute period. The amount of fluorophore liberated was assessed using a standard curve for 4-methylumbelliferone in inhibition buffer. Inhibitor Ki values were determined using the Morrison equation for tight binding inhibition as described previously 39. All curve fitting for enzyme kinetics and inhibition experiments was performed using GraphPad Prism, and error bars correspond to S.D. from three technical replicates (triplicate reads). The experiments were all repeated at least twice to ensure reproducibility of the data.
SEC-MALS
Experiments were conducted on a system comprising a Wyatt HELEOS-II multi-angle light scattering detector and a Wyatt rEX refractive index detector linked to a Shimadzu HPLC system (SPD-20A UV detector, LC20-AD isocratic pump system, DGU-20A3 degasser and SIL-20A autosampler). Work was conducted at room temperature (20 ±2°C). Sample injection volume was 100 µL at a protein concentration of 5 mg/ml. The samples were separated on a Superdex S200 10/300 (GE Healthcare) using 10 mM Tris pH 7, 250 mM NaCl as buffer. Shimadzu LC Solutions software was used to control the HPLC and Astra V software for the HELEOS-II and rEX detectors. Data were analyzed using the Astra V software. MWs were estimated using the Zimm fit method with degree 1. A value of 0.174 was used for protein refractive index increment (dn/dc).
Digestion of HEK293 cell extracts with OGA constructs
HEK293 cells were obtained from ATCC. They were not further authenticated. However, as we are looking at total GlcNAc levels in cell extracts and the digestion of O-GlcNAc by OGA, rather than cell-type specific physiology, the exact cell type is less relevant in our experiments. The cells were tested for mycoplasma contamination in June 2016 (negative result). HEK293 cells were cultured in high glucose (4.5 g/L) Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% (v/v) Fetal Bovine Serum (FBS), 100 units/mL Penicillin and 100 µg/mL Streptomycin in a 5% CO2 humidified incubator at 37 °C. Cells were seeded into a 100 mm cell culture dish and grown to 95% confluency. Cells were washed with ice-cold PBS, harvested by scraping on ice in ice-cold PBS, pooled and centrifuged at 700 g for 10 min at 4 °C. The cell pellet was resuspended in 150 µL of lysis buffer (50 mM NaH2PO4, pH 7.0, 100 mM NaCl, 1% (v/v) NP-40 substitute, 0.5% (w/v) sodium deoxycholate, and 1 mM PMSF) and incubated on ice for 30 min. The cell extracts were then centrifuged at 14,000 g for 10 min at 4 °C and the clear supernatant was collected. The protein concentration, determined by DC™ protein assay (Bio-Rad), was 29 mg/mL. HEK293 cell lysates (5 µL, 150 µg) were mixed with 5 or 25 µM of OGA-Split 1, OGA-L or BtGH84 (5.5 µL 2x solution in 50 mM NaH2PO4, 100 mM NaCl, pH 7.0) in the presence or absence of 250 µM Thiamet-G (0.6 µL 5 mM in PBS). The reactions were incubated at 25 °C for 3 hrs. Under these conditions, enzyme activity, as determined by pNP-GlcNAc hydrolysis, was shown to be stable (see Supplementary Figure 13). After the reaction, 4 µL of binding buffer (500 mM NaH2PO4, 100 mM NaCl, pH 8.0) was added and the mixtures were added to 30 µL of Nickel-NTA Agarose beads (prewashed in binding buffer). The samples were incubated for 1 hr at rt with rotation to remove the His-tagged enzyme. The beads were spun down at 5,000 g for 2 min and the supernatant was collected (15 µL), mixed with 2x Laemmli’s sample buffer containing β-mercaptoethanol and boiled for 5 min at 100 °C. A third (~ 50 µg of protein) of each sample was resolved on a 4-20% Mini-PROTEAN TGX gradient gel (Bio-Rad). The proteins were transferred onto a nitrocellulose membrane using a Bio-Rad wet western blotting system (1 hr at 100V). The membrane was blocked with 2% BSA in PBSt for 1 hr at rt and hybridized with mouse CTD110.6 antibody (BioLegend, 1:3,000) and rabbit anti-β-actin antibody (LI-COR, 1:5,000) in PBS with 2% BSA overnight at 4 °C, followed by IRDye 680LT goat anti-mouse antibody (LI-COR, 1:10,000) and IRDye 800CW goat anti-rabbit antibody (LI-COR, 1:10,000) in PBSt with 2% BSA for 1 hr at rt. Proteins were visualized using a LI-COR Odyssey scanner. The protein ladder used was a PageRuler Prestained Plus Protein Ladder (Thermo Fisher Scientific).
Stability of OGA-Split 1 under reaction conditions
The various OGA constructs were dissolved at 25 µM in 50 mM NaH2PO4, 100 mM NaCl, pH 7.0. The mixtures were incubated at 25 °C for 3 hrs. Before (t = 0 hrs) and after incubation (t = 3 hrs), 2 aliquots of 0.5 µL were taken out and each added to 125 µL of PBS. The diluted enzymes (100 nM) were then mixed 1:1 with of 400 µM 4-Nitrophenyl N-acetyl-D-glucosaminide (pNP-GlcNAc) in PBS and two 45 µL aliquots (for duplicate reads) were transferred to a 384 well Corning clear-bottom plate. pNP-GlcNAc hydrolysis was monitored continuously at a wavelength of 405 nm at 25°C using a SpectraMax i3x multi-mode plate reader (Molecular Devices). Background hydrolysis (the average value in samples without enzyme at the corresponding time point) was substracted from all values, after which reaction velocities were determined by linear regression of the progress curves. These values were then normalized to 100% activity (the average value in samples with the corresponding OGA construct at 0 hrs incubation). Data were analyzed using GraphPad Prism5 software and represent mean values ± standard deviation from two biological replicates with two technical replicates (duplicate reads) each (Supplementary Fig. 16).
Treatment of O-GlcNAcylated TAB1 with OGA constructs
TAB1 was coexpressed with OGT in E.coli and purified as previously described.17 O-GlcNAcylated and HIS-tagged TAB1 (5 µg per sample, 2.5 µL 2 mg/mL in 50 mM NaH2PO4, 100 mM NaCl, pH 7.0) was treated with 5 or 25 µM of OGA-Split 1, OGA-L or BtGH84 (3 µL 2x solution in 50 mM NaH2PO4, 100 mM NaCl, pH 7.0) in the presence or absence of 250 µM Thiamet-G (0.5 µL 3 mM in PBS) at 25 °C for 3 hrs. The reactions were quenched by addition of 4 µL of 5x Laemmli’s sample buffer containing β-mercaptoethanol and boiled for 5 min at 100 °C. A fifth (~ 1 µg of TAB1) of each sample was resolved on a 12% SDS-PAGE gel and analyzed by western blot as described above. Instead of rabbit anti-β-actin antibody, a rabbit anti-HIS antibody was used as loading control (Cedarlane, 1:5,000).
Supplementary Material
Acknowledgements
The authors thank Diamond Light Source for beamtime (proposals mx-1221, mx-7864 and mx-9948), and the staff of beamlines I02, I03 and I24 for assistance. The authors are grateful to Dr. Johan P Turkenburg and Sam Hart for their help in crystal testing and data collection. The authors thank Juliet Borgia, Simon Grist, Andrew Leech and Louise Haigh for technical support. This research was supported by funding from the Biotechnology and Biological Sciences Research Council (BB/K003836/1) and the Canadian Institutes of Health Research (MOP-123341), Brain Canada, Genome British Columbia, the Michael Smith Foundation for Health Research. LIW is supported by the Netherlands Organization for Scientific Research (NWO) and the Banting Postdoctoral Fellowships program are also thanked for financial support. DJV is supported as a Tier I Canada Research Chair in Chemical Glycobiology. GJD is supported by the Royal Society through a Ken Murray research professorship.
Footnotes
Accession codes:
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under the accession codes 5M7R for the apo-structure, 5M7S for the Thiamet-G complex, 5M7T for the PugNAc-imidazole complex and 5M7U for the complex with VV347.
Author contributions:
C.R. designed truncated constructs, cloned, expressed, crystallized and solved the structure. S. C. designed truncated constructs, cloned, expressed, purified and crystallized protein. W.A.O. cloned purified and crystallized protein. G.R.H. designed experiments, cloned and purified protein. L.I.W. performed cell culture and western blot assays. D.T.K. performed the kinetic characterization. V.V. synthesized VV347. R.B. and D.J.V designed the pyrrolidine inhibitors. G.J.D designed cloning and structural experiments. D.J.V designed biochemical and inhibition experiments. C.R., D.J.V. and G.J.D wrote the manuscript with contributions from all authors.
Data Availability:
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under the accession codes 5M7R for the apo-structure, 5M7S for the Thiamet-G complex, 5M7T for the PugNAc-imidazole complex and 5M7U for the complex with VV347. Any other datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Financial Interests
D.J.V. is a co-founder of and holds equity in the company Alectos Therapeutics. D.J.V. serves as CSO and Chair of the Scientific Advisory Board of Alectos Therapeutics of which G.J.D is a member.
References
- 1.Hardiville S, Hart GW. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 2014;20:208–13. doi: 10.1016/j.cmet.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groves JA, Lee A, Yildirir G, Zachara NE. Dynamic O-GlcNAcylation and its roles in the cellular stress response and homeostasis. Cell Stress Chaperones. 2013;18:535–58. doi: 10.1007/s12192-013-0426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrer CM, et al. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol Cell. 2014;54:820–31. doi: 10.1016/j.molcel.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagerlof O, et al. The nutrient sensor OGT in PVN neurons regulates feeding. Science. 2016;351:1293–6. doi: 10.1126/science.aad5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu F, et al. Reduced O-GlcNAcylation links lower brain glucose metabolism and tau pathology in Alzheimer's disease. Brain. 2009;132:1820–1832. doi: 10.1093/brain/awp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuzwa SA, et al. Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat Chem Biol. 2012;8:393–399. doi: 10.1038/nchembio.797. [DOI] [PubMed] [Google Scholar]
- 7.Smith SM, et al. Eearly Clinical Results and Preclinical Validation of the O-GlcNAcase (OGA) Inhibitor MK-8719 as a novel Therapeutic for the Treatment of Tauopathies. Alzheimer's & Dementia: The Journal of the Alzheimer's Association. 2016;12:P261. [Google Scholar]
- 8.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 2011;469:564–567. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, et al. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 2004;11:1001–1007. doi: 10.1038/nsmb833. [DOI] [PubMed] [Google Scholar]
- 10.Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 2001;276:9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- 11.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–8. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis RJ, et al. Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat Struct Mol Biol. 2006;13:365–371. doi: 10.1038/nsmb1079. [DOI] [PubMed] [Google Scholar]
- 13.Rao FV, et al. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 2006;25:1569–78. doi: 10.1038/sj.emboj.7601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macauley MS, Whitworth GE, Debowski AW, Chin D, Vocadlo DJ. O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem. 2005;280:25313–25322. doi: 10.1074/jbc.M413819200. [DOI] [PubMed] [Google Scholar]
- 15.Vocadlo DJ. O-GlcNAc processing enzymes: catalytic mechanisms, substrate specificity, and enzyme regulation. Curr Opin Chem Biol. 2013;16:488–97. doi: 10.1016/j.cbpa.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 16.Butkinaree C, et al. Characterization of beta-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis. J Biol Chem. 2008;283:23557–66. doi: 10.1074/jbc.M804116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krissinel E. Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 2015;43:W314–9. doi: 10.1093/nar/gkv314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells L, et al. Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic beta-N-acetylglucosaminidase, O-GlcNAcase. J Biol Chem. 2002;277:1755–1761. doi: 10.1074/jbc.m109656200. [DOI] [PubMed] [Google Scholar]
- 19.Izumi T, Suzuki K. Neutral beta-N-acetylhexosaminidases of rat brain. Purification and enzymatic and immunological characterization. J Biol Chem. 1983;258:6991–9. [PubMed] [Google Scholar]
- 20.Schimpl M, Schuttelkopf AW, Borodkin VS, van Aalten DM. Human OGA binds substrates in a conserved peptide recognition groove. Biochem J. 2010;432:1–7. doi: 10.1042/BJ20101338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cetinbas N, Macauley MS, Stubbs KA, Drapala R, Vocadlo DJ. Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants. Biochemistry. 2006;45:3835–3844. doi: 10.1021/bi052370b. [DOI] [PubMed] [Google Scholar]
- 22.He Y, Macauley MS, Stubbs KA, Vocadlo DJ, Davies GJ. Visualizing the reaction coordinate of an O-GlcNAc hydrolase. J Am Chem Soc. 2010;132:1807–1809. doi: 10.1021/ja9086769. [DOI] [PubMed] [Google Scholar]
- 23.Cekic N, et al. Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by human O-GlcNAcase. Chemical Science. 2016;7:3742–3750. doi: 10.1039/c6sc00370b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shanmugasundaram B, et al. Inhibition of O-GlcNAcase by a gluco-configured nagstatin and a PUGNAc-imidazole hybrid inhibitor. Chem Commun (Camb) 2006:4372–4. doi: 10.1039/b612154c. [DOI] [PubMed] [Google Scholar]
- 25.Bergeron-Brlek M, et al. A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable OGA Inhibitors. Angew Chem Int Ed Engl. 2015;54:15429–33. doi: 10.1002/anie.201507985. [DOI] [PubMed] [Google Scholar]
- 26.Li MZ, Elledge SJ. SLIC: a method for sequence- and ligation-independent cloning. Methods Mol Biol. 2012;852:51–9. doi: 10.1007/978-1-61779-564-0_5. [DOI] [PubMed] [Google Scholar]
- 27.Fogg MJ, Wilkinson AJ. Higher-throughput approaches to crystallization and crystal structure determination. Biochem Soc Trans. 2008;36:771–5. doi: 10.1042/BST0360771. [DOI] [PubMed] [Google Scholar]
- 28.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J Appl Crystallogr. 2010;43:186–190. [Google Scholar]
- 30.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–11. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cekic N, et al. Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by human O-GlcNAcase. Chem Sci. 2016;7:3742–3750. doi: 10.1039/c6sc00370b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.