Abstract
Understanding the molecular mechanisms underlying coffee-pathogen interactions are of key importance to aid disease resistance breeding efforts. In this work the expression of genes involved in salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) pathways were studied in hypocotyls of two coffee varieties challenged with the hemibiotrophic fungus Colletotrichum kahawae, the causal agent of Coffee Berry Disease. Based on a cytological analysis, key time-points of the infection process were selected and qPCR was used to evaluate the expression of phytohormones biosynthesis, reception and responsive-related genes. The resistance to C. kahawae was characterized by restricted fungal growth associated with early accumulation of phenolic compounds in the cell walls and cytoplasmic contents, and deployment of hypersensitive reaction. Similar responses were detected in the susceptible variety, but in a significantly lower percentage of infection sites and with no apparent effect on disease development. Gene expression analysis suggests a more relevant involvement of JA and ET phytohormones than SA in this pathosystem. An earlier and stronger activation of the JA pathway observed in the resistant variety, when compared with the susceptible one, seems to be responsible for the successful activation of defense responses and inhibition of fungal growth. For the ET pathway, the down or non-regulation of ET receptors in the resistant variety, together with a moderate expression of the responsive-related gene ERF1, indicates that this phytohormone may be related with other functions besides the resistance response. However, in the susceptible variety, the stronger activation of ERF1 gene at the beginning of the necrotrophic phase, suggests the involvement of ET in tissue senescence. As far as we know, this is the first attempt to unveil the role of phytohormones in coffee-C. kahawae interactions, thus contributing to deepen our understanding on the complex mechanisms of plant signaling and defense.
Introduction
Coffee Berry Disease (CBD), caused by the hemibiotrophic fungus Colletotrichum kahawae J.M. Waller & P.D. Bridge, is a major constraint of Arabica coffee production in Africa. This disease may cause up to 50–80% of crop losses, in years of severe epidemics if control measures are not applied [1]. Since the first report in 1922 in Kenya [2], CBD is still restricted to Africa but there are major concerns about the risk of its introduction into Latin America and Asia [1,3]. C. kahawae infects several coffee organs, but major losses result from the infection of green berries. The outbreak of the disease with visible symptoms occurs during the expanding stage of berry development, producing black, sunken anthracnose-like lesions on the green pulp [4,5]. Although the application of fungicides can provide adequate control, the use of coffee resistant varieties is the most appropriate and sustainable management strategy against this disease.
Inheritance studies carried out in Kenya by van der Vossen and Walyaro (1980) [6] and recent molecular studies [7,8] provided evidences that coffee resistance to C. kahawae appears to be controlled by major genes in different loci. Cytological and biochemical studies revealed that coffee resistance to C. kahawae is characterized by restricted fungal growth associated with several host responses, such as hypersensitive-like cell death (HR), formation of cork barriers, callose deposition around intracellular hyphae, accumulation of phenolic compounds (flavonoids and hydroxycinnamic acid derivatives), lignification of host cell walls and increased activity of oxidative enzymes, such as peroxidases [3,9–13]. More recently, differentially expressed genes involved in recognition, signaling and defense responses of coffee to C. kahawae have been identified [14–16].
Plant growth and responses to environmental cues are largely governed by phytohormones. Recent research indicates that an antagonist/synergetic crosstalk among different phytohormones [particularly salicylic acid (SA), jasmonic acid (JA) and ethylene (ET)] play a central role in the regulation of plant immune responses to the pathogen [17,18]. These defense responses are considered to be dependent on the pathogen lifestyle and the genetic constitution of the host [19–22].
In plants, SA can be synthesized via two distinct enzymatic pathways that require the primary metabolite chorismate: phenylalanine ammonia-lyase (PAL)-mediated phenylalanine and isochorismate synthase (ICS)-mediated isochorismate. The NPR1 (non-expressor of pathogenesis-related genes 1) represents a key node in signaling downstream from SA (S1A Fig) [23]. In the absence of SA or pathogen challenge, NPR1 is inactive in cytoplasm as an oligomer. Upon induction, SA accumulation is promoted and NPR1 monomer is released to enter the nucleus where it activates defense gene transcription, such as the PR1 gene [24].
JA biosynthesis starts with the release of α-linolenic acid (α-LA) from membrane lipids and its oxygenation in the chloroplast, followed by the sequential action of allene oxide synthase (AOS) and allene oxide cyclase (AOC), resulting in the synthesis of 12-oxophytodienoic acid (OPDA) (S1B Fig) [25,26]. OPDA migrates to the peroxisome to be reduced by oxophytodienoate redutase 3 (OPR3) and undergo several rounds of β-oxidation to form JA. Then, JA is exported from the peroxisome to cytosol for conjugation to the L-isoleucine (Ile) by jasmonate-resistant 1 (JAR1) resulting in the endogenous bioactive form of JA-Ile [27,28]. JA-Ile then interacts with coronatine insensitive 1 (COI1), an F-box protein which acts as a JA receptor, and targets jasmonate negative regulators [like jasmonate zim domain proteins (JAZ)] for degradation, promoting JA-induced gene transcription such as of PR10 [25,29].
Ethylene is produced from methionine via S-adenosyl-L-methionine (SAM) being the last two biosynthetic steps catalyzed by aminocyclopropane-1-carboxylic synthase (ACS) and ACC oxidase (ACO) (S1C Fig) [30–32]. In the presence of ET, the receptor ethylene resistant 1 (ETR1) binds to the hormone switching off the constitutive triple response 1 (CTR1). Consequently, desphosphorylation of C-terminal end of ethylene insensitive 2 (EIN2) is promoted and the repression of the transcriptional signaling cascade is unlocked [33].
Despite the recent advances achieved on the complex regulation of phytohormones network in plants under different environmental conditions [34–36], this knowledge is still very scarce in plant-Colletotrichum spp. pathosystems, particularly in the coffee-C. kahawae interaction. The aim of this study was to elucidate the possible involvement of the phytohormones SA, JA and ET in the responses of coffee plants resistant (variety Catimor 88) and susceptible (variety Caturra) to C. kahawae. Based on a cytological analysis of the fungal growth and the associated host responses, time-points of the infection process were chosen to evaluate the expression of phytohormones biosynthesis, reception and responsive-related genes by quantitative real-time PCR (qPCR).
Material and methods
Plant material and inoculation
Experimental assays were conducted on coffee hypocotyls, since previous studies shown a correlation between the pre-selection test on hypocotyls and mature plant resistance in the field (r = 0.73–0.80) [37].
For this study, the varieties Catimor 88 (Timor hybrid derivative, which exhibit field resistance to C. kahawae breeding programmes in Kenya) and Caturra (CIFC 19/1 –Coffea arabica L.) were used as resistant and susceptible varieties to C. kahawae isolate Que2 (from Kenya), respectively. Coffee seeds were sown in a mixture of soil:peat:sand (1:1:1) and grown under greenhouse conditions with average temperatures between 16°C and 28°C (minimum and maximum temperatures, respectively) during 8 weeks.
Conidia of C. kahawae isolate Que2, retrieved from a C. kahawae, collection maintained at CIFC/ISA were produced after 7 days at 22°C on malt extract agar (MEA) [38]. Hypocotyls of Catimor 88 and Caturra, were inoculated according to the technique described by van der Vossen et al., (1976) [37] with slight modifications. Briefly, hypocotyls were vertically placed on plastic trays containing a wet nylon sponge and sprayed with a conidia suspension (3x106/ml). The trays were then covered with a plastic bag to simulate a humid chamber and were kept in a Phytotron750 E at 22°C. For the first 24 hours post inoculation (hpi), trays were kept in the dark and afterwards a 12 hours photoperiod was established for the remaining time-course of the experiment. Non-inoculated hypocotyls sprayed with water were kept in the same conditions as the inoculated hypocotyls and used as control.
Light microscopic observation in fresh tissues
Conidial germination and appressorial differentiation were observed on hypocotyl pieces (5cm2) at 3, 6, 9, 12, 15, 18 and 24hpi, as previously described [39]. The hypocotyl pieces were painted with transparent nail polish on the inoculated surface to recreate a tissue surface replica. Once dried, the nail polish was removed, stained and mounted in lactophenol cotton blue. For each experiment, a minimum of six microscope fields, each containing 100 conidia and/or differentiated appressoria on the surface of hypocotyls, were used. To evaluate fungal post-penetration growth stages at 24, 48 and 72hpi, cross sections of inoculated hypocotyl fragments made with a freezing microtome (Leica CM1850), were stained and mounted in lactophenol cotton blue [11,39]. Hyphal length inside hypocotyl tissues were estimated with the aid of a micrometric eyepiece.
To detect autofluorescent cells, cross sections of non-inoculated (control) and inoculated tissues were placed in 0.07M pH 8.9 phosphate solution (K2HPO4) for 5 min, and mounted in the same solution [11,40]. Autofluorescence under epifluorescence blue light is thought to indicate the presence of phenolic-like compounds and cytoplasmic autofluorescence and/or browning is frequently associated with plant cell death [41,42]. Observations were made using light microscopes (LeitzDialux 20 and Leica DM-2500) equipped with a mercury bulb HB 100W blue light (excitation filter BP 450 and 490; barrier filter LP 515).
Data on pre- and post-penetration fungal growth stages and host cell responses are presented as the combined values of three experiments. Fungal growth inside host tissues and plant responses were recorded from 75 to 100 infection sites per experiment at three time-points (24, 48 and 72hpi).
RNA extraction and cDNA synthesis
Hypocotyls (inoculated and non-inoculated) were harvested at 6, 12, 24, 48 and 72hpi, immediately frozen in liquid nitrogen and stored at -80°C prior to RNA extraction. Three biological replicates, each representing a pool of 15 hypocotyls, were used.
Total RNA was extracted with the Spectrum Plant Total RNA kit (Sigma-Aldrich, USA), according to the manufacturer’s instructions. Residual genomic DNA was digested with DNase I (On-columm DNase I Digestion Set, Sigma-Aldrich, USA). RNA purity and concentration was measured at 260/280 nm and 260/230 nm using a spectrophotometer (NanoDrop 1000, Thermo Scientific). RNA integrity was verified by electrophoresis in 1% agarose gel. Genomic DNA (gDNA) contamination was checked by qPCR analysis on the crude RNA [43]. Complementary DNA (cDNA) was synthesized from 2.5μg of total RNA using RevertAid®H Minus Reverse Transcriptase (Fermentas, Ontario, Canada) anchored with Oligo(dT) 23primer (Fermentas, Ontario, Canada), according to manufacturer’s instructions.
Primer design
Within the targeted phytohormone pathways, sixteen genes related with biosynthesis, reception and responsiveness [25,33,44] were selected for expression analysis in Coffea spp. after C. kahawae challenge (Table 1). For SA, the following genes were included: Isochlorismate synthase 2 (ICS2); Phenylalanine ammonia-lyase (PAL); Non-expressor of pathogenesis-related 1 (NPR1); Pathogenesis-related (PR1). For JA the following genes were included: 12-oxoplytodienotae reductase 1-like (OPR3); Coronatine insensitive 1 (COI1); Pathogenesis-related 10 (PR10). For ET the following genes were included: 1-aminocyclepropane-1-carboxylic acid synthase 5 (ACS5); 1-aminocyclepropane-1-carboxylic acid oxidase 2 (ACO2); Ethylene resistant 1 (ETR1); Ethylene insensitive 2 (EIN2); Constitutive triple response 1 (CTR1); Ethylene-responsive factor 1 (ERF1). With the exception of PAL and PR10 genes, that were previously described in coffee [45], the remaining fourteen genes were retrieved from a coffee RNA-seq database [46] as being orthologous of previously described genes in Arabidopsis thaliana (TAIR database:www.arabidopsis.org). Tubulin beta-9 (β-Tub9)/ ribosomal protein S24 (S24) and Insuline Degrading Enzyme (IDE)/S24 were used as reference genes for susceptible and resistant varieties samples, respectively [14]. Coffee specific primers (Table 1) were designed with PrimerSelect version 5.0 (DNAStar, Inc., USA) using the following parameters: amplicon length 70 and 200 bp; size between 17 and 22 bp; annealing temperature (Ta) between 58 and 62°C and GC content± 50%.
Table 1. Primer sequences for qPCR analysis of target and reference genes.
| Gene | Coffee source name |
Name | Primer sequence (5'-3') | Primer length (bp) | Amplicon length (bp) | Ta (°C) | Tm (°C) | PCR efficiency |
|---|---|---|---|---|---|---|---|---|
| ICS2 | Scaffold21359 | Isochorismate synthase 2 | Fw: TGCCATAGTACGAGAAAACA | 20 | 124 | 60 | 79.0 | 94 |
| Rev: CCCAGAAAATCGACCATAAA | 20 | |||||||
| PALa | CaPAL F 13097 | Phenylalanine ammonia-lyase | Fw: GCAGGTCCTACTCATTGTACAAG | 23 | 166 | 60 | 82.0 | 89 |
| Rev: CCATTCCACTCTTTCAAACAATCC | 24 | |||||||
| NPR1 | Scaffold33187 | Non-expressor of PR1 | Fw: AGGGCATTGGATTCTGACGA | 20 | 126 | 60 | 81.5 | 88 |
| Rev: CTCTGTTGTGGTCTTTGCGT | 20 | |||||||
| PR1 | Scaffold170607 | Pathogenesis-related 1 | Fw: GCCCGTAAAGTCACCTGT | 18 | 177 | 60 | 86.5 | 91 |
| Rev: AACTACGCTGCCAAAATC | 18 | |||||||
| OPR3 | Scaffold2739 | 12-oxoplytodienotae reductase 1-like | Fw: ATAACTCCCCACCTTCCAAC | 20 | 198 | 58 | 81.5 | 91 |
| Rev: ACAGCCTTATCCCAACTCTAT | 22 | |||||||
| COI1 | Scaffold40077 | Coronatine insensitive 1 | Fw: CTTAGCATCACCACCCACC | 19 | 157 | 62 | 81.5 | 93 |
| Rev: TCCGATCCCCCATACCAAC | 19 | |||||||
| PR10b | CF589103 | Pathogenesis-related 10 | Fw: GCCACCATCCTTGAAGAGAA | 20 | 151 | 55 | 80.0 | 99 |
| Rev: CAACTCTCTGCTTGGCAGTCT | 21 | |||||||
| ACS5 | Scaffold82864 | 1-aminocyclepropane-1-carboxylic | Fw: AGGGCGTCCTGGTCACTAA | 19 | 148 | 60 | 83.0 | 90 |
| acid synthase 5 | Rev: CTCGGCGAGCTAAAAACTGT | 20 | ||||||
| ACO2 | Scaffold57328 | 1-aminocyclepropane-1-carboxylic | Fw: AAAGTCAGCAATTACCCTCCA | 21 | 144 | 58 | 87.0 | 93 |
| acid oxidase 2 | Rev: ATCCACCCATTCACCATCCT | 20 | ||||||
| ETR1 | Scaffold776 | Ethylene resistant 1 | Fw: GCCCCCAAGATATTCCTAAG | 20 | 92 | 60 | 79.5 | 89 |
| Rev: TGCAAGACCAAGACCACTAC | 20 | |||||||
| EIN2 | Scaffold3828 | Ethylene insensitive 2 | Fw: GTTACTTCTCCAAAACCTACT | 21 | 134 | 60 | 78.5 | 93 |
| Rev: TCCCATTTACCACTCTTATCT | 21 | |||||||
| CTR1 | Scaffold16054 | Constitutive triple response 1 | Fw: GCAGCTGTGGGTTTCAAGG | 19 | 162 | 60 | 83.0 | 90 |
| Rev: AGTGGGGGAGGGTTTAGTC | 19 | |||||||
| ERF1 | C312112 | Ethylene-responsive factor 1 | Fw: TGGCTGGGCACATTTGAC | 18 | 85 | 58 | 84.0 | 90 |
| Rev: GGATTGCTGCTTGACCTC | 18 | |||||||
| IDEc | isotig10635 | Insuline Degrading Enzyme | Fw: TGATCTAAGCTGGTGGAAAGC | 21 | 91 | 55 | 76.3 | 99 |
| Rev: TCAGGTGCATCAGGATGATT | 20 | |||||||
| S24c | SNG-U349723* | Ribosomal protein S24 | Fw: GCCCAAATATCGGCTTATCA | 20 | 92 | 60 | 7.6 | 95 |
| Rev: TCTTCTTGGCCCTGTTCTTC | 20 | |||||||
| β-Tub9c | isotig08544 | Tubulin beta-9 | Fw: ACCCTCCAGCAAACTGATGA | 20 | 100 | 55 | 77.3 | 92 |
| Rev: AGGATGCCACTGCTGATGAT | 20 |
Quantitative real-time PCR
The qPCR experiments were carried out using SYBR Green Supermix (BioRad) in an iQ5 real-time thermal cycler (Bio-Rad, USA). Each 25μl reaction comprised 4μl cDNA template (2.5μg/μl), 12.5μl SYBR Green Supermix (Bio-Rad, USA), 0.4μl of each primer (10μM) and 0.7μl of sterile distilled water. Thermal cycling started with a denaturation step at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15s and annealing at the respective temperature for each gene (Table 1) for 30s. Each set of reactions included a negative control with no template. The amplification efficiency for each gene of interest was determined using the LinRegPCR version 2013.0. Dissociation curves (S2 Fig) and agarose gel electrophoresis were used to analyze non-specific PCR products. Three biological replicates and two technical replicates were used for each sample. Relative gene expression (fold change) was calculated according to Hellemans et al., (2007) [47]. The gene expression data were further visualized using the software MeV viewer (http://www.tm4.org).
Statistical analysis
For statistical analysis of cytological data, Student’s t was applied using IBM®SPSS® Statistics version 20.0 (SPSS Inc., USA) software and arcsine-transformed percentages were used where appropriate. Statistical significance (p≤0.05) of gene expression between the two coffee varieties was determined by the non-parametric Mann–Whitney U test using IBM®SPSS® Statistics version 20.0 (SPSS Inc., USA) software.
Results
Fungal infection and host responses
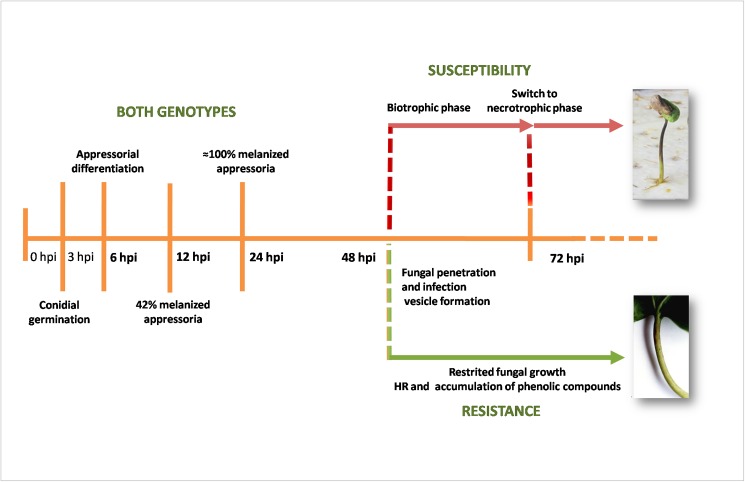
The development of pre-penetration fungal growth stages of C. kahawae was similar in both resistant and susceptible varieties (Fig 1 and Table 2). Conidial germination and appressorial differentiation were initiated at 3 and 6hpi, respectively. At 12hpi, 42% of the appressoria were melanized reaching 89–94% at 24hpi (Fig 2A). In both coffee varieties, the melanized appressoria began to penetrate the epidermal cells with the formation of a globose infection vesicle, at 48hpi (Fig 2B). Hyphae developed from the vesicles grew either intra- and intercellularly colonizing epidermal and cortex cells. In the susceptible variety, the fungus pursued its growth feeding on living host cells (biotrophy) before switching to necrotrophy (72hpi) in the majority of the infection sites (about 90%) (Fig 2C and 2D). Resistance was characterized by restricted fungal growth (fungal hyphae were more frequently confined to the epidermal cells or occasionally to those of the first layer of the cortex cells), as confirmed by the evaluation of hyphal length (Table 3) (Fig 2E and 2F). The entanglement of fungal hyphae originating from different infection sites, together with the increasing number of necrotic host cells in susceptible tissues, did not allow the quantification of the hyphal length beyond 72hpi.
Fig 1. Time-course of Colletotrichum kahawae (isolate Que2) infection in hypocotyls of resistant (Catimor 88) and susceptible (Caturra) coffee varieties.
3hpi and 6hpi–beginning of conidia germination and appressoria differentiation, respectively; 12hpi– 42% of melanized appressoria; 24hpi–almost 100% of melanized appressoria; 48hpi—fungal penetration and biotrophic growth; 72hpi–switch to necrotrophy (susceptible variety) and restriction of fungal growth (resistant variety). Times after inoculation (bold) were selected to collect samples for gene expression studies by qPCR.
Table 2. Percentage of conidial germination and appressorial differentiation of Colletotrichum kahawae on hypocotyls of resistant and susceptible coffee varieties, at different hours post inoculation.
| Hours post inoculation | Coffee varieties | Germinated conidia (%) (x ± SD) | t test* | Appressoria** (%) (x ± SD) |
t test* | Melanized appressoria (%) (x ± SD) |
t test* |
|---|---|---|---|---|---|---|---|
| 3 h | Catimor 88 (R) | 3±3 | 0.15 ns | 0 | - | 0 | - |
| Caturra (S) | 4±4 | 0 | 0 | ||||
| 6h | Catimor 88 (R) | 16±13 | 0.90ns | 43±26 | 0,32ns | 1±1 | 0.37ns |
| Caturra (S) | 16±33 | 46±33 | 2±2 | ||||
| 9h | Catimor 88 (R) | 26±9 | 0.35ns | 65±16 | 0.92ns | 17±12 | 0.06ns |
| Caturra (S) | 29±15 | 65±36 | 9±9 | ||||
| 12h | Catimor 88 (R) | 36±33 | 0.32ns | 69±18 | 0.34ns | 42±11 | 0.92ns |
| Caturra (S) | 33±26 | 76±27 | 42±31 | ||||
| 15h | Catimor 88 (R) | 50±18 | 0.34ns | 78±11 | 0.94ns | 49±17 | 0.34ns |
| Caturra (S) | 57±17 | 79±8 | 56±26 | ||||
| 18h | Catimor 88 (R) | 61±18 | 0.36ns | 87±13 | 0.95ns | 72±20 | 0.95ns |
| Caturra (S) | 65±24 | 88±10 | 71±23 | ||||
| 24h | Catimor 88 (R) | 71±22 | 0.96ns | 89±14 | 1,16ns | 89±11 | 1.16ns |
| Caturra (S) | 72±20 | 94±6 | 94±6 |
R = Resistant; S = Susceptible; X ± SD = mean ± standard deviation
* Student’s t test (ns—non significative)
** Total of appressoria (nonmelanized and melanized)
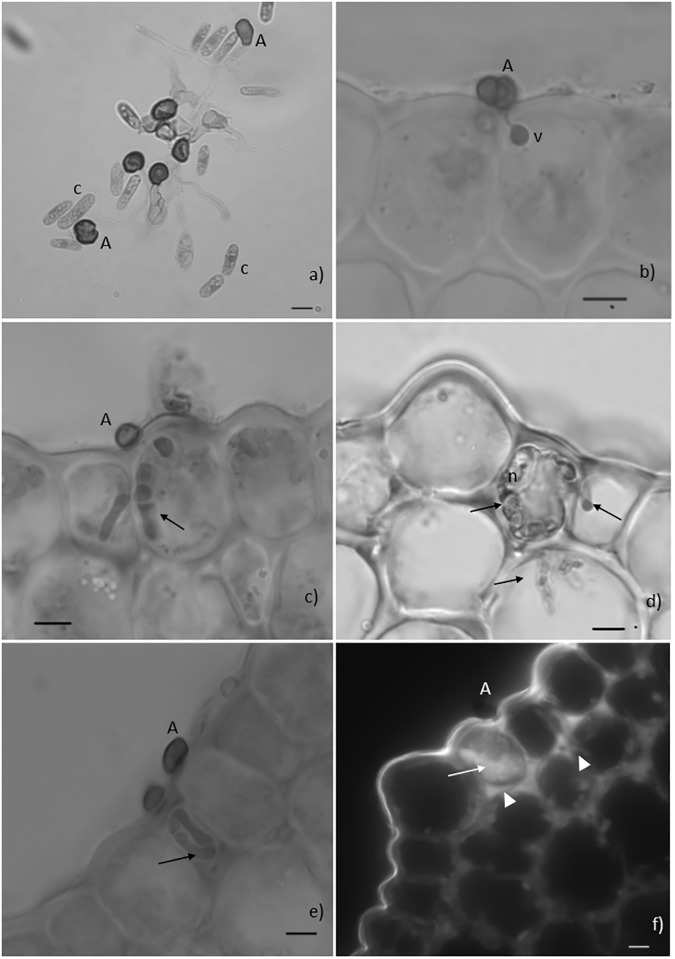
Fig 2. Fungal pre-and post-penetration growth stages and host responses.
Light microscope observations, cotton blue lactophenol staining (2a-2e) and epifluorescence test under blue light (2f). Fig 2a. Conidia (c) germination and formation of melanized appressoria (A) on the surface of a resistant hypocotyl, 24 hours post inoculation (hpi). Fig 2b. Infection site showing a melanized appressorium (A) and an infection vesicle (v) in the epidermal cell of the resistant hypocothyl, 48hpi. Fig 2c. Infection site showing a melanized appressorium (A) and hyphae inside two adjacent epidermal cells of the susceptible hypocotyl (arrow), 48hpi. Fig 2d. Fungal hyphae (arrows) in living and in necrotized (n) cells of the susceptible hypocotyl, 72hpi. Fig 2e. Infection site showing a melanized appressorium (A) and intracellular hyphae (arrow) confined to the epidermal cell of the resistant hypocotyl, 72hpi. Fig 2f. Infection site showing an appressorium (A) associated with autofluorescence of the cytoplasmic content of one epidermal cell (HR-like). Note that the walls of this cell and of adjacent epidermal and cortex cells are also autofluorescent (bars = 10μm).
Table 3. Evaluation of fungal growth in coffee hypocotyls of resistant and susceptible coffee varieties after challenge with Colletotrichum kahawae, at different hours post inoculation.
| Hyphal length (μm)/infection site in hypocotyls of coffee varieties | |||
|---|---|---|---|
| Hours post inoculation | Catimor 88 (R) x± SD | Caturra (S) x± SD | t test* |
| 24 | 0 | 0 | _ |
| 48 | 3.94±2.11 | 8.43±8.22 | 1.49ns |
| 72 | 12.76±8.81 | 42.6±32.52 | 4.25*** |
R = Resistant; S = Susceptible; X ± SD, mean ± standard deviation
* Student’s t-test (ns—non significative
***p≤0.001)
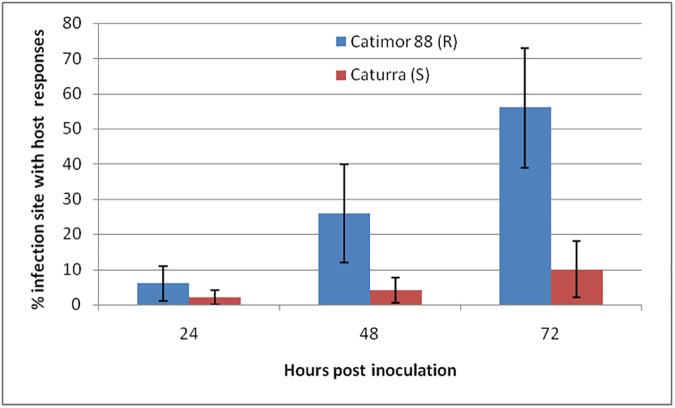
In the resistant variety, the first cytological changes were displayed in the epidermal cells at 24hpi and corresponded to: (i) accumulation of phenolic-like compounds [indicated by autofluorescence (AF) in cell walls only or in cell walls plus the cytoplasmic content]; (ii) deployment of HR (monitored by the AF and/or browning of the cytoplasmic contents) (Figs 2F and 3). During the time course of the infection, these responses spread to adjacent cells of the epidermis and of the first layer of cortex cells. Similar responses were detected in the susceptible variety, but in a significantly lower percentage of infection sites (24hpi: 2%, 48hpi: 4%, 72hpi: 10%) comparatively to the resistant variety (24hpi: 6%, 48hpi: 26%, 72hpi: 56%) (Fig 4). In these infection sites the fungus stopped its growth at the stage of appressoria or infection vesicle.
Fig 3. Percentage of infection sites with host responses.
Accumulation of phenolic-like compounds in the cell walls only, or in both the cell walls and the cytoplasmic contents and deployment of hypersensitive-like cell death (HR) induced by C. kahawae in hypocotyls of coffee varieties Catimor 88 (R-resistant) and Caturra (S-susceptible), at different hours post inoculation. The average percentages were significantly higher in the resistant than in the susceptible coffee variety at 24hpi (t = 2.52; p ≤ 0.05), 48hpi (t = 4.83; p ≤ 0.001) and 72hpi (t = 6.69; p ≤ 0.001).
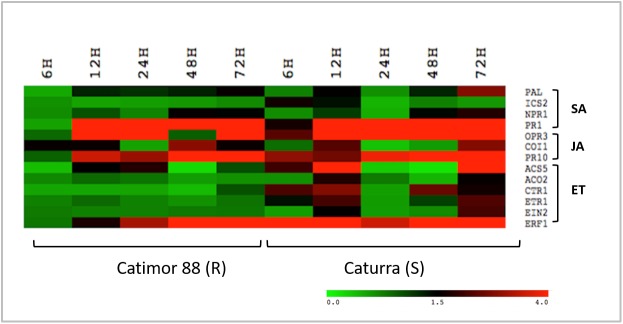
Fig 4. Expression analysis of SA, JA and ET pathway related genes in non-inoculated hypocotyls of Catimor 88 (R-resistant) and Caturra (S-susceptible) vs inoculated hypocotyls with C. kahawae.
Heatmap was colored according to the log2 ratio of expression, where green indicates lower expression, red indicates higher expression and black indicates no expression (see the color scale); in columns are the time-point studied (6, 12, 24, 48 and 72hpi) and in the rows the genes analyzed.
At 5–6 days after inoculation, resistant hypocotyls exhibited scab lesions or, occasionally absence of macroscopic symptoms, whereas susceptible hypocotyls showed typical dark sunken lesions with sporulation (Fig 1).
Expression of genes from SA, JA and ET pathways
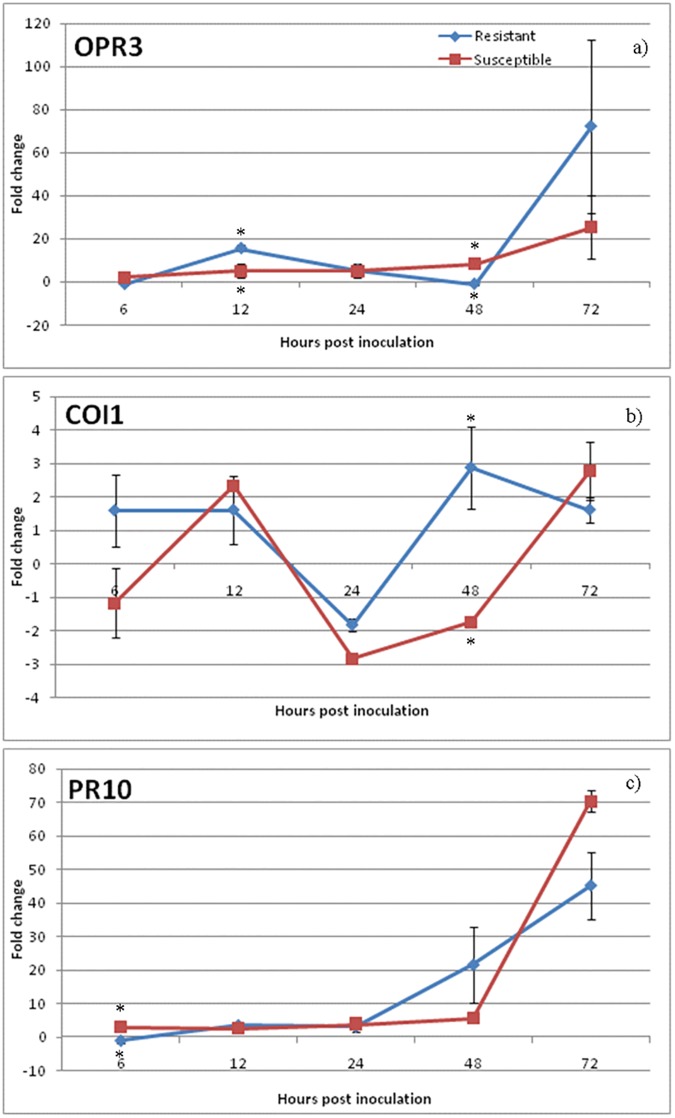
Phytohormone pathway induction during Coffea spp.-C. kahawae interaction was monitored by the expression analysis of sixteen related genes. For a global perspective of the relative expression levels of SA, JA and ET pathway-related genes in the resistant and susceptible varieties along the infection process, a heatmap analysis was performed (Fig 4). The expression patterns of SA pathway-related genes were quite similar for both coffee varieties; being SA biosynthesis and receptor-related genes (ICS2, PAL, NPR1) mostly non-regulated, while PR1 gene was up-regulated from 12hpi onwards (Fig 4 and S3 Fig). On the contrary, the expression level of genes from JA and ET pathways showed differences between the two varieties (Fig 4). The activation of the JA pathway was observed in both coffee varieties, but differently in timing and magnitude (Fig 5). In the susceptible variety, the biosynthesis-related gene OPR3 was up-regulated in all time-points showing the highest level of expression at 72hpi (25.5±14.9). JA-Ile receptor gene COI1 was up-regulated at 12hpi (2.3±0.14) and 72hpi (2.7±0.9), while the responsive–related gene PR10 showed the maximum value of expression at 72hpi (70.3±3.3). In the resistant variety, the up-regulation of the biosynthesis-related gene OPR3 was observed at 12-24hpi (12hpi: 15.5±1.4; 24hpi: 5.5± 3.3) and later at 72hpi (72.2±40.4). The maximum of expression of the JA-Ile receptor gene COI1 was observed at 48hpi (2.8±1.2) which was coincident with the strong increase in expression level of the responsive–related gene PR10 from 48hpi onwards (48hpi: 21.5±11.3; 72hpi:45.2±10.1).
Fig 5. qPCR expression analysis of JA pathway related genes.
Relative expression patterns of OPR3 (biosynthesis), COI1 (receptor) and PR10 (responsive gene) obtained in Catimor 88 (R-resistant) and Caturra (S-susceptible) coffee varieties. Mean and standard deviation of three biological replicates is presented. Fold change as relative expression of gene expression between inoculated and control samples for each of the coffee varieties/inoculation time-points. Asterisks (∗) represent statistical significance (p≤0.05) of gene expression between the two coffee varieties was determined by the non-parametric Mann–Whitney U test using IBM®SPSS® Statistics version 20.0 (SPSS Inc., USA) software.
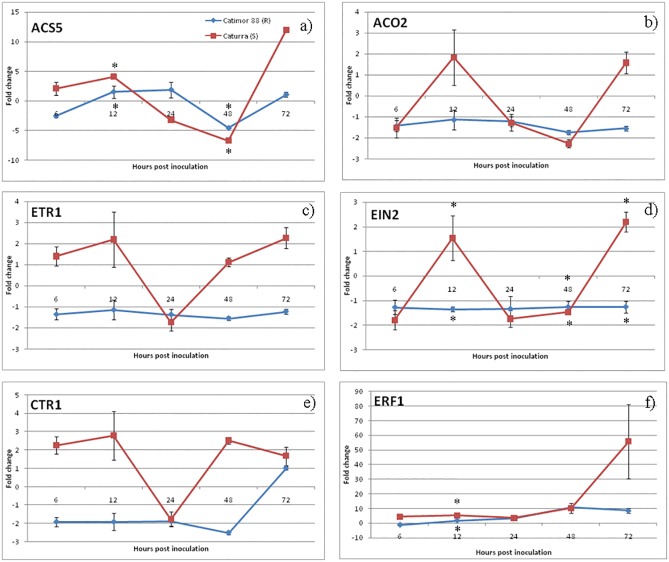
An activation of the ET pathway was also observed in both varieties but with differences in the ET-receptor genes expression profiles (Fig 6). In the susceptible variety, the biosynthesis-related genes ACS5 and ACO2 presented similar expression profiles being both up-regulated at 12hpi (ACS5: 4.1±0.2; ACO2: 1.8±1.3) and 72hpi (ACS5: 12±0.3; ACO2: 1.6±0.5) and down-regulated at 24hpi and 48hpi. ET-receptor gene ETR1 and central ET signaling regulator EIN2, together with the negative ET pathway regulator CTR1 showed similar expression profiles being all up-regulated at 12hpi, and down-regulated at 24hpi followed by an up-regulation at 72hai. ET-responsive gene ERF1 was moderately activated from 6hpi to 48hpi, reaching a maximum value of expression at 72hpi (55.8±25.5). In the resistant variety, the biosynthesis-related gene ACS5 was up-regulated at 12-24hpi (12hpi; 1.5±1.1; 24hpi: 1.8±1.3) while ETR1, EIN2 and CTR1 were down or non-regulated along the infection process. ET-responsive gene ERF1 was moderately activated at all time-points, with an increase in expression level at 48-72hpi (48hpi: 10.7±1.0; 72hpi: 8.5±1.6).
Fig 6. qPCR expression analysis of ET pathway related genes.
Relative expression patterns of ACS5/ACSO2 (biosynthesis), ETR1/EIN2 (receptors), CTR1 (negative regulator) and ERF1 (responsive gene) were obtained in Catimor 88 (R- resistant) and Caturra (S-susceptible) coffee varieties. Mean and standard deviation of three biological replicates is presented. Fold change as relative expression of gene expression between inoculated and control samples for each of the coffee varieties/inoculation time-points. Asterisks (∗) represent statistical significance (p≤0.05) of gene expression between the two coffee varieties was determined by the non-parametric Mann–Whitney U test using IBM®SPSS® Statistics version 20.0 (SPSS Inc., USA) software.
Discussion
In this work, the involvement of phytohormones in the deployment of coffee susceptible and resistance response to C. kahawae was studied. In the two coffee varieties, no differences were observed in fungal development from conidia germination to differentiation of melanized appressoria and fungal penetration. In the majority of the infection sites of the susceptible variety, the fungus pursued its growth without apparent inhibition, first establishing a biotrophic interaction with the host cells and later (at 72pai) switching to a destructive necrotrophic phase, as described for many species of Colletotrichum [48–51]. Conversely, as previously described by Silva et al., (2006) [3] and Loureiro et al., (2012) [11], in the resistant variety the restricted hyphal growth was associated with the hypersensitive-like host cell death (HR), and early accumulation of phenolic compounds both in cell walls and in the cytoplasmic contents. These responses were also observed in the susceptible variety, but in a significantly lower percentage of infection sites and did not prevent the fungal growth, as indicated by the appearance of typical anthracnose symptoms and the presence of acervuli. Time-course experiments carried out by Vargas et al., (2012) [52] also revealed that during the biotrophic growth in susceptible maize leaves, the hemibiotrophic fungus C. graminicola induced classical plant defense responses, such as the accumulation of reactive oxygen species and phenolic compounds. They hypothesized that it is the switch to necrotrophy that enables the fungus to evade the plant immune system and allows pathogenicity.
The classic view on the involvement of phytohormones in plant-pathogen interactions suggests that i) biotrophy is controlled by SA, while necrotrophy/hemibiotrophy is controlled by JA/ET, and that ii) SA is an antagonist of JA /ET [18]. Recent studies challenge these concepts revealing a very complex phytohormone self-regulation and cross-talk [25,53–58]. Indeed, in some reports on plant-hemibiotrophic pathogen interactions, SA signaling seems to be relevant for the outcome of resistance [53,59–63] while in others the role of SA is less clear [64,65]. In the coffee-C. kahawae interactions studied, the low expression of SA biosynthesis and receptor-related genes (ICS2/PAL and NPR1) turns also unclear the role of this phytohormone, although in coffee resistance against the biotrophic fungus Hemileia vastatrix a SA-dependent pathway seems to be involved [66,67]. The hormone-responsive gene PR1 (commonly used as a SA pathway-marker) was unexpectedly induced in both susceptible and resistant coffee varieties when challenged with C. kahawae, but in a greater magnitude in the resistant variety. The apparent lack of coordination between the PR1 expression profile and the other selected SA-pathway markers under study (ICS2, PAL and NPR1) suggests that in this pathosystem, PR1 induction could be mediated by other phytohormones than SA. In accordance, the knockout of OsPAL06I in rice challenged with Magnaporthe oryzae did not influence the expression of PR1a compared to wild-type, although the SA and JA levels were significantly reduced in roots [53]. Tang et al., (2010) [68] reported a differential expression of PR1 and chitinase genes in bananas matured and treated with ethephon (analog to ethylene), demonstrating that benzothiadizole (BTH–analog to SA) failed to induce them. Furthermore, an up-regulation of PR1, among others PRs, was observed after MeJA (methyl jasmonate) treatment [54, 69].
The same authors related the increase of PRs expression with the increase of disease resistance, and identified a high degree of consistency between the concentration fluctuation of endogenous JAs and the expression of PRs, therefore suggesting that accumulation of JAs may be one inner factor for regulation of PRs.
Our results also revealed an induction of the JA pathway in both varieties, however it occurred earlier and/or in a greater magnitude in the resistant than in the susceptible variety. The biosynthesis-related gene OPR3 was induced in both varieties during the appressorial melanization (12-24hpi) and the beginning of host cell responses (24pai). Furthermore, an increase of the expression of JA-responsive gene PR10 was observed at the beginning of fungal penetration (48pai) in the resistant variety, while in the susceptible variety it was coincident with the switch to necrotrophic fungal growth (72pai). In a similar way, Ding et al., (2011) [70] observed that, when wheat was challenged with the hemibiotrophic fungus Fusarium graminearum, only the variety with a high level of resistance, when compared with a susceptible mutant, had an increase in OPR3 activation, concluding that the induction of JA pathway was involved in plant defense. Figueiredo et al., (2015) [25] also observed a significant increase of OPR3 and COI1 expression in the resistant grapevine (Vitis vinifera) cultivar “Regent” at the first hours post inoculation with the biotrophic fungus Plasmopora viticola, when compared with the susceptible cultivar “Trincadeira”, concluding that the timing of JA pathway induction was responsible for a set of efficient defense responses.
In our study, the earlier increase in PR10 expression observed in the resistant variety as early as the fungal penetration stage, may reflect an early attempt of the coffee plant to halt the pathogen development, as is observed in other plant-pathogen interactions [54,71,72]. Although the precise function of PR10 proteins is poorly understood, some exhibit antimicrobial activity, and DNase and/or RNase activity [73,74]. Recent studies also suggest that PR10 may have an important role in the control of phenylpropanoid and flavonoid biosynthesis and their transport to sites where they are needed, such as the reinforcement of the cell wall [75]. This seems to be in line with the accumulation of phenolic-like compounds in the host cell walls found in our study.
The ET pathway seems also to be induced in both coffee varieties however with a remarkable difference in the ET-receptor expression profile, which may reflect the complexity of the role and regulation of this hormone. The ET pathway induction may start at the earliest hours post inoculation, since at least one of the two ET biosynthesis enzymes were found to be up-regulated during the appressorial melanization (12-24hpi) and at the onset of host cell responses (24hpi) in both coffee varieties. It was also observed that at the switch to the nectrophic phase (72hpi), an up-regulation of both ET biosynthesis enzymes occurred but only in the susceptible variety. However, it is not clear which enzyme, ACO or ACS, is the rate-limiting factor in ET biosynthesis. Different reports have suggested that either ACO activity is the key step for controlling ethylene production [76,77] or ACS is the rate-limiting enzyme [78]. In susceptible Nicotiana benthamiana challenged with C. orbiculare, an induction of ACO gene was observed during the biotrophic phase, with a maximum expression coincident with the switch to necrotrophic phase after which expression started to decrease. However, in NbACO1-silenced plants inoculated with C. orbiculare, a higher number of leaf lesions appeared earlier when compared with control plants, suggesting that ET might have some role in plant defense although it was not sufficient to stop the disease progress in this interaction [79]. High induction of ACS and ACO genes was also reported in susceptible citrus flowers inoculated with C. acutatum with a continued increase of expression of both genes up to 7 days after inoculation [80,81]. In this case, it was suggested that the induction of ET biosynthesis, and consequently the activation of ET pathway in susceptibility, was related with senescence of the tissues and promotion of young fruits drops, a characteristic of the Postbloom Fruit Drop disease [80,81]. The regulation of ET-receptors plays a major role in the ET signaling pathway. Ethylene induces receptor degradation through the 26S proteasome at the same time that transcriptional activation of new receptors is promoted. By this way, newly synthesized receptors not yet bonded with ethylene will allow a downstream pathway inhibition as soon as the levels of the hormone decreases [33]. In the susceptible coffee variety, the up/down-regulation profile of ETR1 suggests that also in this pathosystem, ET-receptors may undergo a similar regulation process. However, in the resistant variety, the repression of ETR1 throughout the infection process may suggest a different receptor regulation or even an alternative role for ET. In fact, the ET receptors are largely redundant in the control of ethylene responses but some functional specificity among their different isoforms has recently been uncovered [82], together with evidences of a degree of complexity of ET receptors interaction with one another [83]. Therefore, either other ET receptors or isoforms are activated in response to pathogen attack and/or ET pathway activation is related with other functions rather than pathogen defense only.
ERF1 gene is a transcriptional factor of ET induced defense genes while it is an ET-responsive gene itself, being commonly used as an ET-pathway marker [79,84]. The ERF1 gene was moderately induced in the resistant coffee variety contrasting with the increasing up-regulation in the susceptible variety, which culminates in a strong activation at the beginning of the necrotrophic phase. A comprehensive analysis of the ethylene role in plant response to pathogen attack was undertaken by Chen et al., (2003) [85]. When challenged with C. destructivum, the susceptible variety of N. tabacum produced ET in two distinct moments, the first during the biotrophic phase and the second during the necrotrophic phase (when leaf tissue was severely damaged or dead). It was then suggested that the first peak of ET production was related with the onset of unsuccessful defense responses and the second peak with the senescence of infected tissues. Although no ET production was measured in our assay, our results suggest that ET pathway activation in the susceptible variety may be related with tissue damage promoted by the fungal necrotrophic phase. In fact, it has been suggested that in later infection stages, hemibiotrohic pathogens may produce ethylene and deliver effectors or phytotoxins that manipulate the plant to produce ethylene for entering the necrotrophic stage of infection [86], and thus overcome plant defenses.
Overall, our results suggest that in both coffee varieties, the immune system enable the perception of the pathogen attack as the infection process starts, being the expression of resistance and susceptibility conditioned by the magnitude and/or timing of defense responses. In the resistant variety, the earlier and strong induction of JA biosynthesis and receptor-related genes, together with the activation of PR genes (PR1 and PR10) seems to be important players in the set of defenses that resulted in the arrest of fungal growth. The stronger activation of the ethylene responsive-related gene ERF1 at the switch to the necrotrophic fungal growth, that is coordinated with the activation profile of biosynthesis and receptor-related genes, suggests that this hormone may be relevant in susceptibility, although its possible involvement in defense responses is not discarded. To the best of our knowledge, this work represents the first attempt to unveil the involvement of phytohormones pathways in coffee-C. kahawae interaction making possible to engage on new exploratory functional assays. Future studies focusing on RNA-Seq analysis will provide a better understanding of the molecular mechanisms underlying the defense responses in this pathosystem.
Supporting information
a) SA pathway—SA is synthesized from chorismate through two distinct enzymatic pathways: PAL-mediated phenylalanine and ICS-mediated isochlorismate (IC). SA-induced redox changes lead to the reduction of inactive NPR1 oligomers to active monomers that are translocated into the nucleus, thus activating the defense-related genes (e.g. PR1); b) JA pathway–upon release from the chloroplast membrane, α-linolenic acid is converted into OPDA by sequential steps catalyzed by lipoxygenase (LOX), AOS and AOC. OPDA migrates into the peroxisome where, after reduction by OPR3 and three rounds of β-oxidation, (+)-7-iso-JA and its derivative (−)-JA is formed. By the action of JAR1 these last compounds are converted in the bioactive molecule JA-Ile. JA-dependent gene activation involves the JA-Ile binding to the receptor COI1. JAZ protein, which interacts with the SKP1-Cullin- F-box complex (SCFCOI1) complex, is targeted for degradation by the 26S proteasome, releasing the transcriptional factor MYC2 and promoting the expression of JA-responsive genes (e.g. PR10); c) ET pathway–ET is synthesized from SAM in a two-step reaction catalyzed by ACS and ACO. In the absence of ET, the active CTR1 inactivates EIN2 and the phosphorylation of its C-terminal end is promoted resulting in suppression of the ethylene response. In the presence of ET, receptors like receptor ETR1 binds to the hormone becoming inactivated and, consequently, switching off CTR1. The C-terminal end of EIN2 is then cleaved off and migrates to the nucleus where it activates the expression of ethylene target genes, ERF1 included (adapted from [25,33]).
(TIF)
Dissociation curves for non-specific qPCR products analysis of SA, JA, ET pathways related genes and qPCR reference genes: a) PAL, b) ICS2, c) NPR1, d) PR1, e) OPR3, f) COI, g) PR10, h) ACO2, i) ACS5, j) ETR1, k) EIN2, l) CTR1, m) ERF1, n) IDE, o) β-Tub9, p) S24.
(PUB)
Relative expression pattern of a) PAL/ICS2 (biosynthesis), b) NPR1 (receptors), and c) PR1 (responsive gene) obtained in Catimor 88 (R-resistant) and Caturra (S-susceptible) coffee varieties. Mean and standard deviation of three biological replicates is presented. Fold change as relative expression of gene expression between inoculated and control samples for each of the coffee varieties/inoculation time-points. Asterisks (*) represent statistical significance (p≤0.05) of gene expression between the two coffee varieties was determined by the non-parametric Mann–Whitney U test using IBM®SPSS® Statistics version 20.0 (SPSS Inc., USA) software.
(TIF)
Acknowledgments
The authors wish to acknowledge the technical support provided by Paula Leandro and Sandra Sousa.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by Portuguese national funds through Fundação para a Ciência e a Tecnologia (www.fct.pt/) (PTDC/AGR-GPL/112217/2009), LEAF (UI/AGR/04129/2013) and grants LEAF-AGR/04129/BPD/2015, SFRH/BPD/99712/2014, SFRH/BPD/104629/2014, SFRH/BD/84188/2012 attributed to AL, AF, DB, ID.
References
- 1.van der Vossen HAM, Walyaro DJ. Additional evidence for oligogenic inheritance of durable host resistance to coffee berry disease (Colletotrichum kahawae) in arabica coffee (Coffea arabica L.). Euphytica. 2009; 165:105–111. [Google Scholar]
- 2.McDonald L. A preliminary account of a disease of green coffee berries in Kenya Colony. Trans Br Mycol Soc. 1926; 11:145–154. [Google Scholar]
- 3.Silva MC, Várzea V, Guerra-Guimarães L, Azinheira HG, Fernandez D, Petitot A, et al. Coffee resistance to the main diseases : leaf rust and coffee berry disease. Brazilian J Plant Physiol. 2006; 18:119–47. [Google Scholar]
- 4.Firman ID, Waller JM. Coffee berry disease and other Colletotrichum diseases of coffee 4th ed. K Surrey, England: Commonwealth Mycological Institute; 1977. [Google Scholar]
- 5.Hindorf H, Omondi CO. A review of three major fungal diseases of Coffea arabica L. in the rainforests of Ethiopia and progress in breeding for resistance in Kenya. J Adv Res. 2011; 2:109–120. [Google Scholar]
- 6.van der Vossen HAM, Walyaro DJ. Breeding for resistance to coffee berry disease in Coffea arabica L. II. Inheritance of the resistance. Euphytica. 1979; 29:777–791. [Google Scholar]
- 7.Gichuru EK, Agwanda CO, Combes MC, Mutitu EW, Ngugi ECK, Bertrand B, et al. Identification of molecular markers linked to a gene conferring resistance to coffee berry disease (Colletotrichum kahawae) in Coffea arabica. Plant Pathol. 2008; 57:1117–1124. [Google Scholar]
- 8.Omondi CO, Pinard F. Screening Populations of Arabica Coffee for Molecular Markers Linked to Coffee Berry Disease Resistance. In: Proceedings of 21st International Conference on Coffee Science (ASIC), Montpellier, France; 2006.
- 9.Masaba DM, van der Vossen HAM. Evidence of cork barrier formation as a resistance mechanism to berry disease (Colletotrichum coffeanum) in arabica coffee. Netherlands J Plant Pathol. 1982; 88:19–32. [Google Scholar]
- 10.Gichuru EK. Resistance mechanisms in Arabica coffee to coffee berry disease Colletotrichum kahawae sp. nov.—a review. Kenya Coffee. 1997; 727:2441–2445. [Google Scholar]
- 11.Loureiro A, Nicole MR, Várzea V, Moncada P, Bertrand B, Silva MC. Coffee resistance to Colletotrichum kahawae is associated with lignification, accumulation of phenols and cell death at infection sites. Physiol Mol Plant Pathol. 2012; 77:23–32. [Google Scholar]
- 12.Loureiro A, Figueiredo A, Batista D, Baraldi T, Várzea V, Azinheira HG, et al. New cytological and molecular data on coffee–Colletotrichum kahawae interactions. In: Proceedings of 24th International Conference on Coffee Science (ASIC), San José, Costa Rica. 2013.
- 13.Gichuru EK. Histological comparison of susceptible and resistant interactions of coffee (Coffea arabica and C. canephora varieties) and Colletotrichum kahawae. Agron Africaine. 2007; 19:233–240. [Google Scholar]
- 14.Figueiredo A, Loureiro A, Batista D, Monteiro F, Várzea V, Pais MS, et al. Validation of reference genes for normalization of qPCR gene expression data from Coffea spp. hypocotyls inoculated with Colletotrichum kahawae. BMC Res Notes. 2013; 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diniz I, Figueiredo A, Loureiro A, Batista D, Azinheira H, Talhinhas P, et al. Early expressed genes related to the resistance of coffee to Colletotrichum Kawahae. In: XVI International Congress on Molecular Plant-Microbe Interactions. Rhodes, Greece; 2014.
- 16.Diniz I, Fino J, Loureiro A, Figueiredo A, Azinheira H, Pereira AP, et al. Unveiling the involvement of oxidases in the resistance of Coffea spp. to Colletotrichum kahawae. In: Proceedings of 25th International Conference on Coffee Science (ASIC). Armenia, Colombia; 2015.
- 17.Bari R, Jones JDG. Role of plant hormones in plant defence responses. Plant Mol Biol. 2009; 69:473–488. doi: 10.1007/s11103-008-9435-0 [DOI] [PubMed] [Google Scholar]
- 18.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005; 43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- 19.Andolfo G, Ercolano MR. Plant innate immunity multicomponent model. Front Plant Sci. 2015; 6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazan K, Lyons R. Intervention of phytohormone pathways by pathogen effectors. Plant Cell. 2014; 26:2285–2309. doi: 10.1105/tpc.114.125419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma K-W, Ma W. Phytohormone pathways as targets of pathogens to facilitate infection. Plant Mol Biol. 2016; 91:713–738. doi: 10.1007/s11103-016-0452-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. Networking by small-molecule hormones in plant immunity. Nat Chem Biol. 2009; 5:308–316. doi: 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- 23.Yan S, Dong X. Perception of the plant immune signal salicylic acid. Curr Opin Plant Biol. 2014; 20:64–72. doi: 10.1016/j.pbi.2014.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An C, Mou Z. Salicylic acid and its function in plant immunity. J Integr Plant Biol. 2011; 53: 412–428. doi: 10.1111/j.1744-7909.2011.01043.x [DOI] [PubMed] [Google Scholar]
- 25.Figueiredo A, Monteiro F, Sebastiana M. First clues on a jasmonic acid role in grapevine resistance against the biotrophic fungus Plasmopara viticola. Eur J Plant Pathol. 2015; 142:645–652. [Google Scholar]
- 26.Pieterse MJ, Does D Van Der, Zamioudis C, Leon-reyes A, Wees SCM Va. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012; 28:489–521. doi: 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- 27.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004; 16:2117–2127. doi: 10.1105/tpc.104.023549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009; 5:344–350. doi: 10.1038/nchembio.161 [DOI] [PubMed] [Google Scholar]
- 29.Ahmad P, Rasool S, Gul A, Sheikh SA, Akram NA, Ashraf M, et al. Jasmonates: Multifunctional roles in stress tolerance. Front Plant Sci. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol. 1984; 35:155–189. [Google Scholar]
- 31.Kende H. Enzymes of ethylene biosynthesis. Plant Physiol. 1989; 91:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang KL, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. Plant Cell. 2002; 14:131–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merchante C, Alonso JM, Stepanova AN. Ethylene signaling: Simple ligand, complex regulation. Curr Opin Plant Biol. 2013; 16:554–560. doi: 10.1016/j.pbi.2013.08.001 [DOI] [PubMed] [Google Scholar]
- 34.Das S, Dutta SS, Chowdhury S, Das K. Ethylene signal transduction and signaling roles—A Review. Agric Rev. 2015; 36:133. [Google Scholar]
- 35.Janda M, Ruelland E. Magical mystery tour: Salicylic acid signalling. Environ Exp Bot. 2015; 114:117–128. [Google Scholar]
- 36.Wasternack C. Perception, signaling and cross-talk of jasmonates and the seminal contributions of the Daoxin Xie’s lab and the Chuanyou Li’s lab. Plant Cell Rep. 2014; 33:707–718. doi: 10.1007/s00299-014-1608-5 [DOI] [PubMed] [Google Scholar]
- 37.van der Vossen HAM, Cook RTA, Murakaru GNW. Breeding for resistance to coffee berry disease caused by Colletrotrichum coffeanum Noack (sensu Hindorf) in Coffea arabica L. I. Methods of preselection for resistance. Euphytica. 1976; 25:733–45. [Google Scholar]
- 38.Rodrigues CJ, Várzea V, Hindorf H, Medeiros E. Strains of Colletotrichum coffeanum Noack causing coffee berry disease in Angola and Malawi with characteristics different to the Kenya strain. J Phytopathol. 1991; 131:205–209. [Google Scholar]
- 39.Silva MC, Nicole M, Rijo L, Greiger JP, Rodrigues CJ. Cytochemistry of plant- rust fungus interface during the compatible interaction Coffea arabica (cv. Caturra)-Hemileia vastatrix (race III). Int J Plant Sci. 1999; 160:79–91. [Google Scholar]
- 40.Silva MC, Nicole M, Guerra-Guimarães L, Rodrigues CJ. Hypersensitive cell death and post-haustorial defence responses arrest the orange rust (Hemileia vastatrix) growth in resistant coffee leaves. Physiol Mol Plant Pathol. 2002; 60:169–183. [Google Scholar]
- 41.Bennett M, Gallagher M, Fagg J, Bestwick C, Paul T, Beale M, et al. The hypersensitive reaction, membrane damage and accumulation of autofluorescent phenollics in lettuce cells challenged by Bremia lactucae. The Plant Journal. 1996; 9:851–865. [Google Scholar]
- 42.Heath MC. Apoptosis, programmed cell death and the hypersensitive response. Eur J Plant Pathol. 1998; 104:117–24. [Google Scholar]
- 43.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002; 3:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar D. Salicylic acid signaling in disease resistance. Plant Sci. 2014; 228:127–134. doi: 10.1016/j.plantsci.2014.04.014 [DOI] [PubMed] [Google Scholar]
- 45.Ramiro D, Escoute J, Petitot A-S, Nicole M, Maluf MP, Fernandez D. Biphasic haustorial differentiation of coffee rust (Hemileia vastatrix race II) associated with defence responses in resistant and susceptible coffee cultivars. Plant Pathol. 2009; 58:944–55. [Google Scholar]
- 46.Fino J, Figueiredo A, Loureiro A, Gichuru EK, Várzea V. Transcriptional profiling of compatible and incompatible Coffee—Colletotrichum kahawae interactions through RNA-Seq analysis. In: Proceedings of 25th International Conference on Coffee Science (ASIC), Armenia, Colombia; 2015.
- 47.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007; 8:R19 doi: 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torregrosa C, Cluzet S, Fournier J, Huguet T, Gamas P, Prospéri J-M, et al. Cytological, genetic, and molecular analysis to characterize compatible and incompatible interactions between Medicago truncatula and Colletotrichum trifolii. Mol Plant Microbe Interact. 2004; 17:909–920. doi: 10.1094/MPMI.2004.17.8.909 [DOI] [PubMed] [Google Scholar]
- 49.Munch S, Lingner U, Floss DS, Ludwig N, Sauer N, Deising HB. The hemibiotrophic lifestyle of Colletotrichum species. J Plant Physiol. 2008; 165:41–51. doi: 10.1016/j.jplph.2007.06.008 [DOI] [PubMed] [Google Scholar]
- 50.Damm U, O’Connell RJ, Groenewald JZ, Crous PW. The Colletotrichum destructivum species complex—hemibiotrophic pathogens of forage and field crops. Stud Mycol. 2014;79:49–84. doi: 10.1016/j.simyco.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kubo Y, Harata K, Kodama S, Fukada F. Development of the infection strategy of the hemibiotrophic plant pathogen, Colletotrichum orbiculare, and plant immunity. Physiol Mol Plant Pathol. 2016; 95:32–36. [Google Scholar]
- 52.Vargas W, Martín JMS, Rech GE, Rivera LP, Benito EP, Díaz-Mínguez JM, et al. Plant defense mechanisms are activated during biotrophic and necrotrophic development of Colletotricum graminicola in maize. Plant Physiol. 2012; 158:1342–58. doi: 10.1104/pp.111.190397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duan L, Liu H, Li X, Xiao J, Wang S. Multiple phytohormones and phytoalexins are involved in disease resistance to Magnaporthe oryzae invaded from roots in rice. Physiol Plant. 2014;152: 486–500. doi: 10.1111/ppl.12192 [DOI] [PubMed] [Google Scholar]
- 54.Amil-Ruiz F, Garrido-Gala J, Gadea J, Blanco-Portales R, Muñoz-Mérida A, Trelles O, et al. Partial activation of SA- and JA-defensive pathways in strawberry upon Colletotrichum acutatum interaction. Front Plant Sci. 2016; 7:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Biles CL, Abeles FB, Wilson CL. The role of ethylene in anthracnose of Cucumber, Cucumis sativus caused by Colletotrichum lagenarium. Phytopathology. 1990; 80:732–736. [Google Scholar]
- 56.Guerreiro A, Figueiredo J, Sousa Silva M, Figueiredo A. Linking jasmonic acid to grapevine resistance against the biotrophic oomycete Plasmopara viticola. Front Plant Sci. 2016; 7:565 doi: 10.3389/fpls.2016.00565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sašek V, Nováková M, Jindřichová B, Bóka K, Valentová O, Burketová L. Recognition of avirulence gene AvrLm1 from hemibiotrophic ascomycete Leptosphaeria maculans triggers salicylic acid and ethylene signaling in Brassica napus. Mol Plant Microbe Interact. 2012; 25:1238–50. doi: 10.1094/MPMI-02-12-0033-R [DOI] [PubMed] [Google Scholar]
- 58.Shibata Y, Kawakita K, Takemoto D. Age-related resistance of Nicotiana benthamiana against hemibiotrophic pathogen Phytophthora infestans requires both ethylene- and salicylic acid-mediated signaling pathways. Mol Plant Microbe Interact. 2010; 23:1130–1142. doi: 10.1094/MPMI-23-9-1130 [DOI] [PubMed] [Google Scholar]
- 59.Balmer D, De Papajewski DV, Planchamp C, Glauser G, Mauch-Mani B. Induced resistance in maize is based on organ-specific defence responses. Plant J. 2013; 74(2):213–225. doi: 10.1111/tpj.12114 [DOI] [PubMed] [Google Scholar]
- 60.Grellet-Bournonville CF, Martinez-Zamora MG, Castagnaro AP, Díaz-Ricci JC. Temporal accumulation of salicylic acid activates the defense response against Colletotrichum in strawberry. Plant Physiol Biochem. 2012; 54:10–16. doi: 10.1016/j.plaphy.2012.01.019 [DOI] [PubMed] [Google Scholar]
- 61.He J, Ren Y, Chen C, Liu J, Liu H, Pei Y. Defense responses of salicylic acid in mango fruit against postharvest anthracnose, caused by Colletotrichum gloeosporioides and its possible mechanism. J Food Saf. 2016. [Google Scholar]
- 62.Liu G, Kennedy R, Greenshields DL, Peng G, Forseille L, Selvaraj G, et al. Detached and attached Arabidopsis leaf assays reveal distinctive defense responses against hemibiotrophic Colletotrichum spp. Mol Plant Microbe Interact. 2007; 20:1308–1319. doi: 10.1094/MPMI-20-10-1308 [DOI] [PubMed] [Google Scholar]
- 63.Wanigasekara UWNP, Adikaram NKB, Abayasekara CL. Pre-harvest chemical elicitor treatment enhances induced resistance in harvested banana fruit cv. “Embul” and reduces anthracnose caused by Colletotrichum musae. J Natl Sci Found Sri Lanka. 2014; 42:101–110. [Google Scholar]
- 64.Litholdo CG, Leal GA, Albuquerque PSB, Figueira A. Differential expression of jasmonate biosynthesis genes in cacao genotypes contrasting for resistance against Moniliophthora perniciosa. Plant Cell Rep. 2015; 34:1747–1759. doi: 10.1007/s00299-015-1821-x [DOI] [PubMed] [Google Scholar]
- 65.Prathima PT, Raveendran M, Kumar KK, Rahul PR, Kumar VG, Viswanathan R, et al. Differential regulation of defense-related gene expression in response to red rot pathogen Colletotrichum falcatum infection in sugarcane. Appl Biochem Biotechnol. 2013; 171:488–503. doi: 10.1007/s12010-013-0346-4 [DOI] [PubMed] [Google Scholar]
- 66.Diniz I, Talhinhas P, Azinheira HG, Várzea V, Medeira C, Maia I, et al. Cellular and molecular analyses of coffee resistance to Hemileia vastatrix and nonhost resistance to Uromyces vignae in the resistance-donor genotype HDT832/2. Eur J Plant Pathol. 2011; 133:141–157. [Google Scholar]
- 67.Sá M, Ferreira JP, Queiroz VT, Vilas-Boas L, Silva MC, Almeida MH, et al. A liquid chromatography/electrospray ionisation tandem mass spectrometry method for the simultaneous quantification of salicylic, jasmonic and abscisic acids in Coffea arabica leaves. J Sci Food Agric. 2014; 94:529–536. doi: 10.1002/jsfa.6288 [DOI] [PubMed] [Google Scholar]
- 68.Tang W, Zhu S, Li L, Liu D, Irving DE. Differential expressions of PR1 and chitinase genes in harvested bananas during ripening, and in response to ethephon, benzothiadizole and methyl jasmonate. Postharvest Biol Technol. 2010; 57:86–91. [Google Scholar]
- 69.Duan Z, Lv G, Shen C, Li Q, Qin Z, Niu J. The role of jasmonic acid signalling in wheat (Triticum aestivum L.) powdery mildew resistance reaction. Eur J Plant Pathol. 2014; 140(:169–183. [Google Scholar]
- 70.Ding L, Xu H, Yi H, Yang L, Kong Z, Zhang L, et al. Resistance to hemi-biotrophic F. graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PLoS One. 2011; 6:e19008 doi: 10.1371/journal.pone.0019008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jaulneau V, Cazaux M, Wong Sak Hoi J, Fournier S, Esquerré-Tugayé M- T, Jacquet C, et al. Host and nonhost resistance in Medicago-Colletotrichum interactions. Mol Plant Microbe Interact. 2010; 23:1107–1117. doi: 10.1094/MPMI-23-9-1107 [DOI] [PubMed] [Google Scholar]
- 72.Miles TD, Day B, Schilder AC. Identification of differentially expressed genes in a resistant versus a susceptible blueberry cultivar after infection by Colletotrichum acutatum. Mol Plant Pathol. 2011; 12:463–477. doi: 10.1111/j.1364-3703.2010.00687.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Portal O, Izquierdo Y, De Vleesschauwer D, Sánchez-Rodríguez A, Mendoza-Rodríguez M, Acosta-Suárez M, et al. Analysis of expressed sequence tags derived from a compatible Mycosphaerella fijiensis–banana interaction. Plant Cell Rep. 2011; 30:913–928. doi: 10.1007/s00299-011-1008-z [DOI] [PubMed] [Google Scholar]
- 74.Zubini P, Zambelli B, Musiani F, Ciurli S, Bertolini P, Baraldi E. The RNA hydrolysis and the cytokinin binding activities of PR-10 proteins are differently performed by two isoforms of the Pru p 1 peach major allergen and are possibly functionally related. Plant Physiol. 2009; 150:1235–47. doi: 10.1104/pp.109.139543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Castro A, Vidal S, León IP De. Moss pathogenesis-related-10 protein enhances resistance to Pythium irregulare in Physcomitrella patens and Arabidopsis thaliana. Front Plant Sci. 2016; 7:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ohtsubo N, Mitsuhara I, Koga M, Seo S, Ohashi Y. Ethylene promotes the necrotic lesion formation and basic PR gene expression in TMV-infected tobacco. Plant Cell Physiol. 1999; 40:808–817. [Google Scholar]
- 77.Yu M, Shen L, Fan B, Zhao D, Zheng Y, Sheng J. The effect of MeJA on ethylene biosynthesis and induced disease resistance to Botrytis cinerea in tomato. Postharvest Biol Technol. 2009; 54:153–158. [Google Scholar]
- 78.Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol. 2013; 51:245–266. doi: 10.1146/annurev-phyto-082712-102314 [DOI] [PubMed] [Google Scholar]
- 79.Shan XC, Goodwin PH. Silencing an ACC oxidase gene affects the susceptible host response of Nicotiana benthamiana to infection by Colletotrichum orbiculare. Plant Cell Rep. 2006; 25:241–247. doi: 10.1007/s00299-005-0063-8 [DOI] [PubMed] [Google Scholar]
- 80.Lahey K, Yuan R, Burns JK, Ueng PP, Timmer LW, Kuang-Ren C. Induction of phytohormones and differential gene expression in citrus flowers infected by the fungus Colletotrichum acutatum. Mol Plant-Microbe Interact. 2004; 17:1394–1401. doi: 10.1094/MPMI.2004.17.12.1394 [DOI] [PubMed] [Google Scholar]
- 81.Li W, Yuan RC, Burns JK, Timmer LW, Chung KR. Genes for hormone biosynthesis and regulation are highly expressed in citrus flowers infected with the fungus Colletotrichum acutatum, causal agent of postbloom fruit drop. J Am Soc Hortic Sci. 2003; 128:578–583 [Google Scholar]
- 82.Kevany BM, Taylor MG, Klee HJ. Fruit-specific suppression of the ethylene receptor LeETR4 results in early-ripening tomato fruit. Plant Biotechnol J. 2008; 6: 295–300. doi: 10.1111/j.1467-7652.2007.00319.x [DOI] [PubMed] [Google Scholar]
- 83.Gallie DR. Ethylene receptors in plants—why so much complexity? F1000Prime Rep. 2015; 7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sugano S, Sugimoto T, Takatsuji H, Jiang C-J. Induction of resistance to Phytophthora sojae in soyabean (Glycine max) by salicylic acid and ethylene. Plant Pathol. 2013; 62:1048–1056. [Google Scholar]
- 85.Chen N. The role of ethylene during the infection of Nicotiana tabacum by Colletotrichum destructivum. J Exp Bot. 2003; 54:2449–2456. doi: 10.1093/jxb/erg289 [DOI] [PubMed] [Google Scholar]
- 86.Groen SC, Whiteman NK. The evolution of ethylene signaling in plant chemical ecology. J Chem Ecol. 2014; 40:700–716. doi: 10.1007/s10886-014-0474-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a) SA pathway—SA is synthesized from chorismate through two distinct enzymatic pathways: PAL-mediated phenylalanine and ICS-mediated isochlorismate (IC). SA-induced redox changes lead to the reduction of inactive NPR1 oligomers to active monomers that are translocated into the nucleus, thus activating the defense-related genes (e.g. PR1); b) JA pathway–upon release from the chloroplast membrane, α-linolenic acid is converted into OPDA by sequential steps catalyzed by lipoxygenase (LOX), AOS and AOC. OPDA migrates into the peroxisome where, after reduction by OPR3 and three rounds of β-oxidation, (+)-7-iso-JA and its derivative (−)-JA is formed. By the action of JAR1 these last compounds are converted in the bioactive molecule JA-Ile. JA-dependent gene activation involves the JA-Ile binding to the receptor COI1. JAZ protein, which interacts with the SKP1-Cullin- F-box complex (SCFCOI1) complex, is targeted for degradation by the 26S proteasome, releasing the transcriptional factor MYC2 and promoting the expression of JA-responsive genes (e.g. PR10); c) ET pathway–ET is synthesized from SAM in a two-step reaction catalyzed by ACS and ACO. In the absence of ET, the active CTR1 inactivates EIN2 and the phosphorylation of its C-terminal end is promoted resulting in suppression of the ethylene response. In the presence of ET, receptors like receptor ETR1 binds to the hormone becoming inactivated and, consequently, switching off CTR1. The C-terminal end of EIN2 is then cleaved off and migrates to the nucleus where it activates the expression of ethylene target genes, ERF1 included (adapted from [25,33]).
(TIF)
Dissociation curves for non-specific qPCR products analysis of SA, JA, ET pathways related genes and qPCR reference genes: a) PAL, b) ICS2, c) NPR1, d) PR1, e) OPR3, f) COI, g) PR10, h) ACO2, i) ACS5, j) ETR1, k) EIN2, l) CTR1, m) ERF1, n) IDE, o) β-Tub9, p) S24.
(PUB)
Relative expression pattern of a) PAL/ICS2 (biosynthesis), b) NPR1 (receptors), and c) PR1 (responsive gene) obtained in Catimor 88 (R-resistant) and Caturra (S-susceptible) coffee varieties. Mean and standard deviation of three biological replicates is presented. Fold change as relative expression of gene expression between inoculated and control samples for each of the coffee varieties/inoculation time-points. Asterisks (*) represent statistical significance (p≤0.05) of gene expression between the two coffee varieties was determined by the non-parametric Mann–Whitney U test using IBM®SPSS® Statistics version 20.0 (SPSS Inc., USA) software.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.