Abstract
Intellectual disability affects approximately 2% of the population with males outnumbering females due to involvement of over 300 genes on the X chromosome. The most common form of X-linked intellectual disability (XLID) is fragile X syndrome. We report a family with an apparent XLID pattern with the proband, his mother and maternal half brother having an Xp21.3 deletion detected with chromosomal microarray analysis involving the interleukin 1 receptor accessory protein-like 1 (IL1RAPL1) gene. IL1RAPL1 is highly expressed in the postnatal brain, specifically hippocampus suggesting a specialized role in memory and learning abilities. The proband presented with intellectual disability, a broad face, prominent and wide nasal root, ptosis, a wide philtrum and a small mouth. XLID due to involvement of the IL1RAPL1 gene has been reported to cause nonsyndromic XLID. We report a new family with XLID due to partial deletion of IL1RAPL1, summarize reported literature and describe similar phenotypic similarities among the affected individuals in this family and those reported in the literature proposing that deletion of IL1RAPL1 may cause syndromic XLID. Additional reports are needed to further characterize whether syndromic features are related to disturbances of this gene.
Keywords: X-linked intellectual disability, IL1RAPL1 deletion, Chromosomal microarray analysis, Xp21.3 deletion
1. Introduction
Intellectual disability affects approximately 2% of the population, with affected males outnumbering affected females due to disturbances involving over 300 genes located on the X chromosome [1]. The prevalence of X-linked intellectual disability (XLID) is approximately 1.8 males per 1000 [2, 3]. Two thirds of these subjects have non-specific (nonsyndromic) forms of XLID in which the cognitive impairment is not associated with phenotypic changes such as skeletal abnormalities or dysmorphic facial features [4]. The most common form of XLID is the fragile X syndrome, a syndromic form of XLID with a prevalence of one in 4000–6000 males [5]. We report a family with an apparent X-linked intellectual disability pattern with the proband, his mother and his maternal half brother having an Xp21.3 deletion involving the IL1RAPL1 gene, summarize reported literature and describe phenotypic similarities among the affected individuals in this family and those reported in the literature.
2. Clinical report
The proband was second child born to non-consanguinous parents following an uncomplicated, full term pregnancy. His birth weight was 3033 g (10th centile) and birth length was 45.7 cm (3rd centile). He walked at 16 months and had his first words at 8 months of age. He has worn glasses for hyperopia since childhood and was diagnosed with hypothyroidism at the age of 15 years. Otherwise, he has been physically healthy but with autism, mild intellectual disability, IQ of 55, hyperactivity and short attention span from a young age accompanied by impulsive aggression toward property, people and animals. Self-injury was noted from early childhood, including head-banging, self-hitting, self-poking and head-butting. He was previously placed in residential facilities and a state hospital for care. He has limited expressive language and cognitive abilities to answer abstract questions. Overall his behavior was rated as much improved with a score of 2 [1(very much improved) – 7(very much worse)] while on medications using the Clinical Global Impression (CGI) scale [6]. The CGI scale is widely used in psychiatry and consists of a 3-item observer-rated assessment tool to measure mental illness severity, global improvement or change and therapeutic response.
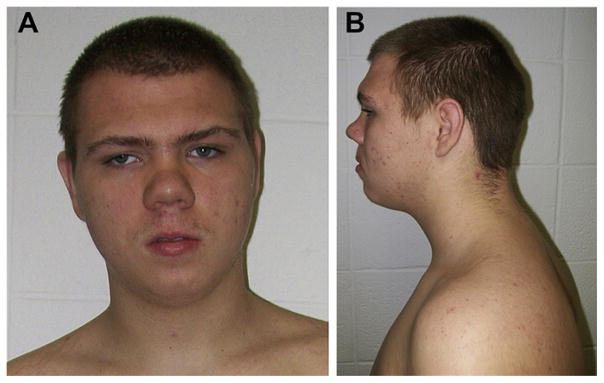
On physical examination at 15 years, 9 months of age, he was cooperative with a height of 165.6 cm (15th centile), weight of 81.3 kg (90th centile) and head circumference of 55 cm (50th centile). Inner canthal distance was 3.2 cm (60th centile), outer canthal distance was 8.8 cm (40th centile) and ear length was 6.5 cm (75th centile). He had prominent supraorbital ridges, ptosis, deep-set eyes, a prominent and sloping forehead with a broad face, synophrys and anterior scalp hair whorls, a short prominent nose with a long and wide philtrum and a small mouth (Fig. 1). Ridging was palpable for several cranial sutures. He had broad hands and feet with a hand length of 18.1 cm (60th centile) and middle finger length of 7.4 cm (50th centile). Vertical, pale striae were noted on his arms and on his protruding abdomen. No heart murmur was detected and lungs were clear to auscultation. External genitalia were that of a normal male. The rest of the examination was within normal range.
Fig. 1.
Frontal (A) and profile (B) views of the proband showing deep-set eyes, ptosis, a prominent sloping forehead with raised supraorbital ridges, a broad face, synophrys, a short prominent nose with a long and wide philtrum and a small mouth.
Fig. 2 shows the pedigree of the family. The proband has one full brother (III-6) without intellectual disability and a maternal half brother (III-8) with a 47, XYY karyotype and intellectual disability. The 47, XYY karyotype in III-8 is not thought to be a significant cause of his condition. Subject III-8 had physical features in common with the proband (III-7) including a broad face, prominent and wide nasal root, ptosis and a small mouth. There is a history of intellectual disability in two maternal uncles (II-2 and II-3) with similar facial findings to the proband (III-7) and his affected maternal half brother (III-8). One maternal aunt (II-5) and three cousins (III-3, III-4, III-5) are known to have intellectual disability. The proband’s mother (II-13) attended special education classes from the first through the third grades. She was mainstreamed in a normal classroom setting from the fourth through the eleventh grades, when she dropped out of school. She had no known physical or health problems. She is engaged in full time employment. Her parents, siblings, nieces and nephews were either deceased or unavailable for study upon repeated requests. The proband’s father is reportedly healthy, but with no additional paternal information.
Fig. 2.
Three generation pedigree of family with X-linked intellectual disability.
3. Results
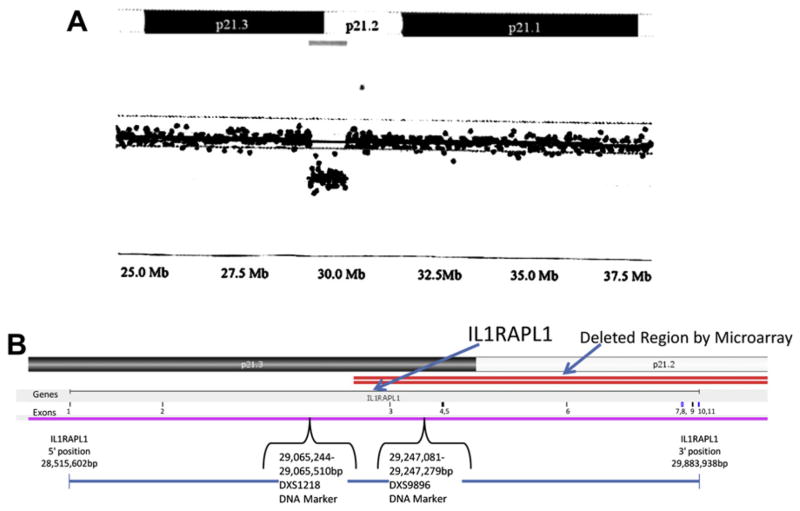
The proband had a cytogenetic study performed at four years of age that showed a 46, XY karyotype. During our clinical genetics evaluation, a chromosomal microarray study was performed commercially by CMDX Laboratory (Irvine, CA) using the 105 K Oligo HD Scan with probe distribution of approximately 30 kb to rule out DNA deletions or duplications. The microarray study showed a 950 kb deletion (located 29.13–30.08 Mb from the p-terminus) involving chromosome bands Xp21.3-p21.2. The interleukin 1 receptor accessory protein-like 1 (IL1RAPL1) gene is localized to chromosome bands Xp22.1-p21.3 at position 28,515,602 to 29,883,938 bp and contains 11 exons. It is partially deleted in our subject confirmed by FISH studies. The proband’s mother was also found to have the same deletion by chromosomal microarray analysis (Fig. 3). IL1RAPL1 is the only gene located within the deleted region.
Fig. 3.
(A) Chromosome microarray hybridization from the mother showing the location of the 950 kb deletion of the Xp21.3-p21.2 region occurring at 29.13–30.08 Mb from the p-terminus which includes a partial deletion of the IL1RAPL1 gene. (B) Expanded view of the X chromosome showing the location of the partial deletion (designated by double line) of the IL1RAPL1 gene and exons.
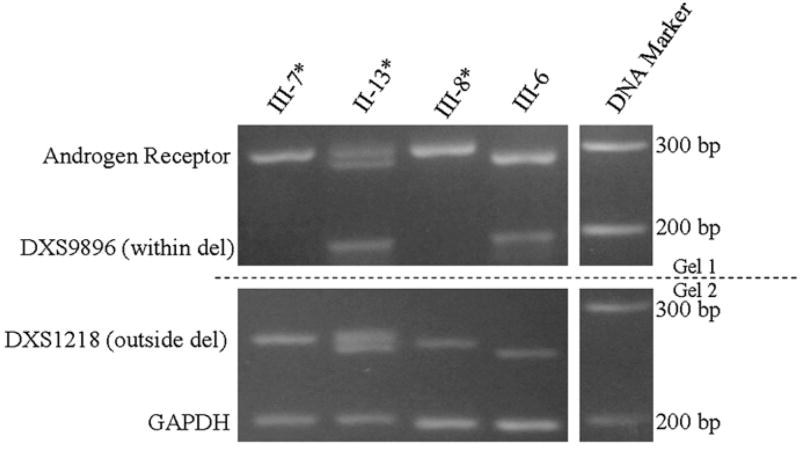
Polymerase chain reaction (PCR) analysis of DNA markers was undertaken on several available family members to examine for deletions of the X chromosome. Specifically, PCR primers (i.e., forward 5′ CCAGCCTGGCTGTTAGAGTA 3′; reverse 5′ ATATTCT-TATATTCCATATGGCACA 3′) for the DXS9896 marker are located between exons 3 and 4 within the deleted IL1RAPL1 gene region at position 29,247,081 to 29,247,279 bp and the DXS1218 locus at position 29,065,244 to 29,065,510 bp between exons 2 and 3 within the non-deleted region of IL1RAPL1 gene (at 28,515,602 to 29,883,938 bp). The PCR primers were used to amplify DNA fragments representing the two DNA markers to further establish the deletion status. All PCR fragments were analyzed using standard agarose 2.5% gel electrophoresis. A PCR fragment for DXS9896 was not amplified for the proband (III-7) and affected brother (III-8) confirming the deletion, while a PCR fragment was amplified for the unaffected brother (III-6) supporting the non-deletion status of the IL1RAPL1 gene for this region. All subjects showed two alleles with different repeat numbers detected for DXS1218 indicating a non-deletion status with the chromosomal microarray analysis performed on two family members (mother, II-13 and proband, III-7) showing a 950 kb deletion extending from 29.13 to 30.08 Mb or beyond the location of the IL1RAPL1 gene. Our PCR studies with DNA markers located between exons 2 and 3 (DXS1218) and exons 3 and 4 (DXS9896) would support a partial deletion of the IL1RAPL1 gene extending from exons 3 through 11 or the 3′ end of the gene in the family (see Fig. 3).
The proband (III-7), his brothers (III-6 and III-8) and mother (II-13) were also analyzed for X chromosome DNA polymorphic patterns of the normal and abnormal X chromosome carrying the partial deletion of the IL1RAPL1 gene contributed by the mother using the polymorphic androgen receptor (AR) gene located at Xq13. Genomic DNA was isolated from peripheral blood or buccal cells using Qiagen DNA Isolation Kits (Qiagen, Inc., Valencia, CA). The isolated genomic DNA was used as a template for PCR amplification of the CAG polymorphic region of the AR gene using the following primers: forward 5′ TCCAGAATCTGTTCCAGAGCGTGC 3′; reverse 5′ GCTGTGAAGGTTGCTGTTCCTCAT 3′. Two separate PCR fragments were amplified representing the two X chromosomes from the mother (II-13) and single alleles for each X chromosome in her three sons (III-6, III-7, III-8) (Fig. 4). Subject III-6 was found to have the mother’s normal X chromosome although the interpretation could be influenced by meiotic recombination. The proband (III-7) had the mother’s X chromosome fragment known to carry the partial deletion of the IL1RAPL1 gene by microarray analysis.
Fig. 4.
PCR primers within the androgen receptor (AR) gene were used to amplify polymorphic DNA fragments which showed that the unaffected Subject III-6 inherited the mother’s (Subject II-13) normal X chromosome, if recombination did not occur, while the affected male siblings (Subjects III-7 and III-8) had the mother’s X chromosome known to carry the partial deletion of the IL1RAPL1 gene. The AR allele 1 (290 bp) is in common with the affected Subjects III-7 and III-8 and represents the X chromosome with the deletion previously identified by chromosomal microarray analysis in the mother. PCR primers for the DXS9896 marker which is located between exons 3 and 4 of the IL1RAPL1 gene and DXS1218 located between exons 2 and 3 were used to amplify DNA fragments. Two DNA fragments representing each of the mother’s X chromosomes were amplified by PCR for DXS1218 and a single fragment was present for the 3 brothers indicating a non-deletion status of this DNA region. A single band was amplified for the unaffected brother, III-6, and the mother, II-13, supporting a non-deletion status for one of the X chromosomes. However, no fragments were amplified for the proband (III-7) or affected brother (III-8) supporting the deletion status for Subjects III-7 and III-8. GAPDH was used as an internal control marker.
* Subjects with partial deletion of the IL1RAPL1 gene identified by either PCR amplification of DNA markers within the gene and/or microarray analysis.
Microarray analysis identified the deletion in the proband (III-7) and the mother (II-13).
Androgen receptor marker location: 66,765,320–66,765,343 bp
DXS1218 location between exons 2–3: 29,065,244–29,065,510 (255–275 bp)
DXS9896 location between exons 3–4 29,247,081 – 29,247,279 (191–232 bp)
GAPDH is a control marker
All marker locations provided using NCBI36/hgl 8 assembly
The X chromosome in females is usually inactivated in a near-equal or random fashion during the last blastocyst stage of embryonic development, but occasionally X chromosome inactivation (XCI) can be non-random or skewed [7,8]. Skewed XCI seems to play a role in the presentation of diseases in females with known X-linked genetic involvement; for example, Rett syndrome [9], X-linked intellectual disability [10] and X-linked adrenoleukodystrophy [11]. To determine the XCI status in our proband’s mother (II-13), which could impact on gene expression, we performed an X chromosome inactivation assay with the methylation-sensitive restriction enzyme HpaII as described previously [12,13]. Skewed XCI is defined as >80% calculated ratio for either one of the AR gene alleles in the digested DNA sample. The XCI status of the proband’s mother (II-13) showed the percent inactivation of 56% for the allele (277 bp) representing the normal X chromosome while the percent inactivation of the allele (290 bp) representing the abnormal X chromosome was 44% (data not shown).
4. Discussion
Carrie et al. [14] first reported non-overlapping deletions and a nonsense mutation in a gene localized to Xp22.1-21.3 region associated in a family with non-specific X-linked intellectual disability and referred to as mental retardation, X-linked 34 (MRX34). This gene encoded a conserved 696 amino acid protein with homology to interleukin 1 receptor accessory proteins. A truncated mutation was found in exon 11 of the IL1RAPL1 gene in three of the affected brothers. The IL1RAPL1 gene is highly expressed in the postnatal brain, specifically the hippocampus. The latter suggests a specialized role for this gene in the physiologic processes underlying memory and learning abilities. Bahi et al. [15] further assessed the role of IL1RAPL1 and suggested that IL1RAPL1 may regulate calcium dependent exocytosis. In 2006, Tabolacci et al. [16] reported a three generation family with non-specific X-linked intellectual disability having mutations in exon 10 of the IL1RAPL1 gene. Later, four affected males in a three generation family segregating for non-specific X-linked intellectual disability showed linkage to Xp22.11-p21.2 and had a deletion of exons 2, 3, 4 and 5 in the IL1RAPL1 gene [1]. Tabolacci et al. [16] suggested that brain neurons may show different X-inactivation patterns after two female mutation carriers with learning disabilities showed blood leukocyte X-inactivation patterns favoring the wild type allele. In addition, Billuart et al. [17] carried out PCR screening of DNA samples from 92 patients with XLID for the presence of deletions in the Xp22.1-p21.3 region using a panel of 12 X-linked DNA micro-satellites in the region and found one case with a deletion confirmed by Southern blot analysis.
Piton et al. [18] reported a family with X-linked intellectual disability and autism spectrum disorder having a deletion of exons 3–7 in the IL1RAPL1 gene. They also described one female (isolated case) with borderline intelligence, Asperger syndrome and anxiety with a mutation (p.1367SX6) in exon 10 of the IL1RAPL1 gene. Later, Behnecke et al. [19] reported a 13-year-old male with mild ptosis, receding forehead, high nasal bridge, broad nasal base, large mouth, maxillary prognathism, macrodontia and developmental delay found with a 300 kb deletion of Xp21.3 involving deletion of exon 2 in the IL1RAPL1 gene identified by Affymetrix 500 K SNP array. Behnecke [19] also reported a 7-year-old male with macrocephaly, downslanting palpebral fissures, deep-set eyes, fullness of the upper eyelids, ptosis, strabismus, short upturned nose with a broad tip, thin upper vermillion, facial hypotonia and developmental delay with a 482 kb deletion involving exons 3 through 5 of the IL1RAPL1 gene identified by microarray analysis. Whibley et al. [20] identified two unrelated families with moderate intellectual disability found to have partial deletions of the IL1RAPL1 gene. Franek et al. [21] reported two families with X-linked intellectual disability and mild dysmorphic features including synophrys, broad and flat nose, prominent lips and prominent jaw with intragenic deletions of the IL1RAPL1 gene. Kozak et al. [22] reported a family with X-linked intellectual disability and evidence of linkage to Xp21.1-Xp22.3 and later Tabolacci et al. [16] characterized a mutation in exon 10 of the IL1RAPL1 gene in the affected individuals. As described by Kozak et al. [22], the affected males in their report had common facial features including a prominent and wide nasal root, abnormal philtrum and nostrils, ptosis, small mouth, broad face, synophrys and palatal problems. In reviewing the facial photographs of affected males reported by Nawara et al. [1], similar features were seen including a broad face, prominent and wide nasal root, ptosis, a wide philtrum and a small mouth.
Hence, our report further documents the presence of a deletion of this gene by chromosomal microarray studies and PCR analysis of probes on the X chromosome within the deleted segment that accounts for intellectual disability in the proband and other family members studied. Although other members in the extended family were not available for analysis, we presume that the cause of their intellectual disabilities was also due to the IL1RAPL1 gene deletion. Deletions involving the IL1RAPL1 gene have previously been thought to cause non-specific XLID; however, in review of the literature and comparing the clinical findings, there appears to be features in common with affected individuals previously reported and with the males in this family (current report) including synophrys, ptosis, small mouth and prominent and wide nasal root as summarized in Table 1. Additional reports are needed to further characterize the apparent phenotype related to specific disturbances of this gene along with intellectual disability. Therefore, the authors encourage the reporting of other individuals with involvement of this gene for better clinical delineation, genetic counseling and research linking the IL1RAPL1 gene to autism and intellectual disability.
Table 1.
Summary of facial features in reported individuals with IL1RAPL1 gene defects.
| Features | Kozak et al. [22] |
Nawara et al. [1] |
Behnecke et al. [19] Subject 1 |
Behnecke et al. [19] Subject 2 |
Franek et al. [21] Family 1 |
Franek et al. [21] Family 2 |
Subject III-7a | Subject III-8a |
|---|---|---|---|---|---|---|---|---|
| Prominent and wide nasal root | + | + | + | − | + | − | + | + |
| Abnormal/wide philtrum | + | + | − | + | − | − | + | − |
| Ptosis | + | + | + | + | − | + | + | + |
| Small mouth | + | + | − | − | − | + | + | |
| Broad face | + | + | − | + | − | − | + | + |
| Synophrys | + | − | − | − | + | + | + | − |
| Prominent jaw | − | − | + | − | − | + | − | − |
| Palatal abnormalities | + | Unknown | Unknown | Unknown | Unknown | Unknown | − | − |
Current report.
References
- 1.Nawara M, Klapecki J, Borg K, Jurek M, Moreno S, Tryfon J, Bal J, Chelly J, Mazurczak T. Novel mutation of IL1RAPL1 gene in a nonspecific X-linked mental retardation (MRX) family. Am J Med Genet Part A. 2008;146A:3167–3172. doi: 10.1002/ajmg.a.32613. [DOI] [PubMed] [Google Scholar]
- 2.Herbst DS, Miller JR. Nonspecific X-linked mental retardation II: the frequency in British Columbia. Am J Med Genet. 1980;7:461–469. doi: 10.1002/ajmg.1320070407. [DOI] [PubMed] [Google Scholar]
- 3.Chiurazzi P, Hamel BC, Neri G. XLMR genes: update 2000. Eur J Med Genet. 2001;9:71–81. doi: 10.1038/sj.ejhg.5200603. [DOI] [PubMed] [Google Scholar]
- 4.Jacquemont S, Des Portes V, Hagerman R. Fragile X and X-linked mental retardation. In: Butler MG, Meaney FJ, editors. Genetics of Developmental Disabilities. 1. Taylor & Francis; Boca Raton: 2005. pp. 247–278. [Google Scholar]
- 5.Crawford DC, Acuna JM, Sherman SL. FMR1 and fragile X syndrome: human genome epidemiology review. Genet Med. 2001;3:359–371. doi: 10.1097/00125817-200109000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guy W. Early Clinical Drug Evaluation Unit Assessment Manual for Psychopharmacology. U.S. Department of Health, Education, and Welfare; Rockville, MD: 1976. [Google Scholar]
- 7.Kay GF, Barton SC, Surani MA, Rastan S. Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell. 1994;77:639–650. doi: 10.1016/0092-8674(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 8.Orstavik KH. Skewed X inactivation in healthy individuals and in different diseases. Acta Paediatr Suppl. 2006;95:24–29. doi: 10.1111/j.1651-2227.2006.tb02385.x. [DOI] [PubMed] [Google Scholar]
- 9.Amir RE, Van den Veyver IB, Schultz R, Malicki DM, Tran CQ, Dahle EJ, Philippi A, Timar L, Percy AK, Motil KJ, Lichtarge O, Smith EO, Glaze DG, Zoghbi HY. Influence of mutation type and X chromosome inactivation on Rett syndrome phenotypes. Ann Neurol. 2000;47:670–679. [PubMed] [Google Scholar]
- 10.Plenge RM, Hendrich BD, Schwartz C, Arena JF, Naumova A, Sapienza C, Winter Rm, Willard HF. A promoter mutation in the Xist gene in two unrelated families with skewed X-chromosome inactivation. Nat Genet. 1997;17:353–356. doi: 10.1038/ng1197-353. [DOI] [PubMed] [Google Scholar]
- 11.Maier EM, Krammerer S, Muntau AC, Wichers M, Braun A, Roscher AA. Symptoms in carriers of adrenoleukodystrophy relate to skewed X inactivation. Ann Neurol. 2002;52:683–688. doi: 10.1002/ana.10376. [DOI] [PubMed] [Google Scholar]
- 12.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 13.Butler MG, Theodoro MF, Bittel DC, Kuipers PJ, Driscoll DJ, Talebizadeh Z. X-chromosome inactivation patterns in females with Prader-Willi syndrome. Am J Med Genet. 2007;143A:469–475. doi: 10.1002/ajmg.a.31506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrie A, Jun L, Bienvenu T, Vinet MC, McDonell N, Couvert P, Zemni R, Cardona A, Van Buggenhout G, Frints S, Hamel B, Moraine C, Ropers HH, Strom T, Howell GR, Whittaker A, Ross MT, Kahn A, Fryns JP, Beldjord C, Marynen P, Chelly J. A new member of the IL-1 receptor family highly expressed in hippocampus and involved in X-linked mental retardation. Nat Genet. 1999;23:25–31. doi: 10.1038/12623. [DOI] [PubMed] [Google Scholar]
- 15.Bahi N, Friocourt G, Carrie A, Graham ME, Weiss JL, Chafey P, Fauchereau F, Burgoyne RD, Chelly J. IL1 receptor accessory protein like, a protein involved in X-linked mental retardation, interacts with neuronal calcium sensor-1 and regulates exocytosis. Hum Mol Genet. 2003;12:1415–1425. doi: 10.1093/hmg/ddg147. [DOI] [PubMed] [Google Scholar]
- 16.Tabolacci E, Pomponi MG, Piertrobono R, Terracciano A, Chiurazzi P, Neri G. A truncating mutation in the IL1RAPL1 gene is responsible for X-linked mental retardation in the MRX21 family. Am J Med Genet Part A. 2006;140:482–487. doi: 10.1002/ajmg.a.31107. [DOI] [PubMed] [Google Scholar]
- 17.Billuart P, Vinet MC, des Portes V, Llense S, Richard L, Moutard ML, Recan D, Bruls T, Bienvenu T, Kahn A, Beldjord C, Chelly J. Identification by STS PCR screening of a microdeletion in Xp21.3-22.1 associated with non-specific mental retardation. Hum Mol Genet. 1996;5:977–979. doi: 10.1093/hmg/5.7.977. [DOI] [PubMed] [Google Scholar]
- 18.Piton A, Michaud JL, Peng H, Aradhya S, Gauthier J, Mottron L, Champagne N, Lafreniere RG, Handan FF, Joober R, Rombonne E, Marineau C, Cossette P, Dube M, Haghighi P, Drapeau P, Barker PA, Carbonetto S, Rouleau GA. Mutations in the calcium-related gene IL1RAPL1 are associated with autism. Hum Mol Genet. 2008;17:3965–3974. doi: 10.1093/hmg/ddn300. [DOI] [PubMed] [Google Scholar]
- 19.Behneck A, Hinderhofer K, Bartsch O, Numann A, Ipach ML, Damatova N, Haaf T, Dufke A, Riess O, Moog U. Intragenic deletions of IL1RAPL1: report of two cases and review of the literature. Am J Med Genet Part A. 2010;155:372–379. doi: 10.1002/ajmg.a.33656. [DOI] [PubMed] [Google Scholar]
- 20.Whibley AC, Plagnol V, Tarpey PS, Abidi F, Fullston T, Choma MK, Boucher CA, Shepherd L, Willatt L, Parkin G, Smith R, Futreal PA, Shaw M, Boyle J, Licata A, Skinner C, Stevenson RE, Turner G, Field M, Hickett A, Schwartz CE, Gecz J, Stratton MR, Raymond FL. Fine-scale survey of X chromosome copy number variants and indels underlying intellectual disability. Am J Hum Genet. 2010;87:173–188. doi: 10.1016/j.ajhg.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franek KJ, Butler J, Johnson J, Siemensen R, Friez MJ, Bartel F, Moss T, DuPont B, Berry K, Bauman M, Skinner C, Stevenson RE, Schwartz CE. Deletion of the immunoglobulin domain of IL1RAPL1 results in nonsyndromic X-linked intellectual disability associated with behavioral problems and mild dysmorphism. Am J Med Genet Part A. 2011;155:1109–1114. doi: 10.1002/ajmg.a.33833. [DOI] [PubMed] [Google Scholar]
- 22.Kozak L, Chiurazzi P, Genuardi M, Pomponi MG, Zollino M, Neri G. Mapping of a gene for non-specific X linked mental retardation: evidence for linkage to chromosomal region Xp21.1-Xp22.3. J Med Genet. 1993;15:8125–8148. doi: 10.1136/jmg.30.10.866. [DOI] [PMC free article] [PubMed] [Google Scholar]