Abstract
Dimorphic human pathogenic fungi interact with host effector cells resisting their microbicidal mechanisms. Yeast cells are able of surviving within the tough environment of the phagolysosome by expressing an antioxidant defense system that provides protection against host-derived reactive oxygen species (ROS). This includes the production of catalases (CATs). Here we identified and analyzed the role of CAT isoforms in Paracoccidioides, the etiological agent of paracoccidioidomycosis. Firstly, we found that one of these isoforms was absent in the closely related dimorphic pathogen Coccidioides and dermatophytes, but all of them were conserved in Paracoccidioides, Histoplasma and Blastomyces species. We probed the contribution of CATs in Paracoccidioides by determining the gene expression levels of each isoform through quantitative RT-qPCR, in both the yeast and mycelia phases, and during the morphological switch (transition and germination), as well as in response to oxidative agents and during interaction with neutrophils. PbCATP was preferentially expressed in the pathogenic yeast phase, and was associated to the response against exogenous H2O2. Therefore, we created and analyzed the virulence defects of a knockdown strain for this isoform, and found that CATP protects yeast cells from H2O2 generated in vitro and is relevant during lung infection. On the other hand, CATA and CATB seem to contribute to ROS homeostasis in Paracoccidioides cells, during endogenous oxidative stress. CAT isoforms in Paracoccidioides might be coordinately regulated during development and dimorphism, and differentially expressed in response to different stresses to control ROS homeostasis during the infectious process, contributing to the virulence of Paracoccidioides.
Keywords: Paracoccidioides spp., dimorphic fungal pathogens, antisense RNA, catalases, peroxide, endogenous and exogenous ROS
1. Introduction
Dimorphic fungal pathogens are the commonest cause of invasive fungal diseases worldwide infecting immunocompetent and immunocompromised hosts and accounting for several million infections each year (Buitrago and Cuenca-Estrella, 2012; Colombo et al., 2011; Sifuentes-Osornio et al., 2012). The largest cluster of thermally dimorphic fungi includes the genera Histoplasma, Blastomyces, Coccidioides, and Paracoccidioides in the order Onygenales. Dimorphism in these species involves the ability to switch between two different morphologies in response to external stimuli, primarily temperature. At 22–25°C, they grow as molds that produce infectious conidia, while at 37°C they grow as yeast-like forms (Histoplasma, Blastomyces and Paracoccidioides) or as spherules (Coccidioides) (Boyce and Andrianopoulos, 2015).
The ability of these pathogens to cause disease entails their capability to survive inside their human host (Missall et al., 2004). They have evolved mechanisms that allow them to adapt and grow within the host and cause disease (de Oliveira et al., 2015; Perez-Nadales et al., 2014). Once inside the host, dimorphic fungi are challenged by a higher temperature that not only induces the transition to the parasitic form, but also increases the generation of Reactive Oxygen Species (ROS), as well as the oxidative damage resulting from the inhibition of the mitochondrial respiration (Martins et al., 2011; Ruiz et al., 2011; Vacca et al., 2004). In addition, the fungus faces other sources of oxidative stress from phagocytic cells, mainly from neutrophils (PMNs) and activated macrophages (Robinson et al., 2004).
These pathogenic fungi are able of surviving within the tough conditions imposed by phagocytic cells, especially when the formation of the phagolysosome takes place. This suggests the presence of an antioxidant defense system that provides protection to the fungus against host-derived ROS (Giles et al., 2006). Oxidative stress has been proven to trigger changes in gene expression, changes that are required for expression of fungal virulence and pathogenicity traits, and include the expression of superoxide dismutases (SODs) (Tamayo et al., 2016; Youseff et al., 2012), glutathione/thioredoxin system, cytochrome C peroxidase (CCP) (Parente-Rocha et al., 2015) and catalases (CATs) (Chagas et al., 2008; Holbrook et al., 2013). CATs are ubiquitous in aerobic organisms and provide protection against the oxidative stress resulting from exogenous and endogenous ROS (Chelikani et al., 2004; Giles et al., 2006). In fungal pathogens such as Blastomyces, Histoplasma, Paracoccidioides, among others, CAT genes encode a small family of proteins including CATA, CATB and CATP, which catalyze the decomposition of hydrogen peroxide (H2O2) into oxygen and water (Chagas et al., 2008; Holbrook et al., 2013; Loew, 1900; Munoz et al., 2015). H2O2 toxicity relies on the products formed during its interaction with ferrous iron (Fe2+) via the Fenton reaction. In the presence of Fe2+, H2O2 is reduced to hydroxyl anion (OH−) and hydroxyl radical (.OH). Although the latter radical has a fleeting nature and a short diffusion distance, it is highly nonselective and is the most active oxidant among the ROS. Due to its transience, living organisms have not developed specific enzymatic systems for its detoxification; thus, the best way to protect the cell against the deleterious effects of this radical is preventing its production (Imlay et al., 1988; Lushchak, 2011). In this way, the presence of CATs allow cells to destroy H2O2 radicals, reducing the probability of converting the peroxide produced by phagocytic cells into more potent and potentially toxic microbicidal reactive oxygen intermediates. Consequently, in the absence of these H2O2-detoxifying enzymes, increased fungal damage can be expected to occur (Henriet et al., 2011).
Paracoccidioides is the etiological agent of paracoccidioidomycosis (PCM), one of the most prevalent systemic mycosis in Latin America (Colombo et al., 2011). In endemic areas, the estimated incidence is approximately one to three cases per 100.000 inhabitants per year (Martinez, 2015). Phylogenetic analyses led to discover that the Paracoccidioides genus compromises two species: P. brasiliensis and P. lutzii (Teixeira et al., 2009). In turn, P. brasiliensis has been classified into different phylogenetic lineages (S1, PS2, PS3 and PS4) (Hahn et al., 2014; Matute et al., 2006; Teixeira et al., 2014), and recently using population genomics, we identified a deep split in the S1 group, breaking it into two lineages named S1a and S1b (Munoz et al., 2016). Isolates among the P. brasiliensis and P. lutzii phylogenetic lineages can infect humans, but they vary in their virulence and induce different host immune responses (Gegembauer et al., 2014; Scorzoni et al., 2015).
Recently, we demonstrated that Paracoccidioides cells produce six SODs isoforms. From these, PbSOD1 and PbSOD3 were involved in the fungus response against oxidative stress, and were required for virulence in a mouse model of infection (Tamayo et al., 2016). In addition to the latter genes, Paracoccidioides genome also encodes three CAT genes. Moreira et al. 2004, Chagas et al. 2008 and 2010 studied the CATs in this dimorphic fungus. These works suggested the possible role of CATP in defense against the oxidative stress produced during the host-pathogen interactions (Chagas et al., 2010; Chagas et al., 2008; Moreira et al., 2004). However, until now, the precise role of this enzyme had remained elusive due to lack of feasible genetic molecular tools that could be applied to the study of the Paracoccidioides genus, allowing specifically target candidate genes. Thus, in the present study, we aimed to better understand the role of these enzymes during adaptation to the host’s environment, specifically CATP.
We initially identified and analyzed the sequences of the CAT gene family members in Paracoccidioides and other dimorphic fungal pathogens from Onygenales. Subsequently, we studied the gene expression of the three isoforms and the susceptibility of Paracoccidioides yeast cells to H2O2. P. brasiliensis strain ATCC 60855 was used to analyze the role of these catalases under different conditions, including the morphological switch, exposure to oxidative stress-inducing agents, and during interaction with human PMNs. Subsequently, the antisense RNA counterpart for CATP (PbCATP-aRNA) was used to facilitate the functional tests performed here. The data here presented shows that PbCATP seems to play an important role in the removal of H2O2 generated via different stimuli. In line with previous studies performed in our laboratory, these results suggest that CATs and other scavenger enzymes such as SODs (Tamayo et al., 2016) are differentially expressed during cell growth and dimorphism, as well as in response to distinct external stresses and within the host.
2. Experimental procedures
2.1. Identification and sequence analysis of CAT isoforms
In order to identify and classify the CAT isoforms present in the genomes of Paracoccidioides and other dimorphic pathogenic fungi, we used bidirectional BLAST analysis v2.2.28+ with default parameters to identify sequence similarities (Altschul et al., 1997), and ortholog matrices generated with OrthoMCL v1.4 with a markov inflation index of 1.5 and a maximum e-value of 1e-5 using whole genome gene sets (Li et al., 2003).
We studied one representative strain from each of the lineages of Paracoccidioides. Pb18, P03 and Pb01 were chosen to represent P. brasiliensis (S1, PS2) and P. lutzii lineages, respectively (Desjardins et al., 2011; Munoz et al., 2014). For the PS3 lineage, and the recently described PS4, we used the gene set of PbCNH and Pb300 respectively (Munoz et al., 2016). In addition, we identified CAT gene family orthologs in related dimorphic fungi such as Coccidioides immitis, C. posadassi and Emmonsia, from dermatophytes Trichophyton rubrum and Microsporum gypseum, and three Aspergillus species as outgroup (Table S1). For all the identified CAT isoforms, protein domain conservation analyses were done using InterProScan (Zdobnov and Apweiler, 2001), by sequence comparison with InterPro collection of protein signature databases in the EMBL-EBI (http://www.ebi.ac.uk/interpro/). Multiple sequence alignments were constructed using Muscle (Edgar, 2004), and phylogenetic trees were constructed employing a distance computation method (Neighbor-joining) (Saitou and Nei, 1987). Other features in the gene/protein sequences and annotations of the CAT isoforms were identified using SignalP 4.1 (Petersen et al., 2011), TargetP 1.1 (Emanuelsson et al., 2000) and PTS1 Predictor (Neuberger et al., 2003).
2.2. Microorganisms and growth conditions
At least one representative isolate from each phylogenetic lineages of Paracoccidioides spp. was used (Table 1). P. brasiliensis knockdown strains employed in this study were derived from the background strain ATCC 60855 (PbWT). Paracoccidioides yeast cells were maintained by sub-culturing in brain heart infusion (BHI) supplemented with 1% glucose (Beckton Dickinson and Company, Sparks, MD), under constant agitation at 36°C, and at 20°C for the mycelia form, unless otherwise indicated. P. brasiliensis conidia were produced and purified using the glass-wool filtration protocol, as described previously by Restrepo et al. (Restrepo et al., 1986). In order to induce and evaluate the transition processes (conidia to yeast (C-Y); mycelia to yeast (M-Y)) and the germination process (yeast to mycelia (Y-M)), cells were incubated at 36°C or at 20°C, respectively, during 3, 12, 48 and 96 h, and kept under constant agitation in a flask containing BHI, as previously reported by us (Garcia et al., 2009; Tamayo et al., 2016). It is important to note that during the morphological switch several fungal morphotypes coexist. Cultures during M-Y transition are characterized by the presence of hyphae, differentiating hyphae (chlamydospore-like cells), transforming yeast (production of multiple buds by the chlamydospore) and mature, multibudding yeast (Nunes, Costa de Oliveira et al. 2005). During C-Y transition, cultures are characterized by the presence of conidia, intermediate cells and yeast cells. After 12 h, intermediate cells are present, from 48 h onwards it is possible to observe the characteristic yeast cells, although, they are more abundant at 72 h. Regarding to the C-M germination, conidia began to germinate approximately at 24 h and the formation of branched mycelia occurs after 96 h (Hernandez et al., 2011a; Hernandez et al., 2011b). Samples were collected for RNA extraction and quantification of gene expression analyses during evaluated time points (Tamayo et al., 2016).
Table 1.
Strains used in this study.
| Species | Strain designation | Phylogenetic lineage |
|---|---|---|
| P. brasiliensis | Pb18 | S1 |
| P. brasiliensis | Pb03 | PS2 |
| P. brasiliensis | Pb60855, PbBAC | PS3 |
| P. brasiliensis | Pb300 | PS4 |
| P. lutzii | Pb01 | Pb01-like |
| P. brasiliensis | PbEV* | PS3 |
| P. brasiliensis | PbCATP-aRNA* | PS3 |
| A. tumefaciens | LBA1100 | N/A |
| E. coli | DH5-α | N/A |
Background strain Pb60855
N/A: Not apply
We used Agrobacterium tumefaciens strain LBA1100 (Beijersbergen et al., 1992) as the recipient for the binary vectors. Bacterial cells were maintained at 28°C in Luria–Bertani (LB) medium containing kanamycin (100 mg/ml). Escherichia coli DH5α was grown at 36°C in LB medium supplemented with kanamycin (50 mg/ml), and used as the host for plasmid amplification and cloning.
2.3. Real-time RT-qPCR analysis
Total RNA was obtained using the Trizol reagent (Invitrogen®) according to manufacturer’s instructions. As formerly described (Tamayo et al., 2016), the total RNA obtained was treated with DNase I (Thermo Scientific®) and tested using a conventional PCR with β-tubulin primers to confirm the absence of chromosomal DNA contamination (Goldman et al., 2003; Tamayo et al., 2016). cDNA was synthesized using 2 μg of total RNA with Maxima® First Strand cDNA synthesis kit for RT-qPCR, according to the manufacturer’s instructions (Fermentas®) (Tamayo et al., 2016).
Real-time PCR was carried out using a Maxima® SYBR Green/Fluorescein qPCR Master Mix (2X; qRT-PCR) kit with SYBR green, according to the manufacturer’s instructions (Fermentas®). The CFX96 real time PCR detection system (Bio-Rad, Hercules, CA) was used to measure gene expression level of CAT isoforms encoded into the Paracoccidioides’ genome. Primers were designed in order to anneal properly to each CAT transcript (Table S2). Additionally, in Pb60855, CAT isoforms were evaluated in cells undergoing the transition (C-Y and M-Y) as well as in the germination processes (C-M and Y-M). We also evaluated gene expression in knockdown strains for PbCATP gene, and in P. brasiliensis cells carrying the empty binary vector as a control (PbEV60855, see below construction of knockdown and empty vector strains). Finally, we performed melting curve analysis after the amplification phase. The β-tubulin gene (TUBE3, housekeeping gene) was used in order to normalize the expression value of each CAT isoform. We also used the elongation factor 3 (TEF3) as a second normalizer control, finding no differences when compared with TUBE3, as previously reported by us (Tamayo et al., 2016). Each experiment was done in triplicate, and the expression level was measured three times.
2.4. Construction of P. brasiliensis PbCATP-aRNA yeast cells
DNA from PbWT was extracted from yeast cells cultures during exponential growth. We employed a Platinum high-fidelity Taq DNA polymerase (Invitrogen®) to amplify antisense RNA (aRNA) fragments, designed on the sequence of PbCATP gene (PADG_00324). Two regions of the gene were selected in order to design six different aRNA fragments and generate the knockdown strains. Five of them were targeted at exon four (80, 120, 92, 106 and 100-bp for PbCATP-AS1, AS2, AS3, AS4 and AS5, respectively) and one at exon three (85-bp for PbCATP-AS6). P. brasiliensis plasmid construction for aRNA and A. tumefaciens-mediated transformation (ATMT) were performed as previously described (Almeida et al., 2007). Briefly, the amplified PbCATP aRNA oligonucleotide was inserted into the pCR35 plasmid under the control of the Calcium Binding Protein 1 (CBP-1) promoter region from Histoplasma capsulatum (Rappleye et al., 2004). The pUR5750 plasmid was used as a parental binary vector to harbor this aRNA cassette within the transfer DNA (T-DNA). The constructed binary vectors were introduced into A. tumefaciens LBA1100 ultra competent cells by electroporation as described previously (den Dulk-Ras and Hooykaas, 1995), and isolated by kanamycin selection (100 mg/ml).
In P. brasiliensis yeast cells, ATMT was done using A. tumefaciens cells harboring the desired binary vector in order to obtain the knockdown strains, and were also transformed with the empty parental vector pUR5750 (PbEV) as a control for the experiments. A 1:10 P. brasiliensis/A. tumefaciens ratio was employed during the 3 days period of co-culture at 28°C. Selection of P. brasiliensis transformants was performed in BHI solid media containing hygromycin B (Hyg; 200mg/ml) over a 15 days incubation period at 36°C. Randomly selected Hyg resistant transformants were tested for mitotic stability and insertion of the T-DNA. The presence of the hygromycin B resistance cassette was confirmed by PCR analysis to detect an HPH 1000-bp amplification product in PbEV and PbCATP-aRNA isolates (Figure S1). Also, PbCATP-aRNA and PbEV strains were successively sub-cultured on BHI without hygromicin B (three consecutive rounds) and later placed again under the selective pressure of the hygromicin B (Tamayo et al., 2016).
2.5. Viability and vitality in P. brasiliensis yeast cells
Growth curves were performed in BHI broth (100 mL). We adjusted the inoculum to an OD of 0.4 for PbWT, PbEV and PbCATP-aRNA yeast cells. Then, cultures were incubated at 36°C and samples were collected at specific time points (0, 3, 6, 12, 24, 48, 72 and 96 h) to determine growth curves by spectrophotometric analysis (OD600 nm; SmartSpec Plus (Bio-Rad, Hercules, CA).
Vitality, defined as the ability of yeast cells to metabolize glucose upon late activation of a cell membrane proton pump and subsequent acidification of the medium due to H+ release (Kara, 1987), was evaluated following the protocol reported by Hernandez et al. (Hernandez et al., 2010). This protocol was detailed in a previous publication by our group (Tamayo et al., 2016). The assays were performed in triplicates.
2.6. Phenotypic analysis of Paracoccidioides strains
For the phenotypic analysis, we tested the sensitivity to hydrogen peroxide in the representative strains of the phylogenetic lineages (P. brasiliensis S1, PS2, PS3, PS4 and P. lutzii) and in PbWT, PbEV and PbCATP-aRNA strains. We used H2O2-saturated filter disks, with an inoculum of 1×105 Paracoccidioides yeast cells dispersed on BHI plates. After inoculation, sterile filter disks were loaded with 20 μl of PBS as a control, and with increasing amounts of H2O2 (0.5, 1, 2, 4 and 8 M). Plates were incubated at 36°C with 5% CO2 and after eight days we measured the clearing area (Holbrook et al., 2013; Tamayo et al., 2016).
We also tested the sensitivity and gene expression profile of the PbWT strain to extracellular induced-oxidative stress, using 0.5 mM riboflavin (R4500, Sigma), 1mM menadione (M5625, Sigma) and 30 mM hydrogen peroxide (02194057, MP Biomedicals). Briefly, 3×106 PbWT yeast cells were inoculated in 20 ml of PBS and incubated for 1 hour at 36°C under constant agitation. For the riboflavin assay, cells were exposed to UV-light in order to induce oxidative stress (Ruane et al., 2004). Following this step, samples were collected for RNA extraction and quantification of gene expression during evaluated conditions was done.
We determined the ability of P. brasiliensis cellular extracts to destroy H2O2 in vitro. PbWT, PbEV and PbCATP-aRNA cultures were grown in liquid BHI at 37°C. Three biological replicate yeast cultures were used. We adjusted the inoculum to an OD of 0.4. For extracellular catalase activity, we separated the cell mass by centrifugation (12,000 rpm, 5 min at 4°C) and subsequent filtration by passage through a syringe filter of 0.22 μm. The pellet was washed twice with PBS and suspended in PBS. Cytoplasmic protein was released by breaking cells with 0.5-mm-diameter glass beads. Later, a step of low speed centrifugation (3000 g, 5 min at 4°C) was applied in order to obtain the cell wall fraction (Lambou et al., 2010). To slow down enzymatic destruction, all steps were carried out at 4°C. Protein concentrations were determined using the Bradford method with bovine serum albumin protein as the standard. Catalase activity assays for extracellular and intracellular samples were performed by spectrophotometrically measuring their ability to reduce the H2O2 concentration over time. For extracellular fractions, we used 200 μl of the culture supernatants with 800 μl of 15 mM H2O2 in 50 mM Na-phosphate buffer (pH 7.2). For extracellular cell-associated fraction, 200 μl of the suspension were added to 800 μl of 15 mM H2O2 in 50 mM Na-phosphate buffer. Finally, intracellular activity was determined using 12 μg of total protein. Hydrogen peroxide destruction was measured at 240 nm with correction at 595 nm every 30 s for 5 min and compared to that in a control containing buffer (Holbrook et al., 2013; Tamayo et al., 2016).
2.7. Interaction of P. brasiliensis yeast cells with human PMNs (ex vivo assay)
Polymorphonuclear neutrophils (PMNs) were isolated from human blood samples taken from healthy volunteers. We used whole blood treated with anticoagulant EDTA. Briefly, a layer 5.0 ml of non-coagulated whole blood was placed over 5.0 ml of Polymorphprep™ in a 12 ml centrifuge tube. Samples were centrifuged for 450 × g for 30 min. The polymorphonuclear fraction was then washed with Hanks’ Balanced Salt Solution and centrifuged for 400 × g for 10 min. Finally, PMNs were re-suspended in Dulbecco’s Modified Eagle Medium (DMEM; Gibco®), enumerated in hemacytometer and cell viability was determined using trypan blue (Mejia et al., 2015). PMNs were seeded into 24-well tissue culture plate and allowed to adhere for 20 min at 36°C with 5% of CO2. For inhibition of NADPH-oxidase, 10μM diphenylene iodinium (DPI; D2926, Sigma) was added to PMNs 20 min before infection.
For all assays, we used a ratio of 1:5 for P. brasiliensis yeasts:host cells. The interaction was kept at 36°C with 5% of CO2, for 3h. CATs gene expression was performed and viability was measured via colony forming units (Kurita et al., 1993; Ruiz et al., 2011).
2.8. In vivo virulence determination
Isogenic 6 to 8-week-old BALB/c male mice, obtained from the breeding colony of the Corporación para Investigaciones Biológicas (CIB), Medellín, Colombia, were used in assays and were kept with food and water ad libitum (Restrepo et al., 1992). All animals were handled according to the national (Law 84 of 1989, Res No. 8430 of 1993) and international (Council of European Communities and Canadian Council of Animal Care, 1998) guidelines for animal research. The CIB research ethics committee approved the experimental protocols.
P. brasiliensis yeast cells were collected from exponentially growing batch cultures in BHI medium and counted using a hemacytometer. Animals (n=5 per isolate) were infected with P. brasiliensis yeast by intranasal delivery of 1.5×106 cells from PbWT, PbEV or PbCATP-aRNA strains suspended in PBS buffer. Mice were monitored daily for survival, weight loss and symptoms of disease. At 12 days post-infection, mice were euthanized and lung, liver and spleen tissues were homogenized in 2 mL PBS. Homogeneous suspensions were diluted (1:10, 1:100 and 1:1000) and 0.1 ml of each dilution was plated on Kurita’s medium (Kurita et al., 1993) in order to determine the fungal burden in each organ. Plates were incubated at 36°C, 5% CO2. CFU counts were assessed ten days after culture. The data was transformed into Log10 CFU/g of tissue.
2.9. Statistical analysis
Data are either the means or representative results of at least three experiments, each performed in triplicate. Statistical analysis and comparisons were performed using paired Student’s t tests.
3. Results
3.1. Catalases and their deduced proteins in Paracoccidioides spp. and other dimorphic pathogens
We identified three catalase isoforms encoded by the genome of five strains of Paracoccidioides spp. representing each of the phylogenetic lineages (S1, Pb18; PS2, Pb03; PS3, PbCNH; PS4, Pb300; P. lutzii, Pb01), which were designated as Catalase A (CATA), Catalase B (CATB) and Catalase P (CATP) based on homology to known catalases reported in fungal reference genomes (Figure 1A and Table S1). Catalases in Paracoccidioides and other dimorphic pathogens from the Ajellomycetaceae such as Histoplasma and Blastomyces have two protein domains, the catalase activity domain (PF00199.14) and the catalase-related immune-responsive domain (PF06628.7) (Figure 1B). In addition, we found that CATB has an N-terminal signal peptide sequence (from 1 to 20 aa), as well as an extracellular location pattern. In Histoplasma, CatB is an immunodominant 90-kDa extracellular catalase highly expressed during yeast phase, also known as the M antigen- (Zancope-Oliveira et al., 1999). CATP, but not CATB and CATA, encodes the C-terminal Peroxisomal Targeting Signal 1 (PTS1) ER(M/L)VASQPQSHL, suggesting a putative localization to peroxisomes for this isoform. Figure 1 shows multiple alignment, conserved domains, and phylogenetic analyses confirming the placement and conservation of PbCATA, PbCATB and PbCATP.
Figure 1. Identification and gene expression of Paracoccidioides spp catalases, and susceptibility to H2O2.
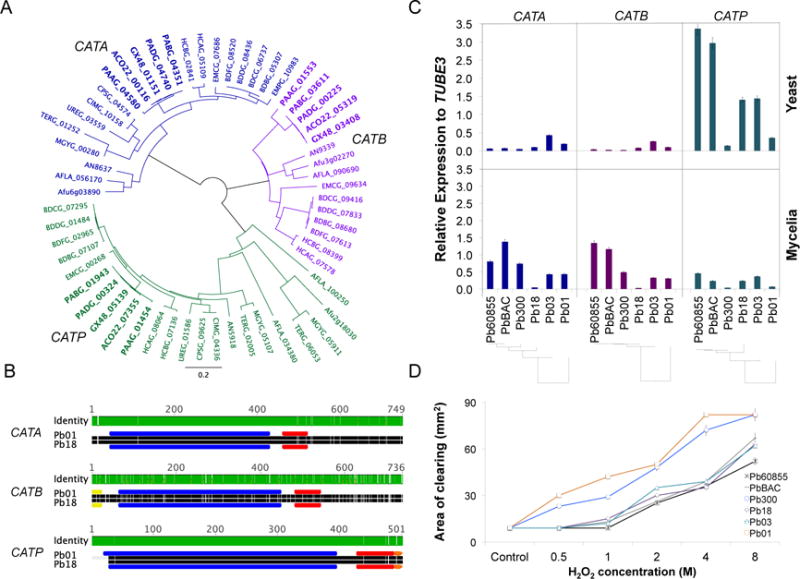
(A) Phylogenetic tree of CAT isoforms (CATA, CATB, CATP) encoded by the genome of Paracoccidioides (gene IDs in bold) and Onygenales such as Histoplasma, Blastomyces, Coccidioides, and dermatophytes; outgroup Aspergillus. (B), Functional annotation and protein domains of identified CATs in Paracoccidioides. The catalase activity domain (PF00199.14) is presented in blue, and the immune-responsive domain (PF06628.7) in red. The predicted secreted signal and the peroxisomal targeting signal are presented in yellow and orange, respectively. Green or white lines across the proteins represent high level of identity or low level of identity for each position of the alignment, respectively. (C) Expression profile of CAT isoforms in yeast phase (36°C) and mycelia phase (20°C) during exponential growth. (D) Susceptibility of Paracoccidioides yeast cells to peroxide (H2O2). Cells were exposed to increasing concentrations of this compound and the inhibition zone was determined as the area lacking yeast cell growth. The chosen strains represent the phylogenetic lineages of Paracoccidioides spp. (PS4: Pb300; PS3: Pb60855 and PbBAC; PS2: Pb03; S1: Pb18, and P. lutzii: Pb01).
Since catalases have been proposed as virulence factors in dimorphic fungal pathogens, and due to the lack of genetic/functional evidence to support this in Paracoccidioides, we examined the molecular evolution more closely, by analyzing single nucleotide polymorphisms (SNP) between the catalases across the Paracoccidioides lineages. We did not find non-sense or frame shift mutations, and found a low number of SNPs in coding regions within P. brasiliensis, an average of 4.75 synonymous and 2 non-synonymous substitutions. However, when compared with P. lutzii, we found that in CATB there were more non-synonymous mutations than synonymous mutations. We calculated the dN/dS ratio as an indicator of selective pressure, and found that CATB has the highest dN/dS across the catalases (0.89) as compared with CATA and CATP (0.23 and 0.04, respectively).
In addition, we analyzed CAT gene family across the order Onygenales, which also includes dimorphic pathogens such as species of Coccidioides (family Onygenaceae) and dermatophytes (family Arthrodermataceae). We found that CATA and CATP are conserved among the Onygenales; however, CATB was present in the Ajellomycetaceae but not in the Onygenaceae or in the Arthrodermataceae (Figure 1A and Table S1).
3.2. Gene expression of CAT isoforms and Paracoccidioides susceptibility to H2O2
We measured the gene expression of the identified CAT isoforms in one representative isolate of each phylogenetic lineage of Paracoccidioides during exponential growth of the parasitic yeast and the saprophytic mycelia phases (Figure 1C). Overall, in yeast cells CATP was highly expressed in PbBAC and Pb60855 (both belonging to PS3), Pb03 (PS2) and Pb18 (S1), but lower levels of gene expression were detected in Pb300 (PS4) and Pb01 (P. lutzii). In the mycelia, gene expression of CATP was generally lower compared to yeast cells for all strains, while CATA and CATB were expressed at slightly higher levels in the PS3 strains tested. The higher expression level of CATP in Pb18, Pb03, PbBAC and Pb60855 was correlated with the degree of susceptibility of each strain to H2O2. Using a disk diffusion assay we found that these strains were the less sensitive to H2O2 treatment in vitro as shown by a smaller area of clearing, which contrasts with Pb01 and Pb300 that were the most sensitive to H2O2 together with the lowest CATP expression (Figure 1D).
3.3. Kinetic expression of CAT isoforms in P. brasiliensis Pb60855 during the morphological switch
The expression profile of CAT isoforms was evaluated in P. brasiliensis cells undergoing transition [conidia to yeast (C-Y) and mycelia to yeast (M-Y)] and germination [conidia to mycelia (C-M) and yeast to mycelia (Y-M)] processes. All isoforms were present throughout the evaluated shifts, but they were differentially expressed. PbCATP presented lower expression levels during transition and germination processes (Figure 2), in contrast to the high expression observed during the exponential phase of growth of yeast cells (Figure 1C, Pb60855); however, PbCATP expression was slightly higher during the M-Y and Y-M morphological shifts. On the other hand, PbCATA and PbCATB showed low expression levels specifically during the M-Y and Y-M morphological shifts; however, the expression level of these genes was increased during the C-Y and C-M morphological shift. PbCATA gene expression was higher during C-M germination, being the main isoform during this process (Figure 2).
Figure 2. Gene expression of CAT isoforms in Pb60855.
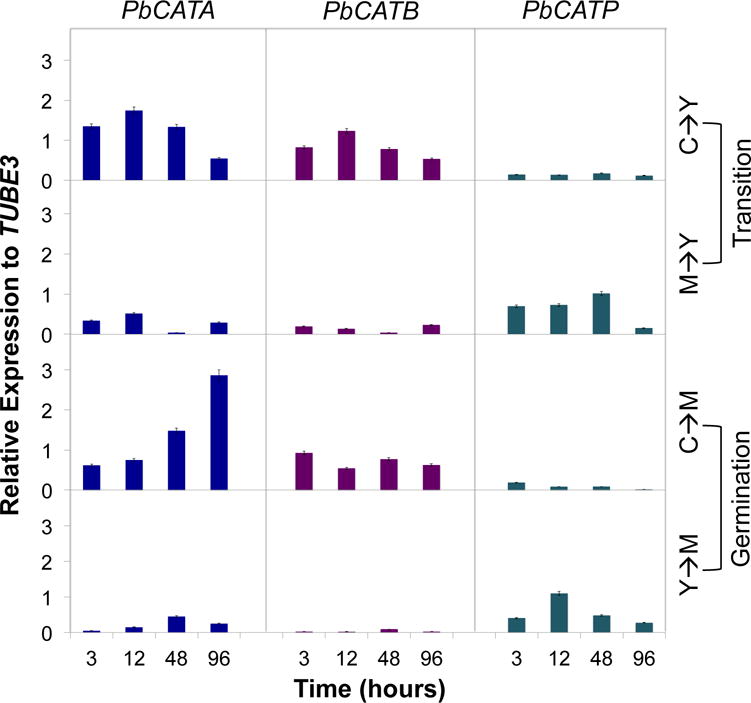
Expression of CAT isoforms was determined during transition (C-Y and M-Y) and also during germination (C-M and Y-M). P. brasiliensis cells were inoculated in BHI liquid medium kept at 36°C and 20°C in order to induce the transition and germination processes, respectively. Gene expression levels of CAT isoforms were determined at 3, 12, 48 and 96 h, and were obtained by RT-qPCR assay. The measurement was normalized with the housekeeping gene β-tubulin.
3.4. PbCAT isoforms are differentially expressed when yeast cells are exposed to distinct oxidative stress-inducing compounds
To determine the effect of oxidative stress-inducing compounds on the expression of CAT isoforms, P. brasiliensis (ATCC 60855) yeast cells were treated with riboflavin, menadione (both superoxide-generating agents) (Goldberg and Stern, 1976) and H2O2; and co-cultured in the presence of human polymorphonuclear neutrophils (PMNs). Menadione induced a significantly increased expression of PbCATA and PbCATB as compared to the control, while riboflavin slightly induced PbCATP. Interaction with human PMNs induced higher expression of PbCATA and PbCATP (Figure 3). However, it is important to state that gene expression levels for all tested conditions were generally low for both PbCATA and PbCATB, as compared to the higher PbCATP levels, (as shown for yeast cells in Figure 1C). Upon exposure to H2O2, PbCATP was the only isoform triggered by this stimulus (up to 10-fold), as well as during interaction with human PMNs (up to 5-fold) as compared to the control (Figure 3).
Figure 3. Expression of PbCAT isoforms during exposure to oxidative stress-inducing compounds.
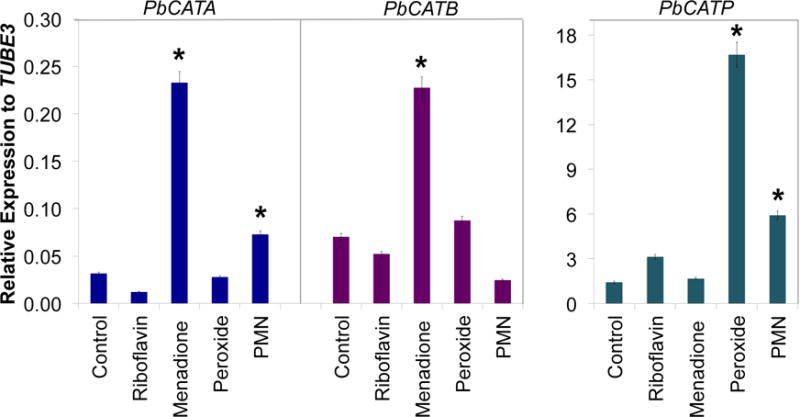
P. brasiliensis yeast cells were exposed to menadione, peroxide (H2O2) and riboflavin for 1 hour. For interaction with human PMNs, yeast cells were co-cultivated during 3 hours. Gene expression levels of CAT isoforms were obtained by RT-qPCR. The measurement was normalized with the housekeeping gene β-tubulin. Results are the mean of three individual experiments. Asterisks denote p ≤0.05 compared to control cells.
3.5. Construction and analysis of a PbCATP-aRNA strain
Based on the signature of differential expression in response to H2O2 and PMN exposure, we constructed a PbCATP-aRNA strain in order to assess the individual contribution of this catalase to the antioxidant defenses in P. brasiliensis. We selected a mitotically stable strain carrying PbCATPAS1 (Figure 4A) with the largest decrease in PbCATP gene expression (±50–70%) (Figure 4B). The insertion of the transfer DNA (T-DNA) into the genome of P. brasiliensis was confirmed via amplification of the HPH cassette (Figure S1). We analyzed more than one PbCATP-aRNA transformant and the same phenotypic features were observed. Therefore, results presented throughout the work refer to one selected aRNA strain denoted as PbCATP-aRNA.
Figure 4. CATP gene silencing in P. brasiliensis.
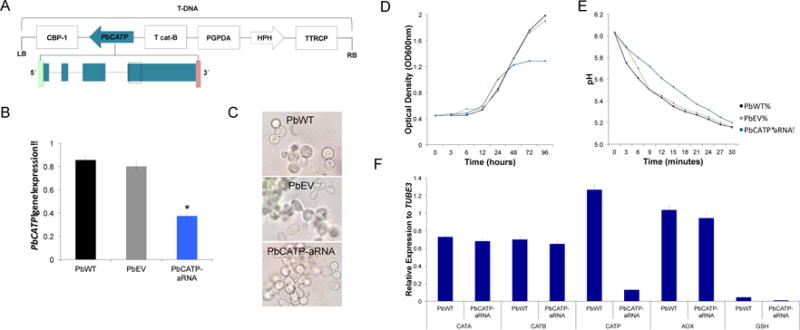
(A) Transfer DNA (T-DNA) inserted into the genome of P. brasiliensis yeast cells via ATMT. The antisense oligonucleotide was targeted to exon four (dashed box), with a length of 80 bp. This AS oligonucleotide was placed under control of the calcium binding protein (CBP-1), the terminator CAT-B and harboring hygromycin B phosphotransferase (HPH) under control of Aspergillus nidulans glyceraldehyde 3-phosphate (PGPDA) and with the terminator TTRCP. (B) PbCATP gene expression levels obtained by RT-qPCR assay. The measurement was normalized with the housekeeping gene β-tubulin in the wild-type (PbWT60855), the empty vector control (PbEV60855) and the antisense RNA strain (PbCATP-aRNA) grown at exponential phase. Mitotic stability was confirmed by sub-culturing P. brasiliensis PbCATP-aRNA yeast cells, checking for low expression levels in this isolate after successive sub-cultures. (C) Light microscopy of P. brasiliensis yeast cells morphology. (D) Growth curve in PbWT60855, PbEV60855, PbCATP-aRNA. Yeast cells were grown in BHI liquid medium at 36°C, OD600 nm was determined at each time point. (E) Vitality in PbWT60855, PbEV60855 and PbCATP-aRNA. The pH change was monitored in yeast suspensions at three minutes intervals, for 30 minutes. Results represent the mean of three individual experiments. (F) Profile expression of CAT isoforms, alternative oxidase (AOX) and glutathione (GSH) in PbWT and PbCATP-aRNA yeast cells during batch culture growth. Gene expression levels were determined by RT-qPCR assay and normalized with the housekeeping gene β-tubulin. Results are the mean of three individual experiments.
Prior to performing additional assays, we analyzed the morphology, growth and vitality during batch culture of PbCATP-aRNA yeast cells. We did not observe morphological alterations neither at microscopic or macroscopic level as compared with PbWT or PbEV (cells transformed with the empty parental vector pUR5750) (Figure 4C). Nonetheless, we observed a decrease in biomass production of the PbCATP-aRNA strain after 48h (mid-exponential phase) as measured by OD and a halt in growth after 72h (Figure 4D). Also, the capacity to metabolize glucose was reduced when compared with PbWT and PbEV yeast cells (Figure 4E).
Furthermore, we analyzed the gene expression profile of other peroxide defense genes in the PbWT and PbCATP-aRNA strains. We evaluated the three CAT isoforms, glutathione (GSH) and the alternative oxidase (AOX) during batch culture growth. No differences were observed for any of the analyzed peroxide defense enzymes during the exponential phase of growth of yeast cells, except as expected for CATP (Figure 4F), suggesting that compensatory transcriptional activation of the evaluated genes did not occur in the PbCATP-aRNA strain.
3.6. PbCATP-aRNA yeast cells are more susceptible to hydrogen peroxide
To determine if CATP plays an important role in the protection of P. brasiliensis yeast cells against H2O2 in vitro, we tested the PbCATP-aRNA strain for its susceptibility to this compound. Paracoccidioides yeast cells expressing CATP (PbWT and PbEV) showed similar high resistance to H2O2 in vitro, while, decreased expression of CATP resulted in a significantly higher susceptibility (ranging from 13.3 to 55%, with the highest difference at 1M of H2O2) (Figure 5A). We also evaluated the ability of extracellular and cell wall (both measuring extracellular activity) and cytoplasmic (intracellular activity) fractions of PbWT, PbEV and PbCATP-aRNA strains to degrade H2O2 in vitro. The PbCATP-aRNA strain had a decreased ability to destroy H2O2 but only in the intracellular fraction (Figure 5B), suggesting a role as an intracellular catalase.
Figure 5. Exposure of CATP knockdown strain to hydrogen peroxide.
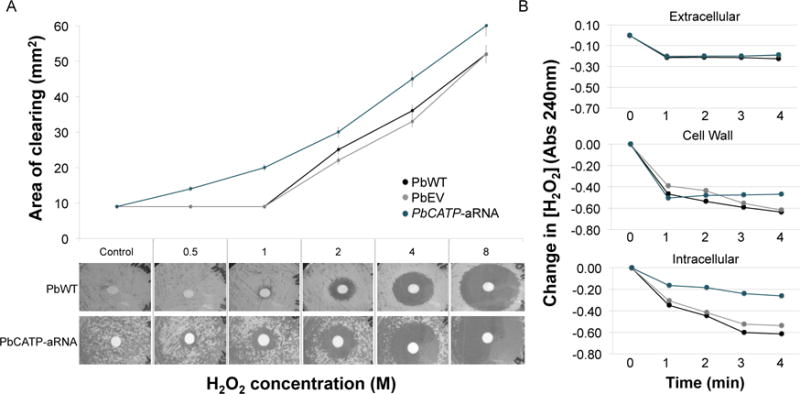
(A) Quantification of the sensitivity to H2O2 in P. brasiliensis yeast cells. Yeast cells were exposed to increasing concentrations of this compound and the inhibition zone was determined as the area lacking yeast cell growth. (Top) Data presented correspond to the average for three replicate tests, with error bars representing standard deviations. (Bottom) Images of P. brasiliensis growth around H2O2 saturated disks. Saturated filter disks without H2O2 as control, and 0.5 to 8 M H2O2. (B) Determination of the ability of P. brasiliensis yeast cells to degrade H2O2. Extracellular, cell wall and cytoplasmic fractions from PbWT, PbEV and PbCATP-aRNA strains were collected. Relative H2O2 destruction was measured as the decrease in the absorbance at 240 nm. Results are the mean of three individual experiments.
3.7. PbCATP’s involvement during interaction with human neutrophils and in a mouse model of infection
As neutrophils are the predominant white blood cells involved in phagocytic killing of several microorganisms (Hoffbrand et al., 2007), we used these cells to test the involvement of CATP in phagocyte-defense. Human PMNs from healthy donors were incubated with both knockdown and wild-type yeast cells. We also inhibited NADPH oxidase from human PMNs with DPI prior to challenging with yeast cells, in order to inhibit their oxidative burst. No differences were detected in survival of PbCATP-aRNA yeast cells exposed to PMNs when compared to the PbWT and PbEV strains (Figure 6A). Further, we determined the gene expression levels of CAT isoforms. PbCATA and PbCATB remained unaltered after the stimulus; while, as expected, PbCATP in the knockdown strain had lower expression levels compared to PbWT, either in the control or upon interaction with PMNs. Additionally, we measured AOX and GSH (peroxide-defense genes), observing a slightly increased induction of PbAOX (Figure S2), suggesting that transcriptional compensation of this gene might occur. In contrast to the high survival of the PbCATP-aRNA strain in ex vivo assay, in a mouse model of infection a significant decrease in fungal burden of this strain was detected in mice lungs compared to the PbWT and PbEV strains (Figure 6B). Also, we recovered CFUs from the liver of mice infected with PbWT and PbEV strains, but not from tissues of mice infected with the PbCATP-aRNA strain (not presented in the figure due to undetected counts in knockdown). CFUs were not recovered from the spleen in any of the infected mice, irrespective of the strain used (data not shown).
Figure 6. Effect of CATP decreased expression in an ex vivo and in vivo models of infection.
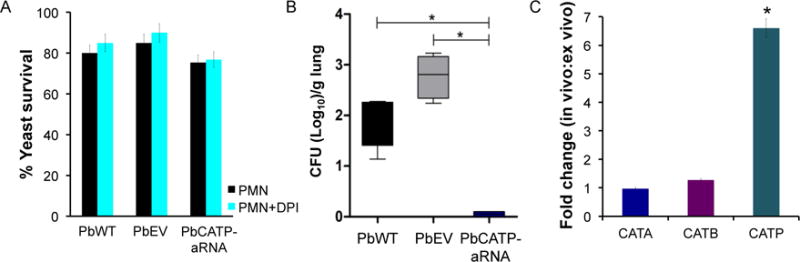
(A) P. brasiliensis yeast cells were incubated in the presence of human PMNs for 3 hours, at 36 °C, with 5% of CO2. Then, yeast cells survival was determined by enumeration of viable CFUs (black bar). Later on, human PMNs were treated with DPI (blue bar) in order to inhibit NADPH-oxidase. Next, yeast survival was determined during the interaction of P. brasiliensis with human PMNs treated with DPI. (B) Virulence of PbCATP-aRNA isolate in an experimental murine model of infection. CFUs recovered from lungs of mice intranasally inoculated with 1.5×106 yeast cells obtained from PbWT, PbEV, and PbCATP-aRNA strains. Mice were euthanized at 12 days post-infection, their organs were harvested and the fungal burden in lung tissue was determined. No significant differences between the PbWT and PbEV strains were found. Asterisks indicate significant differences (p < 0.05) from animals infected with PbWT and PbEV as compared to PbCATP-aRNA strains determined by Student’s t test. (C) Expression of CAT isoforms in yeast cells from PbWT obtained after interaction with PMNs (ex vivo) and in yeast cells recovered from lung tissues (in vivo). Bars represent the fold changes (in vivo compared to ex vivo gene expression) for PbCATA, PbCATB and PbCATP genes, determined through RT-qPCR assay and normalized with the housekeeping gene β-tubulin.
Furthermore, we determined the gene expression of all CAT isoforms in the PbWT strain after interaction with human PMNs (ex vivo) and in yeast cells recovered from lung tissues (in vivo). Strikingly, we found 6.6-fold increased expression in PbCATP between the ex vivo and in vivo models, being significantly higher in yeast cells recovered from mice lungs. In contrast, PbCATA did not change, and PbCATB had a very slight increase (1.27-fold) (Figure 6C).
4. Discussion
Dimorphic fungal pathogens are equipped with a strong antioxidant defense system that provides them with different features and mechanisms required to efficiently establish infection and cause disease (de Oliveira et al., 2015; Perez-Nadales et al., 2014). Among these are the production of proteins that prevent host effector cells from eliciting an oxidative burst, the presence of nonenzymatic antioxidants (melanin) and the utilization of enzymatic antioxidants such as SODs and CATs, which they differentially encode and express. In the present study, we identified and characterized the three members of the CAT gene family in Paracoccidioides and other Onygenales. Unlike Paracoccidioides, Histoplasma and Blastomyces species, Coccidioides and dermatophyte genomes do not encode the extracellular catalase-CATB (Figure 1A). This suggests that oxidative response in the Onygenales may have evolved different mechanisms to counteract oxidative stress via catalases. In Coccidioides, the arthroconidia are susceptible to peroxide treatment in vitro (Galgiani, 1986), suggesting that the absence of CATB could make these cells more susceptible to host-derived ROS. In addition, Coccidioides spherules do not stimulate the oxidative burst of some phagocytic cells (Galgiani, 1986), implying that they would not require a large amount of catalase activity to reduce ROS (specifically peroxide) produced during the host-pathogen interactions. In spite of the absence of CATB in this pathogen, it has been shown that CATP is an up-regulated gene during the spherule parasitic phase (Whiston et al., 2012).
We studied the expression of CAT genes in different fungal strains of Paracoccidioides spp., covering each phylogenetic lineage (Figure 1B). Our data showed that the highest level of gene expression was detected for CATP in yeast cells and particularly in Pb60855, PbBAC (PS3), Pb18 (S1) and Pb03 (PS2), while the lowest levels were detected in P. lutzii and P. brasiliensis Pb300 (PS4) (both in mycelia and yeast phases). In agreement with the obtained expression data, the latter strains were the most susceptible to H2O2 (Figure 1D). In line with the latter results, a previous work performed at our laboratory showed that SODs expression was higher in P. brasiliensis strains than in P. lutzii (Tamayo et al., 2016); correlated with this, Pigosso and colleagues demonstrated that P. lutzii is metabolically more anaerobic than P. brasiliensis (Pigosso et al., 2013), which could be related with results obtained during the course of this work. The fact that these antioxidant gene families are expressed at different levels in the genus raises the possibility that this might have consequences for survival within the host and when facing phagocytic cells and their defensive arsenal. In line with these findings, a transcriptomic analysis performed by Edwards et al. using two different Histoplasma strains from different phylogenetic lineages, demonstrated that both of them rely on different mechanisms to survive inside the host: either the evasion of phagocyte detection or enhanced defense mechanisms (such as higher transcription of oxidative stress defense genes). They also suggested that cells in which expression of antioxidant enzymes is higher would cope and subsequently survive better to the phagocyte oxidative burst (Edwards et al., 2013). Data obtained during the course of this work together with previous studies performed in our laboratory (Tamayo et al., 2016) seems to suggest that these genetic differences in the Paracoccidioides genus could be behind a distinct capability to survive within the host and establish a successful pathogenic process. Future studies should be conducted to further elucidate the role of these genetic variances in the virulence of the Paracoccidioides genus.
CAT genes are differentially regulated during the morphological switch and under oxidative stress-inducing conditions. PbCATA and PbCATB gene expression was higher in conidia and mycelia than in yeast cells, which might be implying that these isoforms could be important during aerobic metabolism present in mycelial cells. Moreover, according to the higher levels of PbCATA transcript in mycelia and during the C-M and C-Y switch, the homologous Aspergillus nidulans and Neurospora crassa CATA have been shown to be up-regulated in spores, protecting them against H2O2 stress and heat shock (Diaz et al., 2001; Navarro et al., 1996). Thus, these data suggest that PbCATA may be important during activation of conidia either in nature or once conidia infect the host and are induced to transform into the yeast pathogenic form. On the other hand, PbCATP transcript was higher in yeast phase (Figure 1C, bar corresponding to Pb60855) and during M-Y transition, as reported by Moreira, 2004 and Chagas, 2008 (Chagas et al., 2008; Moreira et al., 2004), overall suggesting a possible role of this during the host-pathogen interactions.
When exposed to menadione, PbCATA and PbCATB gene expression was significantly increased (Figure 3). Like-minded, menadione, besides generating superoxide anions, also increases the intracellular levels of H2O2 (Goldberg and Stern, 1976), implying, as it had been previously reported, that CATA gene expression is more correlated with endogenous oxidative stress (Chagas et al., 2008; Grossklaus et al., 2013). Further, human PMNs induced both PbCATA and PbCATP gene expression, but not PbCATB. Interestingly, H2O2 only triggered PbCATP gene expression (Figure 3), contrary to what we initially expected (increased gene expression of all isoforms in yeast cells treated with the catalase direct substrate), but similar to previously reported data (Chagas et al., 2008; Grossklaus et al., 2013; Moreira et al., 2004). These behaviors could be linked to the fact that these isoforms might be triggered by a specific ROS response depending on the source of stress, either endogenous (PbCATA and PbCATB) or exogenous (PbCATP) (Chagas et al., 2008). Considering that i) PbCATP gene is expressed at higher levels in exponentially growing yeast cells (Figure 1C), ii) the higher expression during M-Y transition (Figure 2), and that iii) it was the only isoform induced after stimulation of yeast cells with H2O2 (Figure 3), we sought to individually test the contribution of this catalase during events mimicking host-pathogen interactions. To pursue this goal, we developed a knockdown strain for PbCATP, using antisense RNA technology (Almeida et al., 2007; Almeida et al., 2009; Hernandez et al., 2010). We showed that a decrease in PbCATP gene expression significantly increased the susceptibility of P. brasiliensis yeast cells to H2O2 in vitro (Figures 5A and 5B), further supporting that this CAT isoform is important to detoxify this toxic molecule. Moreover, we also observed a decreased ability to destroy H2O2 in the intracellular fraction of the PbCATP-aRNA strain, but not in the extracellular or cell wall fractions. This is in agreement with data reported by Moreira et al. 2004 and our bioinformatic data suggesting a putative cytosolic localization of PbCATP, which also supports a role as an intracellular catalase. Furthermore, during batch culture growth, we observed decreased biomass production evident after 48–72 h of growth, and reduced vitality in PbCATP-aRNA yeast cells. It is important to consider that yeast cells are characterized by an aerobic metabolism, contributing to ROS generation. Additionally, for vitality tests we used a high concentration of glucose. This accelerates glycolysis, stimulating ATP synthesis and leading to an additional source of ROS (S.B. Riis, 1995). The PbCATP-aRNA strain may be impaired and not efficiently counteract these conditions, resulting in both vitality and growth defects.
Interestingly, the decreased expression of PbCATP has no deleterious consequences ex vivo; only minor effects on yeast survival against human PMNs was observed (Figure 6A). One possibility for this result could be the fact that transcript level of PbCATP in the knockdown strain could be enough to maintain its survival when interacting with PMNs. Another possibility raised here is that Paracoccidioides cells synthesize alternative compounds, or express other enzymes in alternative pathways implicated in H2O2 neutralization, which might contribute to its detoxification and, in this way, promote its survival when interacting with phagocytic cells (e.g. melanin and expression of genes such as AOX, glutathione and CCP) (Campos et al., 2005; Hernandez et al., 2015; Parente-Rocha et al., 2015; Ruiz et al., 2011). In fact, we observed that PbAOX had a slightly higher induction in PbCATP-aRNA strain after interaction with PMNs as compared to PbWT (Figure S2), indicating that this gene might be exerting some compensatory effect to protect yeast cells, as a consequence of the decreased PbCATP expression. It is very likely that CCP could be also contributing to this compensatory effect (Parente-Rocha et al., 2015); however, we were not be able to determine the expression levels of this gene. Additionally, it is important to highlight that PMNs produce not only H2O2, but also other reactive species (Dahlgren and Karlsson, 1999) not directly counteracted by catalases, and this knockdown strain seemingly has only a defect to counteract H2O2 stress.
Similarly, although ex vivo experiments showed no-attenuation of the PbCATP-aRNA strain, the reduced gene expression of this isoform had deleterious consequences for Paracoccidioides virulence in vivo. This discrepancy between the ex vivo and in vivo models could be explained by several reasons: i) the mouse represents a much more complex environment for the fungus, and PMNs are not the only active defense mechanisms they will face; ii) in vivo, the oxidative burst is just one of the microbicidal strategies used by the host’s defenses. Physical barriers of the lung together with the presence of other cells of the innate immunity are crucial for defense against fungal infections (Perez-Nadales et al., 2014); iii) the knockdown strain is not able to induce enough PbCATP levels to counteract host’s defenses; iv) the oxidative burst in the granuloma might be more intense; and v) the time of interaction in the ex vivo experiment was just 3 h, meanwhile, in the in vivo model such interaction lasted days, which might have a direct effect on the survival of the fungus.
Interestingly, we found no differences in the expression levels of PbCATA and PbCATB between the ex vivo and in vivo models, but PbCATP gene expression was significantly higher in yeast cells recovered from mice lungs compared to those recovered upon exposure with human PMNs (6.6-fold) (Figure 6C). This fact reinforces the important role of PbCATP in protecting yeast cells against the deleterious effect of H2O2.
The presence, expression and redundancy of different antioxidant genes in Paracoccidioides and in other pathogenic fungi, raises the possibility that differential gene expression in the different isoforms helps to optimize the organisms’ responses to environmental stresses either in nature or once within the animal host (Campos et al., 2005; Giles et al., 2006; Johnson et al., 2002). Our results revealed that PbCATP is the main source of intracellular catalase activity when considering the removal of H2O2 generated under various environmental stresses, but also when the yeast phase expresses its pathogenic potentials. Furthermore, the response against ROS is triggered according to the source of these species, either endogenous or exogenous. Thus, PbCATA and PbCATB are triggered by endogenous signals, with both contributing to ROS homeostasis in Paracoccidioides cells, mainly during the conidia to yeast/mycelia switch and during defense against menadione-induced oxidative stress, while PbCATP is mainly triggered in the presence of exogenous ROS. Although the role of catalases in fungal virulence is not fully understood yet, this work provides new insights concerning this issue. The most significant finding was the relevance of PbCATP during fungal virulence in vivo. Thus, even if CATP is not essential for yeast cells under normal conditions nor during the co-incubation with human PMNs, it plays an important role in the acquisition of tolerance to some conditions set by the host.
Supplementary Material
Acknowledgments
This project was supported by COLCIENCIAS Colombia, grant “Silencing of genes involved in adherence and oxidative stress in Paracoccidioides brasiliensis: Consequences in the infectious process” (project code. 2213-52128253), and by a sustainability grant from the Universidad de Antioquia “Sostenibilidad 2015/16”. The COLCIENCIAS National Doctorate Program Funding supported DT and JFM. JFM has been supported with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Grant Number U19AI110818 to the Broad Institute.
References
- Almeida AJ, et al. Towards a molecular genetic system for the pathogenic fungus Paracoccidioides brasiliensis. Fungal Genet Biol. 2007;44:1387–98. doi: 10.1016/j.fgb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Almeida AJ, et al. Cdc42p controls yeast-cell shape and virulence of Paracoccidioides brasiliensis. Fungal Genet Biol. 2009;46:919–26. doi: 10.1016/j.fgb.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijersbergen A, et al. Conjugative Transfer by the Virulence System of Agrobacterium tumefaciens. Science. 1992;256:1324–7. doi: 10.1126/science.256.5061.1324. [DOI] [PubMed] [Google Scholar]
- Boyce KJ, Andrianopoulos A. Fungal dimorphism: the switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host. FEMS Microbiol Rev. 2015;39:797–811. doi: 10.1093/femsre/fuv035. [DOI] [PubMed] [Google Scholar]
- Buitrago MJ, Cuenca-Estrella M. Current epidemiology and laboratory diagnosis of endemic mycoses in Spain. Enferm Infecc Microbiol Clin. 2012;30:407–13. doi: 10.1016/j.eimc.2011.09.014. [DOI] [PubMed] [Google Scholar]
- Campos EG, et al. Oxidative stress response in Paracoccidioides brasiliensis. Genet Mol Res. 2005;4:409–29. [PubMed] [Google Scholar]
- Chagas RF, et al. Purification of Paracoccidioides brasiliensis catalase P: subsequent kinetic and stability studies. J Biochem. 2010;147:345–51. doi: 10.1093/jb/mvp182. [DOI] [PubMed] [Google Scholar]
- Chagas RF, et al. The catalases of Paracoccidioides brasiliensis are differentially regulated: protein activity and transcript analysis. Fungal Genet Biol. 2008;45:1470–8. doi: 10.1016/j.fgb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Chelikani P, et al. Diversity of structures and properties among catalases. Cell Mol Life Sci. 2004;61:192–208. doi: 10.1007/s00018-003-3206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo AL, et al. Epidemiology of endemic systemic fungal infections in Latin America. Med Mycol. 2011;49:785–98. doi: 10.3109/13693786.2011.577821. [DOI] [PubMed] [Google Scholar]
- Dahlgren C, Karlsson A. Respiratory burst in human neutrophils. J Immunol Methods. 1999;232:3–14. doi: 10.1016/s0022-1759(99)00146-5. [DOI] [PubMed] [Google Scholar]
- de Oliveira HC, et al. Paracoccidioides-host Interaction: An Overview on Recent Advances in the Paracoccidioidomycosis. Front Microbiol. 2015;6:1319. doi: 10.3389/fmicb.2015.01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dulk-Ras A, Hooykaas PJ. Electroporation of Agrobacterium tumefaciens. Methods Mol Biol. 1995;55:63–72. doi: 10.1385/0-89603-328-7:63. [DOI] [PubMed] [Google Scholar]
- Desjardins CA, et al. Comparative genomic analysis of human fungal pathogens causing paracoccidioidomycosis. PLoS Genet. 2011;7:e1002345. doi: 10.1371/journal.pgen.1002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A, et al. Molecular and kinetic study of catalase-1, a durable large catalase of Neurospora crassa. Free Radic Biol Med. 2001;31:1323–33. doi: 10.1016/s0891-5849(01)00637-2. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JA, et al. Histoplasma yeast and mycelial transcriptomes reveal pathogenic-phase and lineage-specific gene expression profiles. BMC Genomics. 2013;14:695. doi: 10.1186/1471-2164-14-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, et al. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–16. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Galgiani JN. Inhibition of different phases of Coccidioides immitis by human neutrophils or hydrogen peroxide. J Infect Dis. 1986;153:217–22. doi: 10.1093/infdis/153.2.217. [DOI] [PubMed] [Google Scholar]
- Garcia AM, et al. Identification of genes associated with germination of conidia to form mycelia in the fungus Paracoccidioides brasiliensis. Biomedica. 2009;29:403–12. [PubMed] [Google Scholar]
- Gegembauer G, et al. Serology of paracoccidioidomycosis due to Paracoccidioides lutzii. PLoS Negl Trop Dis. 2014;8:e2986. doi: 10.1371/journal.pntd.0002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles SS, et al. The Cryptococcus neoformans catalase gene family and its role in antioxidant defense. Eukaryot Cell. 2006;5:1447–59. doi: 10.1128/EC.00098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg B, Stern A. Production of superoxide anion during the oxidation of hemoglobin by menadione. Biochim Biophys Acta. 1976;437:628–32. doi: 10.1016/0304-4165(76)90029-5. [DOI] [PubMed] [Google Scholar]
- Goldman GH, et al. Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot Cell. 2003;2:34–48. doi: 10.1128/EC.2.1.34-48.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossklaus DD, et al. Response to oxidative stress in Paracoccidioides yeast cells as determined by proteomic analysis. Microbes and Infection. 2013;15:347–364. doi: 10.1016/j.micinf.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Hahn RC, et al. Fatal fungemia due to Paracoccidioides lutzii. Am J Trop Med Hyg. 2014;91:394–8. doi: 10.4269/ajtmh.13-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriet SS, et al. Human leukocytes kill Aspergillus nidulans by reactive oxygen species-independent mechanisms. Infect Immun. 2011;79:767–73. doi: 10.1128/IAI.00921-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez O, et al. A 32-kilodalton hydrolase plays an important role in Paracoccidioides brasiliensis adherence to host cells and influences pathogenicity. Infect Immun. 2010;78:5280–6. doi: 10.1128/IAI.00692-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez O, et al. Alternative oxidase plays an important role in Paracoccidioides brasiliensis cellular homeostasis and morphological transition. Med Mycol. 2015;53:205–14. doi: 10.1093/mmy/myu091. [DOI] [PubMed] [Google Scholar]
- Hernandez O, et al. Gene expression during activation of Paracoccidioides brasiliensis conidia. Yeast. 2011a;28:771–81. doi: 10.1002/yea.1902. [DOI] [PubMed] [Google Scholar]
- Hernandez O, et al. Kinetic analysis of gene expression during mycelium to yeast transition and yeast to mycelium germination in Paracoccidioides brasiliensis. Biomedica. 2011b;31:570–9. doi: 10.1590/S0120-41572011000400012. [DOI] [PubMed] [Google Scholar]
- Hoffbrand V, et al. Postgraduate Haematology 2007 [Google Scholar]
- Holbrook ED, et al. Redundant catalases detoxify phagocyte reactive oxygen and facilitate Histoplasma capsulatum pathogenesis. Infect Immun. 2013;81:2334–46. doi: 10.1128/IAI.00173-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA, et al. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–2. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- Johnson CH, et al. Redundancy, phylogeny and differential expression of Histoplasma capsulatum catalases. Microbiology. 2002;148:1129–42. doi: 10.1099/00221287-148-4-1129. [DOI] [PubMed] [Google Scholar]
- Kara BV, Daoud I, Searle B. European Bravery Convention. Proceedings of the 21st Congress. 1987 [Google Scholar]
- Kurita N, et al. An improved culture medium for detecting live yeast phase cells of Paracoccidioides brasiliensis. J Med Vet Mycol. 1993;31:201–5. doi: 10.1080/02681219380000251. [DOI] [PubMed] [Google Scholar]
- Lambou K, et al. Functional analysis of the superoxide dismutase family in Aspergillus fumigatus. Mol Microbiol. 2010;75:910–23. doi: 10.1111/j.1365-2958.2009.07024.x. [DOI] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li L, et al. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loew O. A New Enzyme of General Occurrence in Organismis. Science. 1900;11:701–2. doi: 10.1126/science.11.279.701. [DOI] [PubMed] [Google Scholar]
- Lushchak VI. Adaptive response to oxidative stress: Bacteria, fungi, plants and animals. Comp Biochem Physiol C Toxicol Pharmacol. 2011;153:175–90. doi: 10.1016/j.cbpc.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Martinez R. Epidemiology of Paracoccidioidomycosis. Rev Inst Med Trop Sao Paulo. 2015;57(Suppl 19):11–20. doi: 10.1590/S0036-46652015000700004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins VP, et al. Involvement of an alternative oxidase in oxidative stress and mycelium-to-yeast differentiation in Paracoccidioides brasiliensis. Eukaryot Cell. 2011;10:237–48. doi: 10.1128/EC.00194-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute DR, et al. Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol Biol Evol. 2006;23:65–73. doi: 10.1093/molbev/msj008. [DOI] [PubMed] [Google Scholar]
- Mejia SP, et al. Human neutrophils produce extracellular traps against Paracoccidioides brasiliensis. Microbiology. 2015 doi: 10.1099/mic.0.000059. [DOI] [PubMed] [Google Scholar]
- Missall TA, et al. Mechanisms of resistance to oxidative and nitrosative stress: implications for fungal survival in mammalian hosts. Eukaryot Cell. 2004;3:835–46. doi: 10.1128/EC.3.4.835-846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira SF, et al. Monofunctional catalase P of Paracoccidioides brasiliensis: identification, characterization, molecular cloning and expression analysis. Yeast. 2004;21:173–82. doi: 10.1002/yea.1077. [DOI] [PubMed] [Google Scholar]
- Munoz JF, et al. Genome Diversity, Recombination, and Virulence across the Major Lineages of Paracoccidioides. mSphere. 2016:1. doi: 10.1128/mSphere.00213-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz JF, et al. Genome update of the dimorphic human pathogenic fungi causing paracoccidioidomycosis. PLoS Negl Trop Dis. 2014;8:e3348. doi: 10.1371/journal.pntd.0003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz JF, et al. The Dynamic Genome and Transcriptome of the Human Fungal Pathogen Blastomyces and Close Relative Emmonsia. PLoS Genet. 2015;11:e1005493. doi: 10.1371/journal.pgen.1005493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro RE, et al. catA, a new Aspergillus nidulans gene encoding a developmentally regulated catalase. Curr Genet. 1996;29:352–9. [PubMed] [Google Scholar]
- Neuberger G, et al. Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J Mol Biol. 2003;328:581–92. doi: 10.1016/s0022-2836(03)00319-x. [DOI] [PubMed] [Google Scholar]
- Parente-Rocha JA, et al. Macrophage Interaction with Paracoccidioides brasiliensis Yeast Cells Modulates Fungal Metabolism and Generates a Response to Oxidative Stress. PLoS One. 2015;10:e0137619. doi: 10.1371/journal.pone.0137619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Nadales E, et al. Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet Biol. 2014;70:42–67. doi: 10.1016/j.fgb.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, et al. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Pigosso LL, et al. Comparative proteomics in the genus Paracoccidioides. Fungal Genet Biol. 2013;60:87–100. doi: 10.1016/j.fgb.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Rappleye CA, et al. RNA interference in Histoplasma capsulatum demonstrates a role for alpha-(1,3)-glucan in virulence. Mol Microbiol. 2004;53:153–65. doi: 10.1111/j.1365-2958.2004.04131.x. [DOI] [PubMed] [Google Scholar]
- Restrepo A, et al. A technique to collect and dislodge conidia produced by Paracoccidioides brasiliensis mycelial form. J Med Vet Mycol. 1986;24:247–50. [PubMed] [Google Scholar]
- Restrepo S, et al. Development of pulmonary fibrosis in mice during infection with Paracoccidioides brasiliensis conidia. J Med Vet Mycol. 1992;30:173–84. [PubMed] [Google Scholar]
- Robinson JM, et al. Regulation of the NADPH-oxidase complex of phagocytic leukocytes. Recent insights from structural biology, molecular genetics, and microscopy. Histochem Cell Biol. 2004;122:293–304. doi: 10.1007/s00418-004-0672-2. [DOI] [PubMed] [Google Scholar]
- Ruane PH, et al. Photochemical inactivation of selected viruses and bacteria in platelet concentrates using riboflavin and light. Transfusion. 2004;44:877–85. doi: 10.1111/j.1537-2995.2004.03355.x. [DOI] [PubMed] [Google Scholar]
- Ruiz OH, et al. Alternative oxidase mediates pathogen resistance in Paracoccidioides brasiliensis infection. PLoS Negl Trop Dis. 2011;5:e1353. doi: 10.1371/journal.pntd.0001353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riis SB, Pedersen HM, Sørensen NK, Jakobsen M. Flow cytometry and acidification power test as rapid techniques for determination of the activity of starter cultures of Lactobacillus delbrueckii ssp. bulgaricus. Food Microbiology. 1995;12:245–250. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Scorzoni L, et al. Comparison of virulence between Paracoccidioides brasiliensis and Paracoccidioides lutzii using Galleria mellonella as a host model. Virulence. 2015;6:766–76. doi: 10.1080/21505594.2015.1085277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifuentes-Osornio J, et al. Epidemiology of Invasive Fungal Infections in Latin America. Curr Fungal Infect Rep. 2012;6:23–34. doi: 10.1007/s12281-011-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo D, et al. Identification and Analysis of the Role of Superoxide Dismutases Isoforms in the Pathogenesis of Paracoccidioides spp. PLoS Negl Trop Dis. 2016;10:e0004481. doi: 10.1371/journal.pntd.0004481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MM, et al. Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol Phylogenet Evol. 2009;52:273–83. doi: 10.1016/j.ympev.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Teixeira MM, et al. Paracoccidioides species complex: ecology, phylogeny, sexual reproduction, and virulence. PLoS Pathog. 2014;10:e1004397. doi: 10.1371/journal.ppat.1004397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca RA, et al. Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase, and impairment of mitochondrial metabolism are early events in heat shock-induced programmed cell death in tobacco Bright-Yellow 2 cells. Plant Physiol. 2004;134:1100–12. doi: 10.1104/pp.103.035956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiston E, et al. Comparative transcriptomics of the saprobic and parasitic growth phases in Coccidioides spp. PLoS One. 2012;7:e41034. doi: 10.1371/journal.pone.0041034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youseff BH, et al. Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress. PLoS Pathog. 2012;8:e1002713. doi: 10.1371/journal.ppat.1002713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancope-Oliveira RM, et al. Molecular cloning, characterization, and expression of the M antigen of Histoplasma capsulatum. Infect Immun. 1999;67:1947–53. doi: 10.1128/iai.67.4.1947-1953.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–8. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


