Abstract
Small molecule drugs may complement antibody-based therapies in an immune-oncology setting, yet systematic methods for the identification and characterization of the immunomodulatory properties of these entities are lacking. We surveyed the immumomodulatory potential of 1,402 small chemical molecules as defined by their ability to alter the cell-cell interactions among peripheral mononuclear leukocytes ex vivo, using automated microscopy and population-wide single-cell image analysis. Surprisingly, some 10% of the agents tested affected these cell-cell interactions differentially. The results accurately recapitulated known immunomodulatory drug classes, and revealed several clinically approved drugs that unexpectedly harbor the ability to modulate the immune system, potentially contributing to their physiological mechanism of action. For instance, the kinase inhibitor crizotinib promoted T-cell interactions with monocytes as well as with cancer cells, through inhibition of MST1R/RON (macrophage-stimulating protein receptor) and subsequent upregulation of MHC (major histocompatibility complex) expression. The approach offers an attractive platform for the personalized identification and characterization of immunomodulatory therapeutics.
Introduction
High-content screening using automated fluorescent microscopy has allowed us to objectivize and quantify a great variety of cellular parameters, providing an attractive platform to perform genetic or chemical perturbation screens at unprecedented accuracy and robustness due to the statistical power derived from the large number of events monitored. In particular, the approach has enabled the visualization of the influence of cell-cell and cell-microenvironment interactions, which contribute to population-level phenotypes 1. It has further facilitated the high-throughput screening of co-culture systems, identifying phenotypes of the individual subpopulations, and resolving the interplay between cell populations that may contribute to an integrated drug response.
Automated microscopy has thus far not been adapted for the screening of non-adherent and adherent leukocytes present in blood as a model to study immunology, even as traditional microscopy has proven crucial to understanding the complex interaction dynamics of the immune system 2. As other high-throughput screening technologies have been used to measure the impact of indirect factors on immune response propagation, such as signaling proteins and soluble cytokines, automated microscopy can be expected to provide the unique spatial resolution and throughput required to quantitatively screen the modulation of direct signaling by cell-cell contacts (receptor mediated signaling) that drives an immune response. Such information on cell-cell contacts in blood over large drug libraries is of particular interest as many successful drugs affect properties of cells that are not cell autonomous, but rather rely on the modification of the relationship between cells, best illustrated by the recent success of cancer immunotherapy 3.
Modulation of both the innate and adaptive immune systems is a highly successful strategy in the treatment of systemic diseases such as inflammatory disorders and cancer, where innovation of targeted biological or chemical agents are at the forefront. For instance, the immunomodulatory drug pomalidomide, which induces cytotoxic T-cell and NK-cell activity, and immune check point inhibitors such as the monoclonal antibody ipilimumab (anti-CTLA4) 4, alter the balance of co-stimulatory and co-inhibitory signals that manage self-tolerance and regulate T-cell responses 5. In both of these examples, the drug mediates cellular interactions that result in the death of the target cancer cell, a concept underlying many immunomodulatory drugs that have proven successful in treating a variety of malignant diseases 4.
Here, we describe a high-throughput image-based screening method, and analysis algorithm, which robustly quantifies the immunomodulatory potential of small molecules and other therapeutics, by measuring the changes in the physical interaction of leukocytes. With this method, phenotypic drug screening can be expanded to help realize entities that harness the inherent ability of effector cells, within peripheral blood, to propagate signals and function through direct physical contact - a major goal of immunotherapy.
Results
Systematic quantification of leukocyte cell-cell contacts
We designed a pipeline to directly assay the effects of biologicals and chemical agents for their immunomodulatory properties, through measuring changes in cell-cell contacts of peripheral blood mononuclear cells (PBMCs) ex vivo, by population-wide single cell microscopy of PBMC monolayers (Supplementary Results, Supplementary Fig. 1a, and online methods). Individual cell types were identified using fluorescently tagged antibodies, in various non-overlapping combinations, against extracellular markers unique to the PBMC subpopulations of interest. This revealed extensive cell-cell interactions between the different subpopulations (see close-up in Fig. 1a). As the method requires little material per-test, all assays are performed in the blood of an individual donor, reducing background inflammation due to HLA-mismatching. To assess whether the observed interactions among cells were functionally meaningful and to benchmark the assay, we made use of four biologicals that are known to decrease or increase selected cell-cell contacts: an MHC class II blocking antibody, two clinically used antibodies, rituximab and blinatumomab (Supplementary Fig. 1b), and lipopolysaccharide (LPS).
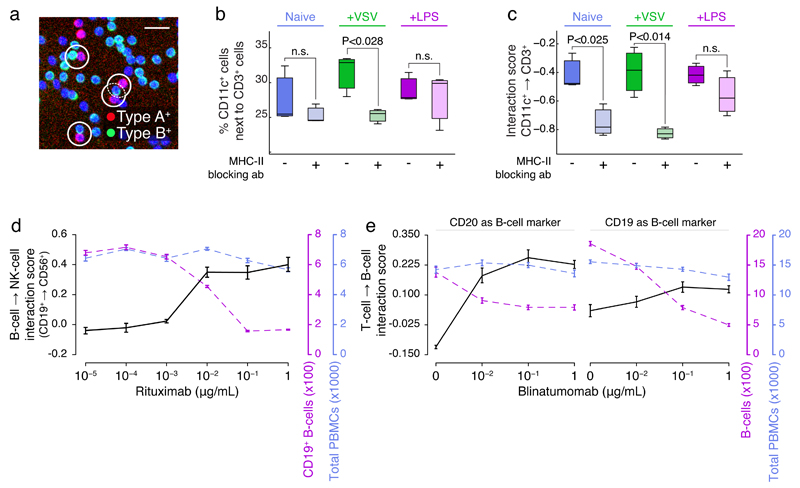
Fig 1. Quantifying PBMC cell-cell interactions perturbed by biologicals.
(a) Example 10x image of cell-cell PBMC subpopulation contacts, with selected contacts highlighted (white circles, scale bar is 25μm). (b) Percent of CD11c+ cells in contact with CD3+ cells, when naive or after stimulation with VSV or LPS, with or without pre-incubation with MHC-II blocking antibody. (c) CD11c+→CD3+ interaction scores corresponding to (d). Interaction score is calculated as the observed percentage of A cells in relation to B cells log2-relative to what is expected if data were randomized. (d) The interaction score of CD19+ B-cells→CD56+ NK cells (black axis; left), CD19+ B-cell counts (purple axis; right), or total PBMC counts (blue axis; far right) as function of increasing rituximab concentration. (e) Interaction scores of (left plot) CD3+ T-cells→CD19+ B-cells or (right plot) CD3+ T-cells→CD20+ B-cells (black axis; left), B-cell counts (purple axis; right), or total PBMC counts (blue axis; far right) as function of increasing blinatumomab concentration. (b-c) were performed in triplicate, and representative of three independent experiments; (d-e) were performed in at least 5 technical replicates, and are representative of (d) 5, or (e) 2 repeats over various healthy donors. Average and standard error of means, or boxplots, over technical repeats shown. A t-test was used to determine significance in (b-c).
The interaction between T-cells and professional antigen presenting cells (APCs), including dendritic cells and macrophages, is an essential step in triggering an adaptive immune response. APCs present foreign antigens on MHC-II receptors to T-cells, which, upon recognition by the T-cell receptors (CD3; TCR), can lead to a targeted immune response 6. Antibodies recognizing the extracellular portion of the MHC-II receptor are known to efficiently obstruct this interaction (Supplementary Fig. 1b, left). When cells were stimulated with vesicular stomatitis virus (VSV), the percentage of CD11c+ cells directly contacting CD3+ T-cells was significantly reduced by incubation with an MHC-II blocking antibody prior to infection, on average from 33% to 25% (P-value < 0.028; Fig. 1b), as measured over a total of 124,059 cell-cell contacts. Such interaction frequencies are however dependent on several variables that directly influence the outcome. In this example they include: the fraction of all cells that are CD11c-positive (xa), the fraction of all cells that are CD3-positive (xb), and the total cell density or overall clustering index, which can be expressed as the fraction of all PBMCs that directly contact one or more PBMCs (xi). Indeed, all three variables showed fluctuations in the MHC control experiment, among others due to stimuli-dependent differences in cellular activation (Supplementary Fig. 1c) 7, which need to be statistically accounted for when interpreting such interaction frequencies. The fraction of cells that are of type A and that interact with cells of type B, if cellular positions and relative abundance of subpopulations were kept equal but cellular identities were shuffled, is given by the formula E = xa · xb · xi . Where xa is the fraction of cells of type A, xb is the fraction of cells of type B, and xi is the fraction of cells with one or more cell contacts. Bootstrap analysis confirmed the equation, consistent with the fact the three variables act as independent probabilities in this context (Supplementary Fig. 1d). Scoring alterations in the interaction frequency relative to E then gives an internally normalized interaction frequency, which we term the ‘interaction score’. Further information on the interaction score can be found in the online methods. The interaction score indicates how much the observed interaction frequency deviates from what would be expected by random, which makes it robust to alterations in the relative abundance of either subpopulation as well as to alterations in overall cell density or cell-cell contacts. We use arrows to indicate the directionality of the interaction score, i.e. relating to the fraction of A cells interacting with B cells, which can deviate from the opposite direction in case of strongly uneven subpopulation sizes or in the case of many-to-one cell-cell contacts. Corrected for these influences, the blocking MHC-II antibody was found to not only reduce the CD11c+→CD3+ T-cell interactions in VSV-stimulated condition, but also in the unstimulated state (Fig. 1c and Supplementary Fig. 1e), likely explained by reduced antigen ‘scanning’ by T-cells 8,9. As expected, both an isotype IgG control antibody and a blocking antibody against CD54, which functions as co-stimulatory signal and is typically not highly expressed on unstimulated monocytes 10, did not significantly alter the CD11c+→CD3+ T-cell interaction score in unstimulated condition (Supplementary Fig. 1f). Further, contact-dependent immune activity has been described as early as 1970, where clustering of CD14+ monocytes stimulated by bacterial lipopolysaccharides (LPS) is an activation-associated signal 11. Accordingly, the interaction score revealed a significant increase in the interaction between CD14+ monocytes resulting from LPS treatment (Supplementary Fig. 1g). In these examples, immune activation and modulation can strongly drive cell proliferation, potentially effecting the number of cells within interacting subpopulations being measured. To additionally confirm that the interaction score is robust to either gain or loss of cellular subpopulations, we simulated fluctuations in the proportion of A and B cells over a wide range (5% to 95%), and then measured the interaction score of A↔B; synthetic data provides a controlled environment without compounding factors such as background inflammation. As expected, we found that the score is inherently robust to modifications in cell number (Supplementary Fig. 2a), enabling us to measure altered cell-cell contacts even in the context of cell proliferation and cell death.
The anti-cancer biologicals rituximab and blinatumomab induce NK-cell-to-B-cell (Supplementary Fig. 1b, middle) and T-cell-to-B-cell (Supplementary Fig. 1b, right) mediated killing, respectively, where the function of the killing depends on direct physical contacts between effector and target cells 12–14. Incubation with these biologicals resulted in not only a dose-dependent increase in respective interaction scores, but also in a concomitant loss of target cells (Fig. 1d-e & Supplementary Fig. 2b). In the case of blinatumomab, the results were independent of the marker used to identify the target B-cell (Fig. 1e). Even with the reduction of the target B-cell population, the interaction score is still increased, due to the score’s normalization to variations in subpopulation numbers (Fig 1d-e), as described above. Thus, the recapitulation of the effects of biologicals with well-defined mechanisms of action, measured by population-wide imaged based screening and spatial analyses, validates this method for use in an immunomodulatory screening campaign.
While biologicals have great precision and efficacy in their mode of action, it would be advantageous to identify small chemical entities with specific immunomodulatory properties that could be more easily handled and manufactured. To determine what classes of drugs, if any, have unknown immunomodulatory effects, we surveyed a collection of 1,402 existing drugs (approved, investigational, and experimental, Supplementary Dataset 1) to see if one can benchmark effects against drugs with known immunomodulatory properties, such as steroids and NSAIDs.
Chemical rewiring of the leukocytic interaction network
Leukocyte interactions across the 1,402 compounds were screened in quadruplicate, resulting in the identification and analysis of the cell-to-cell contacts of over 80 million PBMCs from a single healthy donor blood donation in 7,680 wells. To induce a higher level of cell-cell contacts, alterations of PBMC cell-cell interactions were measured after immune stimulation with vesicular stomatitis virus (VSV), which induces an interferon (IFN)-based inflammatory immune response that is aimed at halting viral replication 15. Pairwise combinations of four major PBMC subpopulations were stained for by immunofluorescence after infection. At the population-level, VSV infection as measured by a single cell GFP-reporter displayed high reproducibility (Fig. 2a and Supplementary Fig. 3a), where VSV preferentially infected myeloid lineage cells (Supplementary Fig. 3b), as expected 7. 80 Compounds decreased VSV infection while 22 increased it (below -2 or above 2 standard deviations, with P<0.05; Fig. 2b and Supplementary Fig. 3c). Importantly, several known anti-inflammatory compounds led to increased VSV infection, including corticosteroids (Fig. 2b and Supplementary Fig. 3d), which may indicate a functional blockage in cellular signaling pathways that rely on soluble factors, and further, that these pathways are active in our model 16.
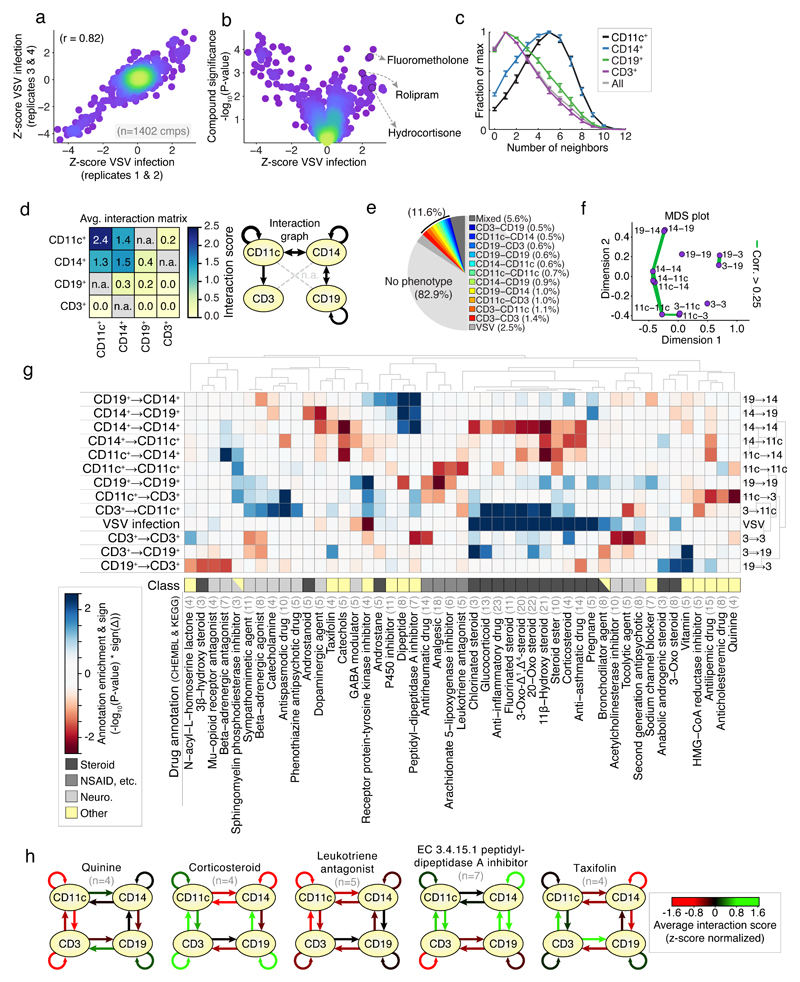
Fig 2. Screening for chemical modifiers of PBMC cell-cell contacts.
(a) reproducibility of VSV-infection over four replicates (average of two replicates per axis); dots correspond to compound; color indicates data density. (b) Average change in VSV infection per compound (z-score normalized) against the significance per compound (-log10(P-value)). Selected anti-inflammatory compounds shown. (c) Distributions of the number of direct contacts per cell type normalized to maximum; values aggregated over screen. (d) Left: Interaction scores of each pairwise combination averaged over screen; grey / n.a. indicates not measured; Right: visualization of the average interaction scores as interaction graph. (e) Percentage of compounds with unique or mixed phenotypes at 2-sigma significance. (f) MDS plot of the similarity between results over each measured interaction. Green lines connect interactions whose screening results are ≥ 0.25. (g) Hierarchical clustering of the enrichment (-log10(P-value) * sign of the phenotype) for selected top-enriched drug classes over all interactions measured, including VSV infection phenotype. Blue and red boxes indicate increased or decreased spatial phenotypes. Manual drug annotation class shown below. Light grey numbers indicate number of compounds. (h) Interaction graphs of average phenotypes for selected annotations from (g); red and green arrows indicate decreased or increased average interaction scores respectively (z-score normalized); black arrows indicate no change. (a-h) represent a large-scale screen performed in replicate or quadruplicate, at single-cell resolution, (13,152 cells per well for 7,680 wells). (a-b) measurements performed in quadruplicate. (c-f) summary statistics combined over 1,402 compounds (mean with standard error of mean shown (c)), (g-h) ≥ 3 compounds per compound annotation. (c-h) Represents 246,650,047 cell-cell interactions.
When analyzing all 246×106 cell-cell contacts measured in the screen, it was observed that the monocyte lineage cells had significantly higher numbers of direct neighbors compared to the lymphocyte lineage cells (Fig. 2c). Evaluating the interaction scores between the distinct subpopulations, the highest scores were observed among and between CD11c+ and CD14+ monocytes (Fig. 2d), and lower, yet higher than random, interaction scores were observed between all measured monocyte-lymphocyte pairs. In contrast, interaction scores of around zero were observed on average between B- and T-cells, and from T-cells to any of the other cell types, indicative of baseline cell-cell contact frequencies equal to those expected by random for T-cells (Fig. 2d). Overall, many more compounds altered only leukocyte cell-cell contacts (11.6%) than altered only virus infection (2.5%) at 2 standard deviations (Fig. 2e). Similarity in the molecular regulation of cell-cell receptor mediated contacts would be expected to lead to similarity in the drug-induced alterations of those cell-cell contacts. Indeed, comparing overall results revealed the highest similarity in the modulation of cell-cell contacts among and with monocyte lineage cells (Fig. 2f).
To analyze the chemical modulation across the entire library, drug annotation enrichments over all interaction scores were calculated and displayed by hierarchical clustering (Fig. 2g, the full list of compounds and their specific immune modulation phenotype is attached as a resource in Supplementary Dataset 2). Inspection of the enriched drug classes altering PBMC cell-cell contacts revealed the presence of four groups, comprising predominant classes of drugs known to modulate the immune system: [1] Steroidal anti-inflammatory compounds such as glucocorticoids that bind to steroid hormone receptors (Fig. 2g, dark grey); [2] Non-steroidal anti-inflammatory drugs (NSAIDs) including arachidonate 5-lipoxygenase, and cyclooxygenase inhibitors (Fig. 2g, grey); [3] Drugs acting on or mimicking the signaling of the sympathetic nervous system, including catecholamines, adrenaline, dopamine, and associated receptor agonist and antagonists (Fig. 2g, light grey); And [4] a fourth group containing compounds described to act on other systems or previously unreported drug classes (Fig. 2g, yellow). Comparing the top 140 drugs with the strongest changed interaction scores to those 140 with strongest cytotoxicity revealed an overlap of only 11 drugs (7%, Supplementary Fig 3e), indicating that the cell-cell contact analysis did not bias towards drugs with strongly altered cell population sizes.
Steroidal anti-inflammatory compounds not only led to strong increased VSV infection, as expected by their mechanism of action, but also displayed significant enrichment for decreased CD14+ cell interactions (Fig. 2g-h). Steroidal anti-inflammatories regulate the immune system via the glucocorticoid receptor, reducing the transcriptional activity of pro-inflammatory transcription factors including NF-κB and IRF317, which may explain the observed decreased clustering of CD14+ cells and increased VSV infection (Fig. 2g-h). CD14+ cell clustering was in fact the cell-cell interaction with the most significant correlation to VSV infection over all measured compounds (Supplementary Fig. 3f, left), with increased infection associated with decreased clustering and vice versa (P<2.3×10-5; Supplementary Fig. 3f, middle). Combined with the increased CD14+ cell clustering upon LPS stimulation (Supplementary Fig. 1g) and previous reports 11, this singled out CD14+ cell clustering as the predominant spatial readout of innate-immune activation and suggested some compounds reduced virus infection by activating an innate immune reaction, which was further confirmed in a second healthy donor (Supplementary Fig. 3f, right).
In contrast to the steroidal anti-inflammatory compounds, several NSAIDs reduced CD11c+ cell clustering, CD19+ B-cell clustering, and CD11c+ cell→CD3+ T-cell interactions (Fig. 2g). NSAIDs act by inhibiting the synthesis of pro-inflammatory signaling molecules and chemo-attractants derived from arachidonic acid 18, including eicosanoids such as prostaglandins and leukotrienes. Interestingly, reduced CD11c+ cell→CD3+ T-cell interactions were observed for sulindic sulfoxide, salicin, celecoxib, tomelukast, and zafirlukast, thus reaching the same phenotype across diverse mode of NSAID action (P<3.8×10-5; Supplementary Fig. 3g) 18. This suggested a strong sensitivity of the CD11c+ cell→CD3+ T-cell interaction to modulation by NSAIDs, consistent with previous reports 19.
Further, several drugs altered leukocyte cell-cell interactions with known mechanism of action not directly linked to the above described steroidal, NSAID, or nervous system related modes of action. Quinine compounds strongly reduced CD11c+ cell→CD3+ T-cell interactions (Fig. 2g-h), in line with their anti-inflammatory effect and interference in MHC presentation, which has led to their current clinical use in management of rheumatoid arthritis and other inflammatory diseases 20. Cholesterol lowering drugs also reduced CD11c+ cell→CD3+ T-cell interactions (Fig. 2g), possibly mediated by the well documented dependency of MHC-II antigen presentation on cholesterol enriched lipid rafts 21,22. A subset of cholesterol lowering drugs, HMG-CoA reductase inhibitors (statins), which also reduced CD11c+ cell↔CD3+ T-cell interactions (Fig. 2g and Supplementary Fig. 3h), have been reported to also act in a cholesterol-independent way via blocking of leukocyte function antigen-1 (LFA-1)-mediated adhesion to and co-stimulation of lymphocytes 23. ACE inhibitors, particularly enalapril and its active metabolite enalaprilat, lead to increased CD14+ cell clustering as well as increased CD14+ cell↔CD19+ B-cell interactions (Fig. 2g and Supplementary Fig. 3h), potentially related to their reported inhibitory effect on the expression of the monocyte chemo-attractant protein-1 24, that inhibits B-cell migration 25. Taxifolins and catechols included plant flavonoids with known antioxidant and anti-inflammatory activity 26, which led to decreased CD14+ cell clustering (Fig. 2g-h). Further, N-acyl-L-homoserine lactones are components of the bacterial quorum sensing machinery, and led to increased CD3+ T-cell clustering and reduced CD19+ B→CD3+ T-cell interactions (Fig. 2g), which have been shown to activate the immune system and increase survival upon a bacterial infection model in pretreated mice 27, and a related compound has been shown to interfere with T-cell differentiation 28, although the mechanism of their recognition and effect on T-cells is as yet unresolved.
Lastly, the screen revealed several neurological modulators, which mimic or interfere with signaling of the nervous system, particularly the sympathetic nervous system that regulates a close physiological connection between the nervous and immune systems, as having immunomodulatory potential 29. For instance, modulators of the neurotransmitter γ-aminobutyric acid (GABA) signaling increased CD19+ B-cell clustering (potentially indicating activation, Fig. 2g). GABA, expressed on most PBMC subpopulations 30, has been shown to have an anti-inflammatory role in mouse models of autoimmune disorders involving B-cells, including multiple sclerosis 31 and rheumatoid arthritis 32, suggesting B-cell mediated effects. Further, agonists and antagonists of the β-adrenergic receptors altered several CD3+ T-cell centered cell-cell contacts (Fig. 2g), consistent with reported expression and function of the β-adrenergic receptors on lymphocytes 33. Agonists increased CD3+ T-cell→CD11c+ cell interactions, and decreased both CD3+ T-cell clustering and CD3+ T-cell→CD19+ B-cell interactions, while antagonists enriched for decreased CD19+ B→CD3+ T-cell interactions (Fig. 2g), likely related to diverse effects of β2-adrenergic receptor signaling on distinct subsets of T-cells reported previously 34.
Taken together, image-based screening of PBMCs enabled the exploration and categorization of the chemical rewiring of the leukocyte cell-cell interaction network, which revealed results in line with a variety of previously reported studies, all from a single phenotypic screen performed on the leukocytes of a single-donor blood sample. The screen here identified small compounds that can modulate and inhibit immune function, implicated novel modes of action for several compounds and existing drugs, and validated numerous effects through imaging.
Crizotinib increases T-cell to APC interactions
An example of drugs with previously unknown immunomodulatory propensities are the inhibitors of receptor protein-tyrosine kinases (RTKi’s), which led to increased interactions between CD11c+ cells and CD3+ T-cells (Fig. 2g and 3a), despite RTKi’s being shown rarely to display immunomodulatory effects 35,36. The observed enrichment resulted from the strong phenotypes obtained for both enantiomers of crizotinib (Fig. 3b). (R)-crizotinib (from here on referred to as crizotinib) is a MET, ALK and ROS 1 kinase inhibitor 37 approved for the treatment of ALK-rearranged non-small cell lung carcinoma (NSCLC) and under investigation for the treatment of several additional solid tumors.
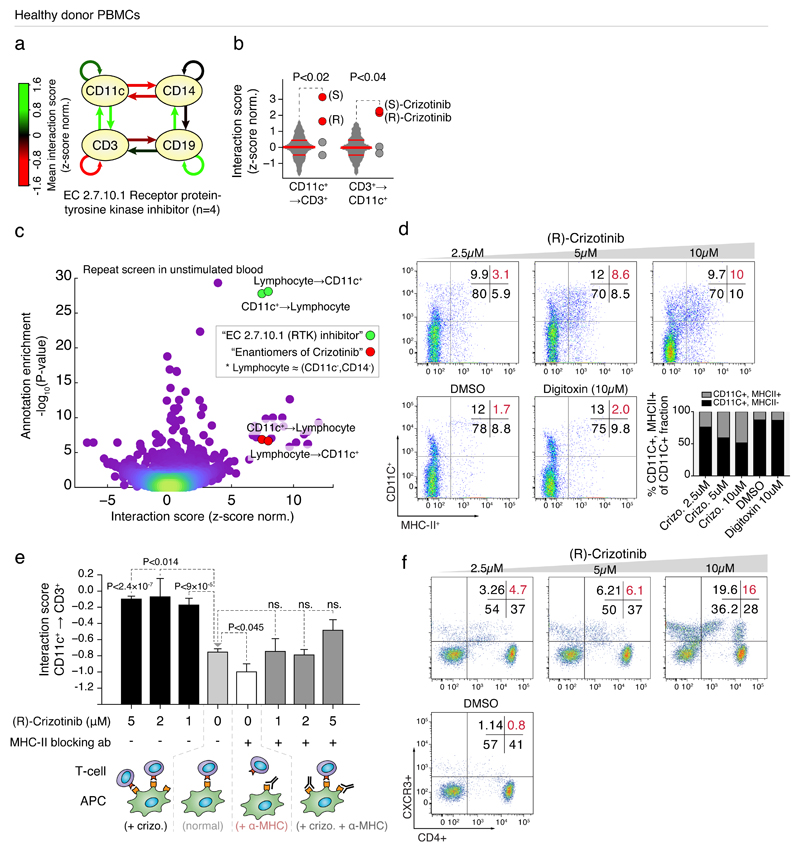
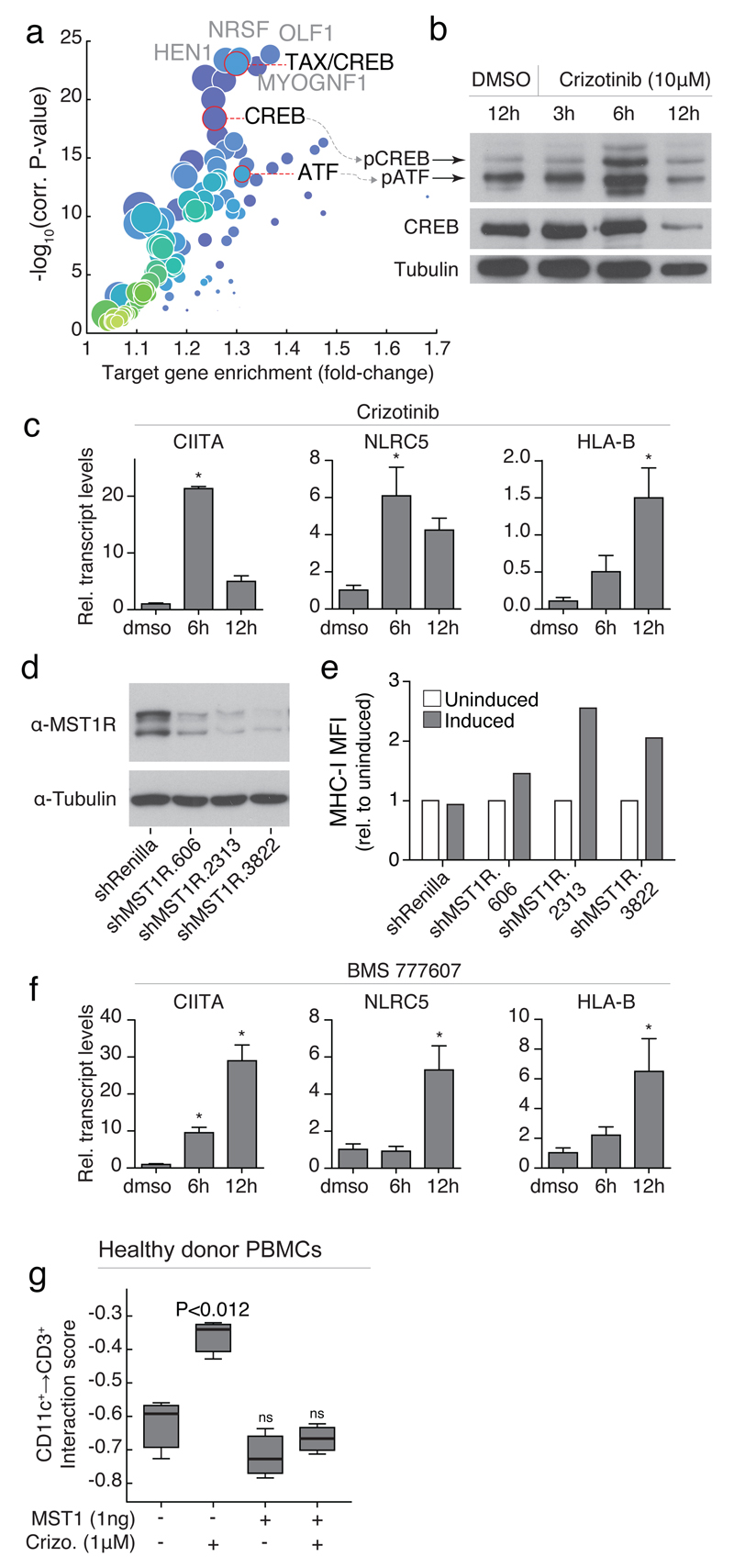
Fig 3. Crizotinib increases T-cell interactions with monocytes through upregulation of MHC-II.
(a) Interaction graph of average interaction scores for compounds annotated as RTKi’s; green and red arrows indicate increased and decreased interaction scores, respectively. (b) Interaction scores of CD11c+→CD3+ cells (left), and CD3+→CD11c+ cells (right) after treatment with (R)- and (S)-crizotinib (red dots) compared to the other RTKi’s (grey dots), as well as respective interaction score distributions over the entire screen (grey plots, with 25, 50, and 75-percentiles indicated as horizontal lines). (c) Annotation enrichment analysis over all interaction scores measured in naive PBMCs. Dots indicate individual annotations and their cell-cell interactions. Y-axis indicates significance of annotation enrichment for compounds with a certain phenotype. X-axis indicates average z-score normalized interaction score per annotation. Green dots: results for the RTKi’s and lymphocytes↔CD3+ interactions; red dots: ‘enantiomers of crizotinib’ annotation and lymphocytes↔CD3+ interactions. (d) Flow cytometry of MHC-II expression on CD11c+ cells of naive PBMCs after overnight incubation with (R)-crizotinib, digitoxin, or DMSO. Bar graph (gray; lower right panel) represents percentage of double-positive (CD11c+/MHC-II+) cells in the CD11C+ fraction. (e, top) Interaction score of CD11c+→CD3+ after crizotinib treatment without (left) or with (right) MHC-II antibody. (e, bottom) Schematic of the interpretation of (e, top). Significance values compare to “normal”. (f) Flow cytometry CD4+ CXC3R(CD183)+ Th1 T-cells after overnight incubation with (R)-crizotinib. (a-c) Data from screens performed in (a-b) quadruplicate or (c) replicate, (d-f) representative of at least three repeats. (e) Results of 164,569 measured cell-cell interactions. T-test was used to determine significance in (e), and standard error of means is shown.
A repeat of the leukocyte interaction screen over the entire drug library, using a blood donation from a second healthy donor but without background stimulation by VSV infection, reproduced the specific crizotinib effect: the RTKi drug class was the strongest enriched drug class over all cell-cell interactions (Fig. 3c), with significantly increased interactions between CD11c+ cells and lymphocytes (i.e. monocyte marker-negative cells) observed upon crizotinib treatment (Supplementary Fig. 4a). As MHC-to-TCR contact dependent signaling drives APC→T-cell interactions and downstream activation, we assayed for altered MHC expression upon crizotinib treatment of PBMCs from additional healthy donors by flow cytometry, and observed a dose-dependent increase of MHC-II surface levels on CD11c+ cells not observed for an unrelated cytotoxic compound (digitoxin) or DMSO (Fig. 3d). Furthermore, an MHC-II blocking antibody strongly decreased the crizotinib-enhanced interaction between CD11c+ monocytes and CD3+ T-cells, indicating the MHC-II upregulation mediates the observed immunomodulatory effect (Fig. 3e). Comparison of the T-cell compartment after ex vivo crizotinib incubation of PBMCs indicated a crizotinib-induced CD4+ Th1, but not Th2, response, which is indicative of an inflammatory and cytotoxic immune milieu (Fig. 3f and Supplementary Fig. 4b).
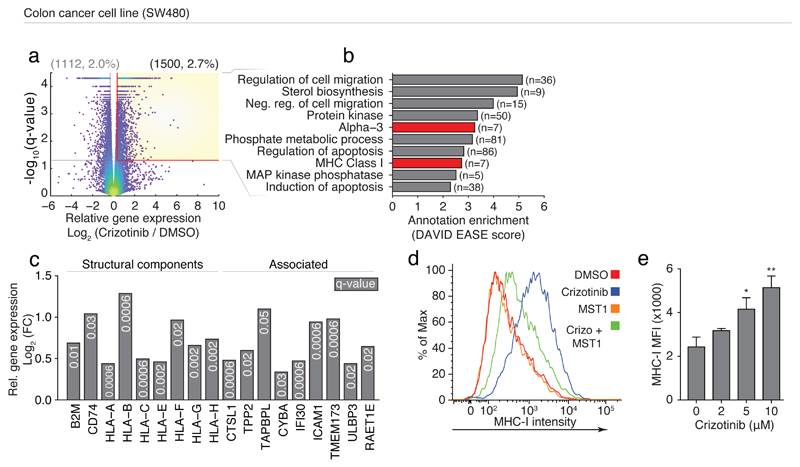
MHC-I and II gene expression is partially under control of overlapping transcription factors 38, and MHC-I self-antigen presentation is a crucial factor in mounting a successful anti-cancer immune response 3, where increased MHC expression and antigen presentation on cancer cells is a desirable effect in the treatment and maintenance of cancer 39. We therefore measured the transcriptional response using RNA sequencing in the colorectal adenocarcinoma-derived cell line (SW480 cells) after treatment with 2μM crizotinib (Fig. 4a and Supplementary Dataset 3). The genes upregulated by crizotinib treatment showed significant enrichment for MHC-I annotations (Fig. 4b-c), including all three major MHC-I cell surface receptors (HLA-A, B, and C) as well as the invariant chain β2 microglobulin (B2M) (Fig. 4c), and genes involved in antigen presentation and peptide processing, loading, and trafficking (Fig. 4c). While MHC-II expression is typically restricted to professional APCs, the invariant chain of MHC-II (CD74) was also significantly upregulated on SW480 cells after crizotinib treatment (Fig. 4c). Crizotinib-enhanced expression of MHC-I on the SW480 cell surface was confirmed by flow cytometry (Fig. 4d-e).
Fig 4. Crizotinib drives MHC-I expression in SW480 cells.
(a) Significance (y-axis) and log2(fold change) (x-axis) of RNAseq gene expression in SW480 cells after 72h of 2μM (R)-crizotinib treatment compared to DMSO; dots indicate individual genes; dot color indicates local data density. Yellow box indicates significantly upregulated genes (n=1500). (b) Annotation enrichment analysis of the genes significantly upregulated by crizotinib treatment (yellow box, (a)); selected top-enriched annotations are shown, MHC-related classes indicated in red. (c) Log2(fold-change) (y-axis) for selected MHC-related genes (from (a-b)). (d-e) Flow cytometry measured (d) Intensity or (e) MFI of MHC-I on SW480 cells after incubation with (d) 3μM crizotinib (blue), 1μg/ml MST1 (orange), together (green), or DMSO (red), or (d) increasing concentrations of crizotinib, overnight. (a-c) RNAseq was performed in technical triplicates. (d-e) are representative experiments of at least three repeats. Significance in (b) calculated by one-way ANOVA.
Transcription factor binding site enrichment analysis on the upregulated genes revealed strong enrichment for binding sites of CREB and ATF (Fig. 5a), both important transcription factors for MHC-I and –II class genes, that further regulate and cooperate with the MHC class-specific transcription factors CIITA and NLRC5 38,40. Indeed, we observed increased phosphorylation of both CREB and ATF upon crizotinib treatment of SW480 cells (Fig. 5b, and Supplementary Fig. 5a), along with increased transcript levels of CIITA and NLRC5 (Fig. 5c).
Fig 5. Immunomodulatory effect of crizotinib is mediated by MST1R inhibition.
(a) Transcription factor binding site enrichment from genes upregulated in (Fig 4a). (b) Western blot of phospho- and total-CREB/ATF after incubation of SW480 cells with crizotinib. (c,f) QPCR of indicated genes incubated with (c) 10μM crizotinib or (f) 1μM BMS-777607, normalized to GAPDH. (d-e) (d) Western blot of MST1R in lysates of SW480 cells after induction of siRNA and (e) MFI of MHC-I after overnight siRNA induction. (g) Interaction scores of CD11c+→CD3+ cells after overnight incubation with crizotinib and/or MST1. (b-g) are representative experiments of at least three repeats. (c, f ,g) t-test, and boxplots or standard error of means are shown.
We recently identified the target spectrum of crizotinib in SW480 cells with chemical proteomics 41, and among the most abundant interactors in these cells was the Macrophage Stimulating 1 Receptor (MST1R, also known as RON), a close c-MET homolog. Crizotinib is described to have a dissociation constant of 25nM to MST1R 42, to inhibit MST1R with an IC50 of approximately 200nM 43, and KINOMEscan results have showed 100% binding of MST1R at 1μM (R)-crizotinib 41. MST1R is a known negative regulator of MHC-II expression and immune function in mice, as activation by its ligand, Macrophage Stimulating 1 (MST1), leads to decreased STAT1 phosphorylation and CIITA expression44,45. Furthermore, naive MST1R knockout mice show increased immune cell infiltration in lungs46, thus we speculated that crizotinib could increase MHC expression by inhibition of MST1R/RON.
Using inducible shRNA targeting MST1R in SW480 cells, the loss-of-function coincided with increased MHC-I surface expression over independent short hairpins after 72h of efficient MST1R knockdown (Fig. 5d-e and Supplementary Fig. 5b-d), while no such increase was observed for the negative control shRNA (Fig. 5e). Furthermore, treatment of SW480 cells with a more potent and specific MST1R-inhibitor, BMS-777607 47, also led to strong upregulation of CIITA, NLRC5 and HLA-B (Fig. 5f), while, as expected, the ligand MST1 did not induce expression of these genes (Supplementary Fig. 5e). Increased MHC-I surface expression upon BMS-777607 was confirmed by flow cytometry (Supplementary Fig. 5f), while incubation with MST1 showed no change in MHC-I surface levels by flow cytometry (Fig. 4d and Supplementary Fig. 5f). Importantly, combined incubation of SW480 cells with MST1 and crizotinib led to reduced MHC-I surface levels compared to crizotinib treatment alone (Fig. 4d), and further competition experiments using crizotinib and MST1 in healthy donor blood revealed a full reversal of the crizotinib-increased interactions between CD3+ T-cells and CD11c+ monocytes (Fig. 5g).
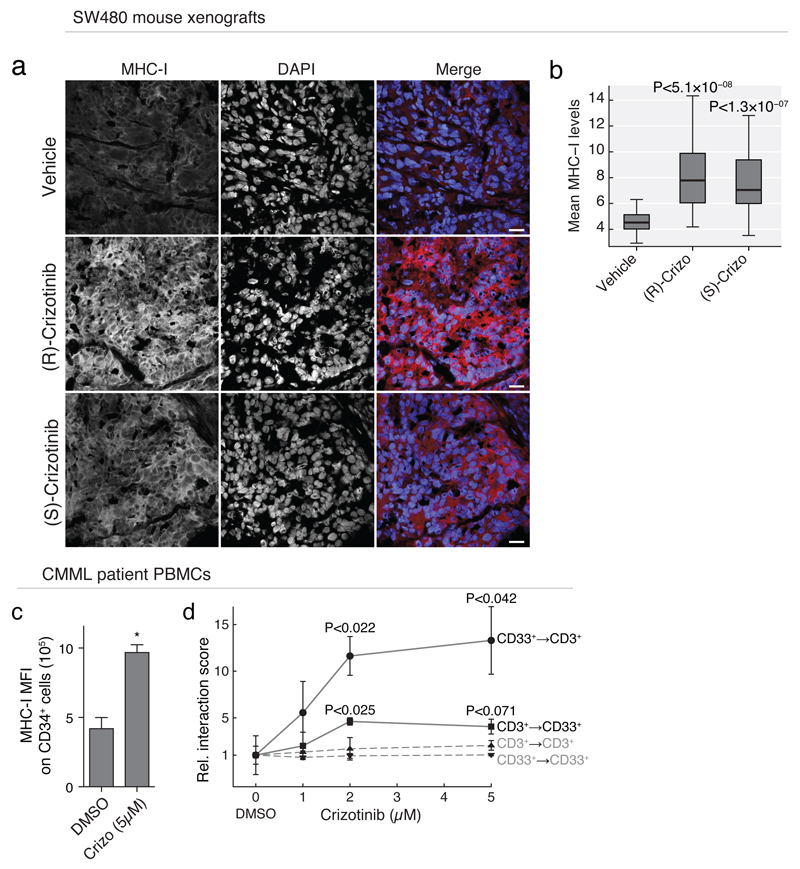
To test if crizotinib induced MHC-I expression also in vivo, we injected crizotinib or vehicle alone into SCID deficient mice with a SW480 xenografted tumor. Comparison of the tumors by immunohistology indicated significantly higher levels of MHC-I expression in the crizotinib treated mice compared to vehicle control (Fig. 6a-b), recapitulating the ex vivo immunomodulatory effect of crizotinib in vivo.
Fig 6. In vivo assessment of the immunomodulatory potential of crizotinib.
(a) Immunohistochemistry imaging of SW480 xenograft tumors stained with anti-human MHC-1 (40x images, scale bar 20μm) and (b) image quantification of MHC-I expression levels for 24-images per group, after treatment with 50mg/kg p.o. vehicle, (R)- or (S)-crizotinib. (c) MFI of MHC-I expression on CD34+ blasts and (d) DMSO-relative interaction scores between CD33+ blasts and CD3+ T-cells after crizotinib treatment in PBMCs of a patient diagnosed with a post-CMML AML. (a-b) Staining was performed on 3 tumors per group, 8 images each; selected images are representative of average. (c-d) are representative experiments of at least three repeats. (b-d) Significance was calculated by a t-test, and boxplots or standard error of means are shown.
The immunomodulatory effect of crizotinib was not limited to healthy blood or the SW480 colon-carcinoma cell line: analysis of publicly available transcriptomics data of an ALK-positive NSCLC-derived cell line made resistant to crizotinib by prolonged incremental exposure to the drug (up to 1μM), compared to its parental H3122 cell line 48, also revealed increased expression of MHC-I and II genes (Supplementary Fig. 6a-c). We confirmed this by qPCR, measuring strong upregulation of transcript abundance of CIITA, NLRC5 and HLA-A upon crizotinib treatment of the parental H3122 cells (Supplementary Fig. 6d). Further, we performed chemical proteomics with the racemic mixture of crizotinib in H3122 cells, which revealed extensive binding of MST1R also in this setting (Supplementary Table 1). Thus, crizotinib interacted with MST1R and led to an increased expression of MHC genes also in this model cell line for NSCLC, for which crizotinib is clinically approved. Intriguingly, MST1R may be a relevant crizotinib target in NSCLC, as a meta-analysis of 15 studies measuring gene expression in lung cancers showed consistent and significant upregulation of MST1R specific to NSCLC (Supplementary Fig. 6e).
The presentation of self-antigens on MHC-I by cancer cells is a key step in the anti-cancer immune response of the host, and is essential for several checkpoint inhibitor treatments currently under investigation 39. To test the immunomodulatory potential of crizotinib in primary patient-derived material, PBMCs of a patient diagnosed with chronic myelomonocytic leukemia, with over 70% CD33+ and CD34+ blast cells in peripheral blood, were incubated ex vivo with crizotinib. Crizotinib-treated peripheral blasts showed a doubling in MHC-I surface expression levels as measured by flow cytometry (Fig. 6c), as well a concentration-dependent increase in T-cell↔blast interactions as measured by automated microscopy (Fig. 6d). The anti-cancer effects of genetic ablation and chemical inhibition of MST1R have been shown in vivo in mouse models for lung cancer, revealing increased CD8+ T-cell tumor infiltration resulting in reduced metastasis 45. Combined, our data suggest that the crizotinib-mediated MST1R inhibition and subsequent increase in MHC-I surface levels on colon, lung, and blood cancer cells, may aid an anticancer immune response and be beneficial for clinical use in combination with immunotherapy in a range of malignancies (Supplementary Fig. 6f) 8. A clinical trial combining crizotinib with CTLA-4-blockade by ipilimumab for the treatment of NSCLC may reveal if clinical anti-cancer immune responses benefit from the here discovered immunomodulatory effect of crizotinib (NCT01998126 clinicaltrials.gov).
Discussion
The work presented here defines the possibility to systematically quantify and identify the immunomodulatory potential of drugs and biologicals over the complex multi-lineage populations in PBMCs of one individual. Prior to this, the technical options and models for measuring system-wide immunomodulatory events at high-throughput were limited, driving the need for such a screening tool. The dataset provides evidence of immunomodulatory potential for some 150 chemical agents, many of which have not been reported to display effects on the immune system and my thus become object of further investigation. As for crizotinib, it is conceivable that the immunomodulatory potential of some of these agents unknowingly contributes to the clinical success of individual drugs. While the library here was screened at single concentrations, it is feasible that additional effects may become apparent at higher, but still safe, concentrations. This large-scale functional characterization of the current collection of “common” drugs represents a new dimension and a potential future standard assay in the characterization of existing and new drugs. It remains to be seen, how often the effects reported here result in altered cell-cell interactions, cell infiltrations and altered immune functions in other tissues. Certainly, the platform can easily be adapted to screen for new agents aiming at specific immunomodulatory effects. Equally exciting is the perspective to perform “personalized” assessments on individual patients or healthy donors on a routine basis. The systematic perturbation of the immune system of an individual over such a varied array of pathways and processes may thus provide a unique functional “profile” of the health status of a person, likely to depend on age, gender, diet and transient and chronic infections. We illustrate how the approach may be useful to identify agents that act immunomodulatory in cancer, but other diseases characterized by increased inflammation, such as rheumatoid arthritis or autoimmune diseases, may be particularly interesting to consider in further studies. The finding that crizotinib, a drug in clinical use, may exerts some of its anti-tumor activity through an immunomodulatory effect may help sharpen its ideal therapeutic window. In general, modulating the immune system by small compounds may be a desirable integrated positive effect that is more common than previously expected, and a result of their full polypharmacological properties 49,50.
Online Methods
Collection and purification of peripheral blood and bone marrow cells
Peripheral blood was obtained from healthy donors, from patients with CMML who were treated at the Medical University of Vienna/General Hospital Vienna, and blood from the Austrian Red Cross. Austrian Red Cross donations were enriched buffy coats. Blood coagulation was prevented with EDTA or heparin. The Ethics Commission at the Medical University of Vienna/General Hospital Vienna approved the collection of samples from healthy donors and patients. All donors and patients provided written informed consent after nature and consequences of the study were explained. All donors were blind to end users. Peripheral blood was diluted 1:1-1:3 in PBS and mononuclear cells were isolated with lymphoprep density gradient from Axis-Shield following manufacturer directions. Purified cells were suspended in RPMI supplemented with 10% FBS and penicillin-streptomycin. For all experiments involving primary cells, cells were cultured in RPMI supplemented with 10% FBS and penicillin-streptomycin.
Non-adherent PBMC monolayer formation, small-molecule screening, viruses, reagents, cell lines
50nl of selected screening compounds in DMSO, and DMSO controls, were transferred to Corning 384-well, tissue-culture treated clear bottom plates by a Labcyte Echo liquid handler attached to a Perkin Elmer high-content cell::explorer workstation. Screening compound libraries were obtained from the NIH Molecular Libraries Program, gifts from various groups (see acknowledgements), or designed in-house. Selected compounds were routinely quality controlled by mass spectrometry. Compounds screened are outlined in Supplementary Dataset 1. 50μl of culture media containing approximately 40x104 cell / ml is pipetted into each well of a 384-well plate, and incubated at 37°C with 5% CO2, allowing cells to settle to the bottom; the erythrocyte concentration within the sample after purification should be limited to allow PBMCs space on the well bottom. For virus screens, cells were incubated with compound for 3 hours before VSV-GFP was added in 10μl at MOI=10 for 18 hours. Screen in Figure 2 was performed in quadruplicate (global level) and duplicate (population level) over 384-well plates from a single healthy donor; See figure legend for technical replicate information for other experiments. For the healthy donor naive spatial screen, PBMCs were incubated with compound for 36 hours. For blocking-antibody experiments anti-human HLA-DR (G46-6) (eBiosciences), anti-human CD54 (HA58) (BDbiosciences) or mouse-IgG2a isotype control (C1.18.4) (BioXCell) were incubated for 3 hours prior to overnight stimulation with VSV or 10ng/ml LPS (Invivogen). HLA-DR blocking antibody was incubated overnight prior to treatment with crizotinib for 3 hours. Rituximab (anti-human CD20, Absolute antibody), or Blinatumomab (Amgen, Vienna General Hospital in-patient Pharmacy) were incubated at indicated concentrations in healthy donor blood overnight or 48 hours, respectively. Recombinant MST1 from RnD Systems. All screens were stopped by fixing and permeabilizing the cells with 10μl of a solution of 4% formaldehyde and 0.01% triton-X114 in PBS for 10 minutes at room temperature. Fixative containing medium was removed and 30 μl of an experiment-dependent antibody cocktail in PBS was added to the cells one hour at room temperature, all antibodies at 1:300 dilution. Antibodies were selected for their ability to identify specific populations of interest: Anti-human CD19 (HIB19, APC, B-cells), CD11c (3.9, APC, majority DCs and monocytes), CD3 (HIT3a, PE, T-cells), CD14 (61D3, PE, macrophages and monocytes), and CD34 (4H11, APC, hematopoietic progenitors) from eBiosciences, CD20 (2H7, GFP, B-cells) from BD Biosciences, and CD56 (A07788, PE, NK-cell) from Beckman Coulter, were all used in various combinations to identify populations of interests in each experiment, as described in the text. DAPI (Sigma) was used for nuclear detection at 10 𝜇M. SW480 and H3122 cells were cultured in DMEM or RPMI respectively, supplemented with 10% FBS and penicillin-streptomycin. H3122 cells were a kind gift from Eric Haura, and SW480 from Walter Berger. Cells used in experiments regularly undergo ELISA and PCR tests for mycoplasma.
Flow cytometry
PBMCs or SW480 cells were kept naive or incubated with small molecules compounds for indicated time points. Antibodies used for flow cytometry are listed above as used for imaging, as well as anti-Human HLA-DQ (SK10, FITC, MHC-II major population), HLA-ABC (G46-2.6, Pe-Cy5, MHC-I major population), CD4 (SK3, PE, T-cell subset), CD183(CXCR3-173, FITC, T-cell subset), and CD194(CCR4-D8SEE, APC, T-cell subset) from eBiosciences. Samples were run on BD LSRFortessa with Diva software and analyzed with FlowJo.
Western Blots
SW480 cells were plated overnight and stimulated at the indicated time points. Pellets were lysed using IP lysis buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 5 g/ml TPCK) and protease inhibitor mixture for 10 min on ice before high-speed clearance, and quantified with Bradford. Proteins were detected with rabbit anti-Phospho-CREB (Ser113)(87G3), mouse anti-CREB (86B10), or rabbit anti-RON (C81H9) (Cell Signaling) and mouse anti-tubulin (Abcam), and detected using a goat anti-mouse or -rabbit (H+L) HRP conjugated secondary antibody (Jackson ImmunoResearch Laboratories), and exposed on film.
RNA isolation and qPCR
RNA was purified from SW480 or H3122 cells using and RNeasy Kit (Qiagen) and was reverse transcribed using oligo dT primers and RevertAid Reverse Transcriptase (Fermentas). qPCR was performed using SensiMix SYBR Green (Bioline) analyzed on a Rotor-Gene Q from Qiagen. Genes CIITA (F: GGCTGGAATTTGGCAGCAC, R: GCCCAACACAAGGATGTCTC), NLRC5 (F: CTGGCCAGTCTCACCGCACAA, R: CCAGGGGACAGCCATCAAAATC), HLA-A (F: AAAAGGAGGGAGTTACACTCAGG, R: GCTGTGAGGGACACATCAGAG), and HLA-B (F: CTCATGGTCAGAGATGGGGT, R: TCCGCAGATACCTGGAGAAC) were normalized to the housekeeping gene GAPDH (F: GAAGGTGAAGGTCGGAGT, R: GAAGATGGTGATGGGATTTC).
MST1R inducible knockdown
97-mer shRNA sequences were obtained as ultramer oligonucleotides (IDT), PCR amplified and cloned into the inducible retroviral miR-E shRNA vector pRT3GEN (pSIN-TRE3G-turboGFP-miR-E-PGK-NeoR) as described previously using standard cloning techniques 51. The Tet-on competent SW480 cells were generated by virally transducing vector pMSCV-RIEP (pMSCV-rtTA3-IRES-EcoR-PGK-PuroR) and standard retroviral packaging. After virus infection, transformed cells were selected using 5μg/mL puromycin (Sigma-Aldrich), and SW480 RIEP cells were transduced in a similar way using the retroviral shRNA vector pRT3GEN. The following 97mer target sequences were used: Renilla at position 713, TGCTGTTGACAGTGAGCGCAGGAATTATAATGCTTATCTATAGTGAAGCCACAGATGTATAGATAAGCATTATAATTCCTATGCCTACTGCCTCGGA, MST1R at position 606, TGCTGTTGACAGTGAGCGATCCCGGTGACACAGACACAAATAGTGAAGCCACAGATGTATTTGTGTCTGTGTCACCGGGAGTGCCTACTGCCTCGGA, MST1R at position 2313, TGCTGTTGACAGTGAGCGACCAGTGCTGATAGCAGTGCAATAGTGAAGCCACAGATGTATTGCACTGCTATCAGCACTGGCTGCCTACTGCCTCGGA, and MST1R at position 3822, TGCTGTTGACAGTGAGCGAAGGGAGTACTATAGTGTTCAATAGTGAAGCCACAGATGTATTGAACACTATAGTACTCCCTGTGCCTACTGCCTCGGA. Following virus transduction and selection using 2mg/mL G418 (Roth), shRNA expression was induced by addition of 2μg/mL doxycycline to the culture media. MHC-I expression was analyzed by flow cytometry after 72 hours of doxycycline addition. pRT3GEN-shRen.713 targeting Renilla Luciferase coding sequence served as negative control shRNA. MFI of MHC-I (PE-Cy5 channel) was normalized to uninduced levels. Knockdown efficiency was determined by western blot as described above.
RNA Sequencing
SW480 cells were seeded in 6-well plates 24h before (R)-crizotinib was added to a final concentration of 2μM. After incubation for 72h, medium was aspirated and cells washed with PBS. Total RNA was isolated using the RNeasy kit (Qiagen) and RNA amount was measured using Qubit 2.0 Fluorometric Quantitation system (Life Technologies), both following manufacturer protocols. RNA integrity number (RIN) was determined using Experion Automated Electrophoresis System (Bio-Rad). RNA-seq libraries were prepared with TruSeq Stranded mRNA LT sample preparation kit (Illumina) using Sciclone and Zephyr liquid handling robotics (PerkinElmer). Library amount was quantified using Qubit 2.0 Fluorometric Quantitation system (Life Technologies) and the size distribution was assessed using Experion Automated Electrophoresis System (Bio-Rad). Sequencing libraries were pooled, diluted and sequenced on Illumina HiSeq 2000 using 50 bp single-read chemistry. Base calls provided by the Illumina Realtime Analysis software were converted into BAM format using Illumina2bam and demultiplexed using BamIndexDecoder (https://github.com/wtsi-npg/illumina2bam). Transcriptome analysis was performed using the Tuxedo suite. TopHat2 (v2.0.10, http://genomebiology.com/2013/14/4/R36/abstract) was supplied with reads passing vendor quality filtering (PF reads) and the Ensembl transcript set (Homo sapiens, e73, September 2013) as reference. TopHat2 analyses were run independently for each replicate. Cufflinks (v2.1.1, http://www.nature.com/nbt/journal/v31/n1/full/nbt.2450.html) was used to assemble transcripts from spliced read alignments, using the Ensembl e73 transcriptome as the reference as well as de novo assembly of transcript models. Differential expression was assessed with Cuffdiff v2.1.1 (http://www.nature.com/nbt/journal/v28/n5/full/nbt.1621.html). Transcriptome sets of all replicates for each sample group were combined with Cuffmerge. Finally, cummeRbund (http://www.bioconductor.org/packages/release/bioc/html/cummeRbund.html) and biomaRt (http://www.bioconductor.org/packages/release/bioc/html/biomaRt.html) were used in combination with custom R scripts to perform quality assessment and further refine analysis results. All sequencing was performed by the Biomedical Sequencing Facility at CeMM (http://medical-epigenomics.org). SW480 RNA sequencing results are available in Supplementary Dataset 3. Transcription data from ALK-positive H3122 cells resistant to crizotinib treatment was obtained from NCBI Gene Expression Omnibus (GEO) accession number GSE49508.
Drug affinity chromatography and protein mass spectrometry
The crizotinib affinity probe was prepared in two steps from commercially available 3-[(1RS)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]pyridin-2-amine (Selleckchem) according to literature and drug-affinity matrices were prepared as described previously (Huber et al., 2014). Briefly, affinity chromatography and elution were performed in duplicate using 25nmol of compound immobilized on 50μL NHS-activated Sepharose 4 Fast Flow beads (GE Healthcare Bio-Sciences) and 10mg total cell lysate as protein input per replicate. For competition experiments, cell lysates were pretreated with 20μM 3-[(1RS)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]pyridin-2-amine for 30 minutes. Eluates were labeled with iTRAQ and quantitative protein mass spectrometry and bioinformatics analysis utilizing the R isobar package were performed as previously reported 48,52.
Xenograft mouse model and immunohistochemistry
All animals were acclimatized for one week, and had free access to water and food during the experiment. Animals were under a 12-hour light cycle, and temperature, humidity and housing per laboratory animal guidelines and regulations. The group size was based on previous experience on variability of tumor growth within control groups. Animals were grouped based on body weight, exclusion/inclusion criteria were pre- established in the ethical permit, and outliers in body weight were excluded; although none for this study; groups were not randomized. SCID mice (female, 5-6 weeks, Scanbur, n = 3/group) were injected s.c. with 1×106 SW480 cells together with a matrix gel (1:1) in the sacral area. Treatment was initiated one day after cell inoculation. Vehicle or (S)- or (R)-crizotinib was administered subcutaneously once daily at 25mg/kg for 35 days. (S)- or (R)-crizotinib was diluted in 1% DMSO, 10% ethanol, 10% cremaphore, 10% Tween 80, 69% PBS. Animals were sacrificed 2 hours after last administration of drugs on day 26. Tumors were quickly dissected, snap-frozen, and kept at -80°C until further analysis. Tumor sections were fixed in acetone for 10 min, and rinsed in TBS. The sections were blocked in 3% BSA in TBS. MHC-I antibody (MHC class I clone W6/32, Biolegend) was diluted in BSA in TBS and incubated overnight at 4°C in humid atmosphere. After incubation, the slides were rinsed in TBS with 0.025% Triton-X. AlexaFlour555-conjugated secondary antibody (Invitrogen) was diluted 1:500 in BSA, and incubated for 1 hour at room temperature. The slides were counter stained with DAPI, and mounted with prolong gold. Images were taken on an LSM780 confocal (Zeiss) with equal laser line power settings over all samples. 8 random images were taken from 3 tumors per group; image analysis was performed blind to the experimenter. All experiments involving animals followed protocols approved by Stockholms Norra djurförsöksetiska nämnd (laboratory animal ethical committee Stockholm) and were in compliance with 2010/63/EU directive.
Image and computational analysis
Imaging
Each well of a 348-well plate was imaged at 10x with 2x2 non-overlapping images, so that within the 2x2 square was approximately 95% of the well surface. The images were taken sequentially from DAPI, GFP, PE, and APC channels, with lasers and bandpass filter sets set so the channels were non-overlapping; a PerkinElmer Operetta or Opera Phenix automated spinning disk confocal microscope were used. Fluorophores were tested for channel separation on each machine. The raw .tiff images were exported from the microscope for analysis, below and stored for subsequent data mining.
Illumination correction
Illumination correction was performed based on a novel algorithm that depends on the observation that PBMCs in the monolayer never fully cover the entire well, but always leave adequate space in between individual cells or clusters of cells to estimate regional background intensities. As we imaged each well entirely (including regions of wells outside of images), we first detect regions of each image that contains areas outside of the well, based on full-well image reconstruction and plate-wide analysis of where outside well areas are most likely. For each individual single-channel image a 2D polynomial with two degrees of freedom in each dimension was fit on log10-transformed(1 + intensity) values, excluding regions of images containing areas outside the well, excluding the brightest 30% of the signal (working under the assumption that bright signal is not background signal, which does not hold for brightfield images), and weighted to a combination of the inverse signal intensity (to weigh dark regions heavier) and a smooth function that weights edges of images heavier, as those are relatively underrepresented compared to the typically brighter center of the image. As the illumination correction function for each individual image is described in the efficient form of a polynomial function, goodness of fit statistics and background patterns and intensities could be compared across each image from the same channel from the entire plate, for outlier detection (3 x interquartile range (IQR) from median statistics). For outliers, fits are retried with different parameters (excluding larger regions of potential outside-well area), or if that fails, corrected with plate-average background correction functions for images of the same channel and at the same site within the other wells. Remaining outlier wells are flagged as potential technical problems. The algorithm has been optimized for robustness over hundreds of imaged 384-well plates containing PBMCs, and improvements in separation of negatively-, singly, and doubly-stained cells have been confirmed.
Background correction
Image-based small compound screens suffer from the fact that small compounds themselves can be sources of fluorescence signal, overlapping in unpredictable ways with the wavelengths of the imaged channels. To correct for small (within 3xIQR from median) background intensity variations, also stemming from other sources such as light source fluctuations, we further corrected for global background differences within all images of the same channel and of each well, and across all wells of the same plate. Outlier background values per well are flagged as potentially auto-fluorescent compound containing or otherwise problematic wells, and discarded from final screen statistics. Finally, illumination- and background-corrected images are reconstructed and stored independently for further image analysis. Improvements at the level of image quality and image analysis results were confirmed, and robustness of the algorithm was improved over hundreds of plates.
Image quality evaluation
All (illumination- and background-corrected) images of each plate were automatically rescaled and merged per well and per plate, allowing for quick visual evaluation of the quality over the entire dataset of a single plate.
Single cell image analysis using CellProfiler (code availability)
Single cell image analysis was performed using CellProfiler v2, based on DAPI-stain derived nucleus (‘Nuclei’) detection, expansion from nucleus for cell outline detection (‘Cells’), and a second and third set of expansions from Cells, to identify a ring outside of each cell for a local background (‘LocalBackground’) area sampling. Standard CellProfiler intensity, texture and shape features were measured from Nuclei, Cells and the LocalBackground over all channels where applicable. Differences between Nuclear and Cell log-transformed intensities over LocalBackground were used for the plate-wide identification of marker-positive cells, and different stains were thresholded in a supervised manner for the large-scale drug screens. Unsupervised thresholding was performed based on the predominant marker-negative cell population over the entire plate. Thresholds were regularly visually controlled, at the plate-wide population distribution level, as well as at the single cell level, relating images and image analysis results.
SVM-based machine learning for quality control
Iterative SVM-based machine learning was applied at the level of individual cells to identify poorly segmented cells, cells covered by contaminants or artifacts (‘blobs’), or false-positively identified cells, as described in 4,53.
Plate-effect correction
Plate-effect correction was performed using a weighted 2D polynomial fit, excluding positive control wells and 5% of the strongest outlier wells, and weighted based on a function that weights outside wells stronger than inside wells and outlier wells weaker compared to wells closer to the plate median. Globally, improvements in reproducibility across replicate wells coming from plates with individually randomized plate layouts, as well as in the separation of positive and negative control wells were confirmed.
Significance calculation & hit selection for large-scale screens
For the large-scale screens, datasets were plate effect normalized, normalized to the median of the DMSO-controls of each plate, and significance of the hits was calculated based on a Student’s t-test of mean equals 0, over all replicated wells (n=4 for the virus screens). Final infection and viability scores are calculated as the average over all replicates. Hits were selected based on p-value < 0.01 and based on an absolute average z-score of at least 2. All other significance scores in figures were calculated using a two-tailed t-test, unless otherwise noted.
Compound annotation enrichment analysis
Compounds were annotated based on their annotations as stored in the database and ontology of Chemical Entities of Biological Interest (CHEBI) from the European Bioinformatics Institute (http://www.ebi.ac.uk/chebi/) as well as from the KEGG Compound database (http://www.genome.jp/kegg/compound/). Overall, this retrieved 136 unique annotations for our library. Enrichment scores were calculated as two-tailed t-test’s between the phenotypes of compounds with a given annotation compared to the phenotypes of compounds without that given annotation.
Cell-cell interaction analysis
Pairwise distances (measured in pixels) between all cells in a well were calculated over a reconstruction of nuclear positions over the different images per well. Cells were considered pairwise if the pairwise distance between nuclear centroids was equal to or less than 15 pixels. The observed fraction of A→B interacting cells equals the fraction of (all A-type cells interacting with one or more B-type cell). This fraction was log2-transformed against the (EAB) value calculated for that well, i.e. (the fraction of A-type cells of all cells) * (the fraction of B-type cells of all cells) * (the fraction of all cells that have at least one or more contacts within 15 pixels). Results were confirmed to be insensitive to reasonable increments of the ‘15 pixel’ threshold. Although the reference value (EAB) is independent of the direction of interaction, A→B and B→A can diverge as a consequence of the interaction conformation of the two subpopulations. So called ‘rosettas’ existing for instance of an A-cell binding to many B-cells can induce strongly divergent results between A→B and B→A. Readouts were corrected for plate effects as described above, and analyzed further by various means including drug class enrichment analyses and hit selection strategies as also described above.
Supplementary Material
Additional Information:
Any supplementary information, chemical compound information, and source data are available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
Acknowledgments
We are grateful to donors and patients for their part in this study. CeMM is supported by the Austrian Academy of Sciences. We acknowledge funding from the ERC i-FIVE advanced investigator grant and Austrian Science Fund grant F4711-B20 (G.S.F.). The Austrian Federal Ministry of Science, Research, and Economy, and the National Foundation for Research, Technology, and Development (S.K.). The Swedish Cancer Society, the Knut and Alice Wallenberg Foundation, and the Torsten and Ragnar Söderberg Foundation (T.H.). Swiss National Science Foundation fellows to B.S. (P300P3_147897) and N.K. (P2EZP3_159114), an EMBO long-term Fellowship to G.I.V. (1543-2012), and an Marie-Sklodowska Curie Action fellowship to N.K. (SLIM). Our screening compound libraries are from the NIH clinical collection, gifts from F. Bracher, T. Nielsen, S. Nijman, J. Bradner, The Broad Institute, and Haplogen GmbH. JQ1 was provided by S. Knapp. We thank Manuele Rebsamen, Astrid Fauster, Giorgia Jurisic, Adrián César-Razquin, Cara C. West, Enrico Girardi, Georg Winter for assistance and critical reading of the manuscript, and the G.S.F. laboratory for scientific discussion.
Footnotes
Author Contributions
G.I.V., B.S., N.K. J.W.B., K.V.M.H, C.-H.L., K.S., A.R., U.W.B., M.S. performed the experiments, P.K., U.J. organized clinical samples, S.K., O.L.d.l.F. provided reagents and intellectual contributions, P.K., U.J. W.R.S., T.H., G.S.F., were responsible for human and animal ethical guidelines, G.S.F. oversaw the project, B.S., G.I.V., G.S.F. analyzed the data and wrote the manuscript.
Data Availability:
The RNA sequencing data from Supplementary Dataset 3 can be found on the Gene Expression Omnibus (accession number GSE93124). The protein interactions from this publication, in Supplementary Table 1, have been submitted to the IMEx (http://www.imexconsortium.org) consortium through IntAct and assigned the identifier IM-25603. The datasets integrated for the meta-analysis of MST1R expression in lung cancers (Supplementary Figure 6e) was retrieved from www.oncomine.com (exact data sets are referenced below the figure). The transcription data from ALK-positive H3122 cells resistant to crizotinib treatment was previously published and obtained from NCBI Gene Expression Omnibus (GEO) accession number GSE49508.
Competing Financial Interests:
The spatial screening and interaction score for use in immunomodulatory drug discovery is patent pending with G.I.V., B.S., N.K., G.S.F. listed as inventors, WO2016046346. The patents are licensed to Allcyte GmbH (Vienna, Austria), which G.I.V., B.S., N.K., and G.S.F. have co-founded.
References
- 1.Snijder B, et al. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 2009;461:520–523. doi: 10.1038/nature08282. [DOI] [PubMed] [Google Scholar]
- 2.Germain RN, Robey EA, Cahalan MD. A decade of imaging cellular motility and interaction dynamics in the immune system. Science. 2012;336:1676–81. doi: 10.1126/science.1221063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14:561–84. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 4.Hoos A. Development of immuno-oncology drugs - from CTLA4 to PD1 to the next generations. Nat Rev Drug Discov. 2016;15:235–47. doi: 10.1038/nrd.2015.35. [DOI] [PubMed] [Google Scholar]
- 5.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 6.Roche PA, Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat Rev Immunol. 2015;15:203–16. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou W, et al. Viral infection triggers rapid differentiation of human blood monocytes into dendritic cells. Blood. 2012;119:3128–3131. doi: 10.1182/blood-2011-09-379479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Germain RN, Stefanova I. The dynamics of T cell receptor signaling: complex orchestration and the key roles of tempo and cooperation. Annu Rev Immunol. 1999;17:467–522. doi: 10.1146/annurev.immunol.17.1.467. [DOI] [PubMed] [Google Scholar]
- 9.Wu LC, Tuot DS, Lyons DS, Garcia KC, Davis MM. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 2002;418:552–6. doi: 10.1038/nature00920. [DOI] [PubMed] [Google Scholar]
- 10.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 11.Lolekha S, Dray S, Gotoff SP. Macrophage aggregation in vitro: a correlate of delayed hypersensitivity. J Immunol. 1970;104:296–304. [PubMed] [Google Scholar]
- 12.Maloney DG, Smith B, Rose A. Rituximab: mechanism of action and resistance. Semin Oncol. 2002;29:2–9. doi: 10.1053/sonc.2002.30156. [DOI] [PubMed] [Google Scholar]
- 13.Loffler A, et al. Efficient elimination of chronic lymphocytic leukaemia B cells by autologous T cells with a bispecific anti-CD19/anti-CD3 single-chain antibody construct. Leukemia. 2003;17:900–9. doi: 10.1038/sj.leu.2402890. [DOI] [PubMed] [Google Scholar]
- 14.Topp MS, et al. Safety and activity of blinatumomab for adult patients with relapsed or refractory B-precursor acute lymphoblastic leukaemia: a multicentre, single-arm, phase 2 study. Lancet Oncol. 2015;16:57–66. doi: 10.1016/S1470-2045(14)71170-2. [DOI] [PubMed] [Google Scholar]
- 15.Honke N, et al. Enforced viral replication activates adaptive immunity and is essential for the control of a cytopathic virus. Nat Immunol. 2012;13:51–7. doi: 10.1038/ni.2169. [DOI] [PubMed] [Google Scholar]
- 16.Hu X, Li WP, Meng C, Ivashkiv LB. Inhibition of IFN-gamma signaling by glucocorticoids. J Immunol. 2003;170:4833–9. doi: 10.4049/jimmunol.170.9.4833. [DOI] [PubMed] [Google Scholar]
- 17.Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365–76. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- 18.Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015;15:511–23. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harizi H, Corcuff JB, Gualde N. Arachidonic-acid-derived eicosanoids: roles in biology and immunopathology. Trends Mol Med. 2008;14:461–9. doi: 10.1016/j.molmed.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Ben-Zvi I, Kivity S, Langevitz P, Shoenfeld Y. Hydroxychloroquine: from malaria to autoimmunity. Clin Rev Allergy Immunol. 2012;42:145–53. doi: 10.1007/s12016-010-8243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson HA, Hiltbold EM, Roche PA. Concentration of MHC class II molecules in lipid rafts facilitates antigen presentation. Nat Immunol. 2000;1:156–62. doi: 10.1038/77842. [DOI] [PubMed] [Google Scholar]
- 22.Komaniwa S, et al. Lipid-mediated presentation of MHC class II molecules guides thymocytes to the CD4 lineage. Eur J Immunol. 2009;39:96–112. doi: 10.1002/eji.200838796. [DOI] [PubMed] [Google Scholar]
- 23.Weitz-Schmidt G, et al. Statins selectively inhibit leukocyte function antigen-1 by binding to a novel regulatory integrin site. Nat Med. 2001;7:687–92. doi: 10.1038/89058. [DOI] [PubMed] [Google Scholar]
- 24.Schindler R, Dinarello CA, Koch KM. Angiotensin-converting-enzyme inhibitors suppress synthesis of tumour necrosis factor and interleukin 1 by human peripheral blood mononuclear cells. Cytokine. 1995;7:526–33. doi: 10.1006/cyto.1995.0071. [DOI] [PubMed] [Google Scholar]
- 25.Flaishon L, et al. Expression of the chemokine receptor CCR2 on immature B cells negatively regulates their cytoskeletal rearrangement and migration. Blood. 2004;104:933–41. doi: 10.1182/blood-2003-11-4013. [DOI] [PubMed] [Google Scholar]
- 26.Rice-Evans CA, Miller NJ, Bolwell PG, Bramley PM, Pridham JB. The relative antioxidant activities of plant-derived polyphenolic flavonoids. Free Radic Res. 1995;22:375–83. doi: 10.3109/10715769509145649. [DOI] [PubMed] [Google Scholar]
- 27.Khajanchi BK, Kirtley ML, Brackman SM, Chopra AK. Immunomodulatory and protective roles of quorum-sensing signaling molecules N-acyl homoserine lactones during infection of mice with Aeromonas hydrophila. Infect Immun. 2011;79:2646–57. doi: 10.1128/IAI.00096-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritchie AJ, et al. The Pseudomonas aeruginosa quorum-sensing molecule N-3-(oxododecanoyl)-L-homoserine lactone inhibits T-cell differentiation and cytokine production by a mechanism involving an early step in T-cell activation. Infect Immun. 2005;73:1648–55. doi: 10.1128/IAI.73.3.1648-1655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elenkov IJ, Wilder RL, Chrousos GP, Vizi ES. The sympathetic nerve--an integrative interface between two supersystems: the brain and the immune system. Pharmacol Rev. 2000;52:595–638. [PubMed] [Google Scholar]
- 30.Alam S, Laughton DL, Walding A, Wolstenholme AJ. Human peripheral blood mononuclear cells express GABAA receptor subunits. Mol Immunol. 2006;43:1432–42. doi: 10.1016/j.molimm.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 31.Bhat R, et al. Inhibitory role for GABA in autoimmune inflammation. Proc Natl Acad Sci U S A. 2010;107:2580–5. doi: 10.1073/pnas.0915139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian J, Yong J, Dang H, Kaufman DL. Oral GABA treatment downregulates inflammatory responses in a mouse model of rheumatoid arthritis. Autoimmunity. 2011;44:465–70. doi: 10.3109/08916934.2011.571223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders VM. The beta2-adrenergic receptor on T and B lymphocytes: do we understand it yet? Brain Behav Immun. 2012;26:195–200. doi: 10.1016/j.bbi.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loza MJ, Foster S, Peters SP, Penn RB. Beta-agonists modulate T-cell functions via direct actions on type 1 and type 2 cells. Blood. 2006;107:2052–60. doi: 10.1182/blood-2005-08-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heine A, Held SA, Bringmann A, Holderried TA, Brossart P. Immunomodulatory effects of anti-angiogenic drugs. Leukemia. 2011;25:899–905. doi: 10.1038/leu.2011.24. [DOI] [PubMed] [Google Scholar]
- 36.Santoni M, et al. Role of natural and adaptive immunity in renal cell carcinoma response to VEGFR-TKIs and mTOR inhibitor. Int J Cancer. 2014;134:2772–7. doi: 10.1002/ijc.28503. [DOI] [PubMed] [Google Scholar]
- 37.Cui JJ, et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK) J Med Chem. 2011;54:6342–63. doi: 10.1021/jm2007613. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi KS, van den Elsen PJ. NLRC5: a key regulator of MHC class I-dependent immune responses. Nat Rev Immunol. 2012;12:813–20. doi: 10.1038/nri3339. [DOI] [PubMed] [Google Scholar]
- 39.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–14. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno CS, Beresford GW, Louis-Plence P, Morris AC, Boss JM. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–51. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- 41.Huber KVM, et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature. 2014;508:222–227. doi: 10.1038/nature13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis MI, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–51. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 43.Christensen JG, et al. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther. 2007;6:3314–22. doi: 10.1158/1535-7163.MCT-07-0365. [DOI] [PubMed] [Google Scholar]
- 44.Wilson CB, et al. The RON receptor tyrosine kinase regulates IFN-gamma production and responses in innate immunity. J Immunol. 2008;181:2303–10. doi: 10.4049/jimmunol.181.4.2303. [DOI] [PubMed] [Google Scholar]
- 45.Eyob H, et al. Inhibition of ron kinase blocks conversion of micrometastases to overt metastases by boosting antitumor immunity. Cancer Discov. 2013;3:751–60. doi: 10.1158/2159-8290.CD-12-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mallakin A, et al. Gene expression profiles of Mst1r-deficient mice during nickel-induced acute lung injury. Am J Respir Cell Mol Biol. 2006;34:15–27. doi: 10.1165/rcmb.2005-0093OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroeder GM, et al. Discovery of N-(4-(2-amino-3-chloropyridin-4-yloxy)-3-fluorophenyl)-4-ethoxy-1-(4-fluorophenyl )-2-oxo-1,2-dihydropyridine-3-carboxamide (BMS-777607), a selective and orally efficacious inhibitor of the Met kinase superfamily. J Med Chem. 2009;52:1251–4. doi: 10.1021/jm801586s. [DOI] [PubMed] [Google Scholar]
- 48.Lovly CM, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med. 2014;20:1027–34. doi: 10.1038/nm.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10:130–7. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 51.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–13. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 52.Winter GE, et al. Systems-pharmacology dissection of a drug synergy in imatinib-resistant CML. Nature chemical biology. 2012;8:905–912. doi: 10.1038/nchembio.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rämö P, Sacher R, Snijder B, Begemann B, Pelkmans L. CellClassifier: supervised learning of cellular phenotypes. Bioinformatics (Oxford, England) 2009;25:3028–3030. doi: 10.1093/bioinformatics/btp524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.