Abstract
Incorporation of variant histone sequences, in addition to post-translational modification of histones, serves to modulate the chromatin environment. Different histone chaperone proteins mediate the storage and chromatin deposition of variant histones. Although the two non-centromeric histone H3 variants, H3.1 and H3.3, differ by only five amino acids, replacement of histone H3.1 with H3.3 can modulate transcription for highly expressed and developmentally required genes, lead to the formation of repressive heterochromatin, or aid in DNA and chromatin repair. The human HIRA complex composed of HIRA, UBN1, CABIN1 and transiently ASF1a, forms one of two complexes that binds and deposit H3.3/H4 into chromatin. A number of recent biochemical and structural studies have revealed important details underlying how these proteins assemble and function together as a multi-protein H3.3-specific histone chaperone complex. Here we present a review of existing data and present a new model for assembly of the HIRA complex and for HIRA-mediated incorporation of H3.3/H4 into chromatin.
Graphical Abstract
Introduction
About 3 billion base pairs of DNA are packaged into the nucleus of every cell in the human body1. To allow for the storage and regulation of this great length of genetic material the negatively charged DNA is compacted through binding to positively charged histone proteins for the formation of nucleosomes, the basic repeating units of chromatin2,3. The nucleosome core particle is built from 147 base pairs of DNA wrapped around an octamer of core histone proteins composed of two copies each of H2A, H2B, H3, and H44–6. While the majority of nucleosomes are composed of canonical histones, there are also histones with variant amino acid sequences that are incorporated at specific regions of chromatin7. Targeted incorporation of these variant histones7 and post-translation modification to a variety of histone residues8 are the two major ways that histones contribute to modulation and regulation of the chromatin environment to mediate distinct DNA-templated activities such as DNA repair, transcriptional regulation or DNA replication.
Nucleosome assembly is coordinated by histone chaperone proteins, which bind specific histones to mediate their storage, eviction, or deposition from/or into chromatin9–11. Histone chaperones are generally dedicated to binding and deposition of either the H3/H4 unit or the H2A/H2B unit although there are examples of proteins capable of chaperoning both H3/H4 and H2A/H2B12,13. Some histone chaperons are able to bind several different variants of a particular histone, while others are dedicated to specifically binding just one variant sequence11. The histone chaperone ASF1a binds to either H3.1/H4 and H3.3/H4 and associates with the CAF-1 complex for H3.1/H4 deposition or the HIRA complex for H3.3/H4 deposition14,15. Although H3.1 and H3.3 differ by only 5 amino acids, the HIRA complex specifically mediates replication-independent H3.3/H4 deposition, while the CAF-1 histone chaperone carries out H3.1/H4 deposition coupled to DNA replication and damage repair14,16.
The HIRA complex is composed of the proteins HIRA, Ubinuclein-1 (UBN1), and CABIN114,17,18. These three proteins function with ASF1a to mediate deposition of H3.3/H4 primarily at the bodies of actively transcribed genes19,20, gene regulatory regions17,18,21, developmentally regulated genes22,23, and areas of DNA and chromatin damage and repair24–27. The ATRX/DAXX histone chaperone complex also binds specifically to H3.3/H4 but functions independently of ASF1a and the HIRA complex to deposit H3.3/H4 mainly into areas of heterochromatin at telomeres, pericentromeres, and endogenous retroviral elements19,28–31. While the HIRA and ATRX/DAXX complexes have largely separate functions, they employ similar structural features to bind H3.3/H432, both complexes may have some overlapping function in H3.3/H4 deposition during cellular senescence17,18,31,33,34, and ATRX/DAXX has been reported to deposit H3.3/H4 to activate gene transcription in neuronal cells 35.
In recent years, a number of biochemical and structural studies have contributed to a greatly increased understanding of the molecular assembly and function of the HIRA histone chaperone complex. In this review, we focus on detailing these developments in order to present an updated model of HIRA complex assembly for H3.3-specific binding and deposition functions.
HIRA Complex Subunits
The HIRA complex is assembled from HIRA, UBN1 and CABIN1 and coordinates with ASF1a to carry out H3.3-specific deposition. All three subunits and ASF1 were identified and characterized individually before it was discovered that they work in concert to mediate deposition of H3.3/H414. In this section we will review the known biological functions of the individual proteins that comprise the HIRA complex.
ASF1a
ASF1 (anti-silencing function 1) was originally identified in S. cerevisiae and named for the observed de-repression of silent mating type loci when overexpressed36. ASF1 was later characterized as a novel H3/H4 binding protein involved in histone deposition during DNA replication and repair37. In human cells, ASF1 has two isoforms, ASF1a and ASF1b14,15. ASF1a/b bind to both H3.1/H4 and H3.3/H4 with similar affinity14. The CAF-1 complex cooperates with both ASF1a/b for H3.1/H4 deposition, while only ASF1a serves to deliver H3.3/H4 to the HIRA complex14,15,34. Both HIRA and CAF-1 associate with the ASF1 N-terminal core using a small but conserved B-domain15,34. ASF1a/b have also been shown to cooperate with MCM2 to coordinate H3/H4 deposition. While ASF1a and ASF1b are highly conserved in their N-terminal core domains, they are divergent in their C-terminal tails; it has been shown that the C-terminal tail of ASF1b in combination with some region of HIRA outside the B-domain work together to inhibit HIRA binding, leading to a preferential association of HIRA with ASF1a15.
HIRA
Human HIRA was identified within a genomic region lost in chromosome 22q11 deletion syndrome (DiGeorge syndrome)38. Depletion of HIRA was initially thought to be a possible contributing factor to the symptoms observed in patients with DiGeroge syndrome38, although deletion of the Tbx1 gene was later identified as responsible for the symptoms of DiGeorge syndrome 39. Previous to the identification of HIRA within the 22q11 deletion, two S. cerevisiae proteins were identified as transcriptional regulators of the histone genes and named Hir1 and Hir2 for their histone regulatory function40,41. When the open reading frame in the 22q11 deletion was analyzed it was discovered that it was homologous to Hir1 and Hir2 and the protein was named HIRA. The histone chaperone function of HIRA was not discovered until later when the HIRA complex was purified from HeLa cells using a tagged H3.3 to pull down a complex containing ASF1a, HIRA, UBN1, and CABIN114. The transcriptional regulation of histone genes observed for S. cerevisiae Hir1/Hir2 has not been observed for human HIRA, although histone deposition by the HIRA complex has been linked to transcriptional activation for a number of highly expressed and developmentally regulated genes19,22,42,43. After the histone chaperone function of the human HIRA complex was characterized, S. cerevisiae Hir1/Hir2 were shown to form a complex with Hpc2 and Hir3, the S. cerevisiae orthologs of UBN1 and CABIN1, respectively, to carry out replication independent H3/H4 deposition44,45.
UBN1
UBN1 was originally identified as a ubiquitously expressed protein in the nuclei of human cells and shown to associate with endogenous and viral transcription factors to compete with binding to their DNA target sites46,47. Although this transcription factor binding activity of human UBN1 is not well characterized at the biochemical level, the HIRA complex member CABIN1 has also been reported to bind a number of transcription factors48–52 so it is possible that the original observation was mediated through CABIN1. Similarly to HIRA, UBN1 was identified as a member of the HIRA complex when pulled down with H3.3. HIRA and UBN1 together appear to form a functional core of the HIRA complex as knock down, mutation, and depletion experiments suggest that both ASF1a and CABIN1 are dispensable for proper H3.3 deposition activity of the complex26,53–55. Although the two proteins are weakly conserved, UBN1 was identified as an ortholog of the S. cerevisiae Hir complex subunit Hpc2 through alignment of the most conserved domain between the two proteins referred to as the Hpc2-related domain (HRD)17, or less commonly as the Hpc2-Ubinuclein-1 domain (HUN)56. Recently UBN1 was shown to have H3.3-specific binding activity and likely mediates the specific deposition of H3.3/H4 over H3.1/H4 by the HIRA complex32.
CABIN1
The largest member of the HIRA complex at 2220 amino acids, CABIN1, is predicted to have around thirty tetratricopeptide repeats (TPR)56,57. TPR domains are built from repeats of bi-helical pairs and often serve to mediate protein-protein interactions in large complexes58. Outside of binding to the C-terminal domain of HIRA18, the role of this TPR containing protein in the H3.3 deposition process remains unknown. Several functions of CABIN1 independent of the HIRA complex have been well studied. CABIN1 was identified as a binding partner of calcineurin and acts as a regulator of the calcineurin mediated signaling pathway55. CABIN1 has also been shown to physically associate with transcription factors p5351 and MEF248,50,52,59 to modulate their transcriptional regulatory activity. Further, modulation of MEF2 activity by CABIN1 is also dependent on HIRA and ASF1a histone chaperone activity60; so it is possible that the role of CABIN1 is to bind to these, and potentially other, transcription factors and recruit the HIRA complex to deposit H3.3/H4 at targeted areas of chromatin. This HIRA mediated H3.3/H4 deposition may serve to modulate the local chromatin architecture and alter levels of transcription. Similar to CABIN1, the Hir3 S. cerevisiae ortholog of CABIN1 forms a complex with Hir1, Hir2, and Hpc2 to regulate transcription of histone genes and carry out replication independent histone deposition41,44,45.
HIRA Complex Assembly and Structure
In recent years, several studies have focused on determining many of the key protein-protein contacts involved in assembly of the HIRA complex and its specific association with H3.3/H4 over H3.1/H4. Here we present a summary of current biochemical and structural data in an effort to describe the most accurate model of HIRA complex assembly and function as an H3.3-specific histone chaperone.
HIRA as a molecular scaffold
From several early studies, it became clear that the protein HIRA was the central member of this H3.3-specific histone chaperone complex. The histone chaperone activity of HIRA was described independently of ASF161,62, and its direct interaction and cooperation with ASF1a was identified shortly thereafter14,15,34. The obvious functional significance of the HIRA protein in H3.3 deposition led to the current nomenclature of referring to the entire HIRA/UBN1/CABIN1 complex, and transiently ASF1a, as the HIRA complex. The undefined C-terminal domain of HIRA has been reported to bind H3/H4 with phosphorylation at H4S4763, although this data has not been expanded upon and several other studies suggest that UBN1 may be responsible for the H3.3-specific binding and deposition by the HIRA complex17,26,32,55. Most of the existing data suggests that HIRA likely serves as a molecular scaffold for assembly of the complex, as it is known to directly associate with every other member of the HIRA complex (Fig. 1). Interestingly the stability of the core HIRA/UBN1/CABIN1 complex is dependent on the presence of all three proteins, as knockdown or removal of any individual subunit results in severely decreased protein expression levels for the remaining subunits17,18,55.
Figure 1. Domain structures and interactions of HIRA complex members and H3.3/H4.
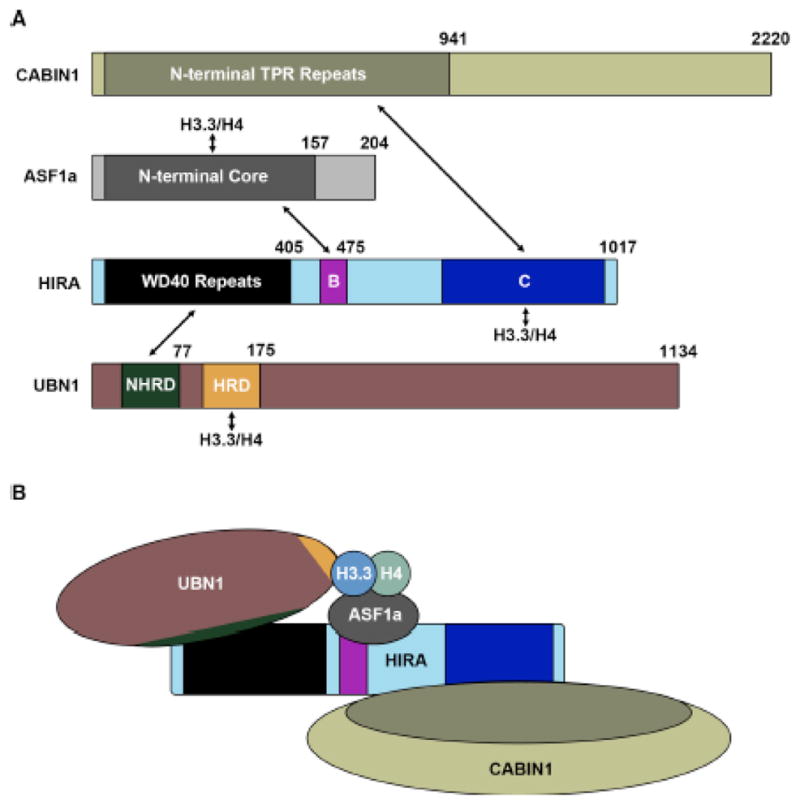
(A) Domain structures of HIRA, UBN1, CABIN1 and ASF1a with arrows depicting all reported interactions between the complex members and H3.3/H4. (B) Proposed model for assembly of the HIRA complex based on the reported interacting domains.
ASF1a/H3/H4
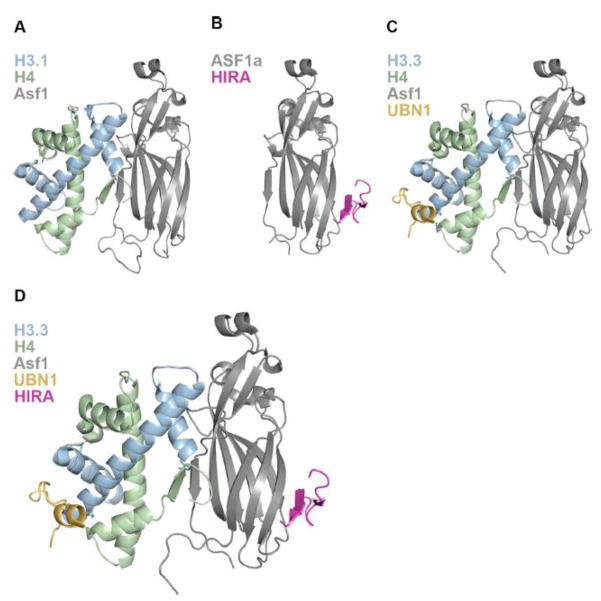
Binding and deposition of H3/H4 by the ASF1 core domain has been extensively studied using biochemical and structural techniques35,42,58–60 (Fig. 1). While there is less structural information for the ASF1a/H3/H4 complex64, the ASF1 core domain is so conserved that information from the S. cerevisiae structure is likely applicable to the human ASF1a system37,45,65–67. It is clear that ASF1 binds to one H3/H4 dimer and has extensive contacts with H3/H4 along the (H3/H4)2 tetramer interface5,66–68 (Fig 2a) thus inhibiting the formation of the (H3/H4)2 tetramer, which in the absence of ASF1 exists in dynamic equilibrium with the H3/H4 dimer12,69. ASF1a does not make any contacts to or near H3 residues 87–90, which are the major differentiating amino acids between H3.3 and H3.1, and is capable of supplying H3.1/H4 to the CAF-1 complex or H3.3/H4 to the HIRA complex14,15,34.
Figure 2. Structural basis for HIRA complex assembly and function.
(A) 1.7 Å crystal structure of S. cerevisiae Asf1 bound to X. laevis H3.1/H4 depicting the interaction of Asf1 with one H3/H4 dimer. (B) 2.7 Å crystal structure of the human HIRA/ASF1a complex showing association of the HIRA B-domain with the ASF1a core on the surface opposite to where ASF1a binds H3/H4. (C) 2.3 Å crystal structure of the UBN1/H3.3/H4/Asf1 complex revealing the molecular contacts between the UBN1 HRD and histone H3.3. (D) Structural model of the HIRA B-domain superimposed onto the UBN1/H3.3/H4/Asf1 complex depicting a hypothetical intermediate in the pathway of HIRA-mediated H3.3/H4 deposition.
HIRA/ASF1a
The interaction between ASF1a and HIRA was discovered upon the initial pull-down of the HIRA complex form HeLa cells with tagged H3.314 and the HIRA B-domain spanning residues 439–475 was identified as the region responsible for contacting the ASF1a N-terminal core34 (Fig. 1). The specific amino acid contacts between HIRA and ASF1a were then identified through determination of a 2.7 Å crystal structure of HIRA B-domain residues 446–466 bound to the ASF1a core domain15 (Fig. 2b). Importantly the structure reveals that HIRA associates with the ASF1a core domain on the ASF1 surface opposite to where ASF1 binds the H3/H4 dimer, indicating that another domain of HIRA or member of the complex is likely capable of binding to H3.3/H4 on the outer surface, where the differing residues between H3.3 and H3.1 reside, to potentiate H3.3/H4 specific binding and chromatin deposition (Fig. 2a,b).
HIRA/UBN1
The interaction between HIRA and UBN1 is essential for the stability of the HIRA complex as endogenous knockdown of UBN1 results in decreased HIRA expression, and the same is observed for UBN1 expression when HIRA is knocked down17. Additionally UBN1 is essential for H3.3-specific deposition as depletion of UBN1 leads to decreased H3.3/H4 incorporation into chromatin26. As the interaction between HIRA and UBN1 is essential for the stability and H3.3/H4 deposition function of the HIRA complex17,26, understanding the molecular basis the HIRA/UBN1 interaction is of great interest. It was initially reported that the N-terminal WD40 repeat domain in HIRA interacted with the HRD in UBN117. As the HRD is the most evolutionarily conserved domain in UBN1 this result initially made biological sense17,56. Although upon further biochemical dissection of the HIRA/UBN1 complex it was discovered that a weakly conserved domain slightly N-terminal to the HRD was essential for HIRA binding activity by UBN1, while the HRD was dispensable, this new UBN1 domain N-terminal to the HRD was called the NHRD70 (Fig. 1).
HIRA/CABIN1
CABIN1 was shown to be dispensable for H3.3/H4 deposition by the HIRA complex26, although similarly to UBN1, it appears to be important for the structural integrity of the HIRA complex. Depletion of endogenous CABIN1 levels in cells results in decreased expression of endogenous HIRA, and depletion of HIRA has the same effect on CABIN1 expression18. Additionally, in the yeast Hir complex, removal of Hir1, Hir2 or Hpc2 results in decreased expression of Hir355. Together these data suggest an important role for CABIN1 in the stability of the HIRA complex. While the entire 2220 amino acids of CABIN1 are spanned by about 30 TPR repeats57, deletion experiments have mapped the N-terminal TPR segment of CABIN1, encompassing residues 1–941, as the minimal region required for HIRA binding18 (Fig. 1). Deletions probing HIRA have identified the structurally undefined C-terminal domain of HIRA composed from residues 736–963 as responsible for contacting CABIN118 (Fig 1).
UBN1/H3.3/H4
After the NHRD of UBN1 was identified as required for complex formation with the HIRA WD40 repeats, two studies implicated the more strongly conserved UBN1 HRD in H3.3-specific binding and chromatin deposition32,55. Experiments probing function of the yeast Hir complex in H3.3-specific deposition in a human cell system showed that deletion of the HRD in Hpc2 abrogates H3.3/H4 deposition specificity55. Following this, a 2.3 Å crystal structure of UBN1 HRD residues 122–142 bound to H3.3/H4/Asf1 was determined and together with biochemical studies revealed that the highly conserved UBN1 HRD binds to H3.3 in the proximity of residues 87–90 to mediate specificity for binding to H3.3/H4 over H3.1/H432 (Fig. 2c). The crucial H3.3 residue for UBN1 binding specificity appears to be G90, as swapping M90 from H3.1 into H3.3 strongly decreases binding affinity32.
Structural model for the HIRA complex
Based on the existing structural data for the HIRA sub-complexes of HIRA/ASF1a and UBN1/H3.3/H4/Asf1 we can infer a structural model where ASF1a associates with histones H3.3/H4 and delivers them to the HIRA complex for deposition through association of ASF1a with the HIRA B-domain, and H3.3/H4 with the UBN1 HRD (Fig. 2d). As the order of assembly for the HIRA complex is not known, we propose that the UBN1 HRD binds to H3.3/H4 in complex with ASF1a at some point during ASF1a/HIRA complex formation. As the UBN1 HRD has greatly reduced binding affinity for H3.1/H4 in comparison with H3.3/H432, UBN1 likely functions to reject any ASF1a/H3.1/H4 complexes and ensure that only ASF1a bound to H3.3/H4 will be fully accepted for chromatin deposition by the HIRA complex.
Model for histone deposition by the HIRA complex
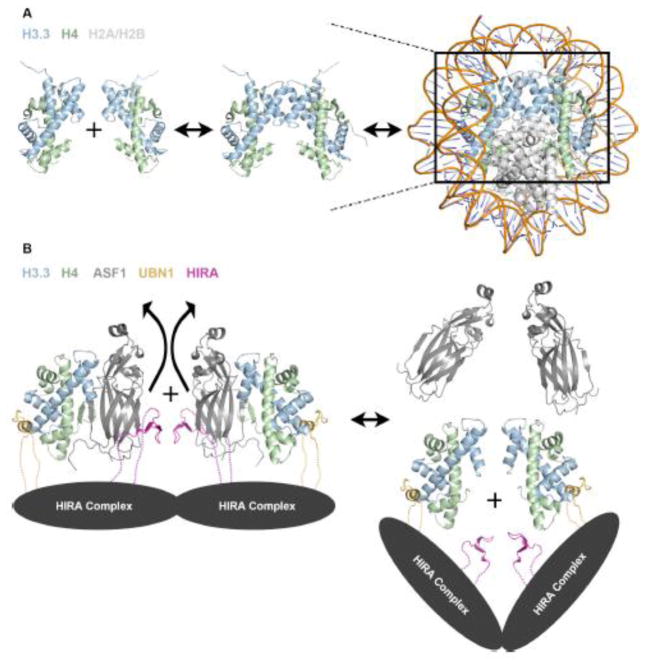
A compilation of recent findings on HIRA complex assembly and H3.3-specific binding and deposition has led to our proposed structural model for the HIRA complex represented by a HIRA/UBN1/ASF1a/H3.3/H4 sub-complex (Fig. 2d). While all existing data currently point to our proposed molecular assembly, this complex may represent an intermediate in the formation of the (H3/H4)2 tetramer to be deposited into nucleosomes. ASF1a binds to a dimer of H3/H4 along the tetramerization interface and prevents the formation of the (H3/H4)2 tetramer67. In the absence of ASF1, H3/H4 dimers exist in equilibrium with (H3/H4)2 tetramers12,69,71 (Fig 3a). Stable isotope labeling with amino acids in cell culture (SILAC) mass spectrometry data indicates that the majority of H3/H4 is deposited into nucleosomes in the (H3/H4)2 tetramer form and tetramer splitting between nucleosomes is relatively rare72 (Fig 3a).
Figure 3. Model for incorporation of H3.3/H4 into nucleosomes by the HIRA complex.
(A) In solution, histones H3/H4 exist in equilibrium between dimer and tetramer populations, although strong evidence suggests that they are deposited into the nucleosome in the tetramer form. (B) Model for HIRA complex mediated formation of an (H3.3/H4)2 tetramer prior to nucleosome deposition based on existing structural and biochemical data. The dotted lines represent presumed contacts lacking structural insight.
Many histone chaperones (CAF-112, MCM273,74, SPT275, Rtt10676, Vps7577, Nap177, and SPT1613) have been shown to stably bind an H3/H4 tetramer. MCM2 has been shown to associate with H3/H4/ASF1 or an (H3/H4)2 tetramer during DNA replication12,73, and similarly to the UBN1 HRD, a loop of MCM2 interacts with the histone surface between the α1 and α2 helices of H332,73. One molecule of MCM2 is capable of binding to an H3/H4 dimer associated with ASF1 while two MCM2 molecules can also bind to an (H3/H4)2 tetramer; indicating that MCM2 may act as an intermediate in the formation of (H3.1/H4)2 tetramers for deposition and stabilization of (H3.1/H4)2 tetramers for recycling during DNA replication73. Additionally, several studies indicate that the HIRA complex may be able to still deposit H3.3/H4 even in the absence of ASF1a26,53,54. These data indicate that the ASF1a is likely not a permanent member of the complex and may serve to bind and protect H3.3/H4 before delivering them to the HIRA complex for tetramer formation and chromatin deposition. Based on these observations we propose a model for H3.3/H4 deposition by the HIRA complex where H3.3/H4/ASF1a associates with HIRA/UBN1/CABIN1 complex followed by eviction of ASF1a and UBN1-mediated formation of an (H3.3/H4)2 tetramer prior to deposition onto DNA (Fig 3b).
Although crystallographic evidence shows that residues 122–148 of the UBN1 HRD bound to an H3.3/H4 dimer in complex with ASF132, in solution ITC data in the absence of ASF1 indicates that fragments of UBN1 as large as 92–175 can bind an (H3.3/H4)2 tetramer with a stoichiometry of 2 molecules or UBN1 to one (H3.3/H4)2 tetramer32. As the UBN1 ITC experiments were conducted with H3/H4 concentrations where the histones should exist almost entirely in the tetramer form12,32, it can be concluded that the data represents two molecules of UBN1 associating with identical binding sites on one (H3.3/H4)2 tetramer. Considering both the ITC and crystallographic evidence, it seems possible that UBN1 may function, similarly to MCM2, to bind H3/H4 dimers associated with ASF1 and mediate tetramer formation (Fig 3b).
The molecular mechanism of how two UBN1 molecules cooperate to form the (H3.3/H4)2 tetramer is not known. Although there is no existing evidence for oligomerization of the human HIRA complex, the S. cerevisiae Hir complex has been reported to form an assembly of one copy each of Hir1 and Hir3 and two copies each of Hir2 and Hpc244. With two copies of the UBN1 ortholog Hpc2 found in the S. cerevisiae complex, it is possible that two HIRA complex molecules may come together for the formation of the (H3/H4)2 tetramer (Fig 3b).
Additionally, it is unclear if the HIRA complex cooperates with ATP-dependent chromatin remodelers to mediate deposition of H3.3/H4 onto DNA similarly to the complex formed by the histone chaperone DAXX and chromatin remodeler ATRX. It has been reported that S. cerevisiae Hir complex likely functions to recruit the SWI/SNF complex to histone promoters 78 but a direct interaction between the Hir and SWI/SNF complexes has not been conclusively shown. In mammalian cells the HIRA and CAF-1 complexes have been shown to work in concert with the chromatin remodeler INO80 during DNA double-strand break repair, although there is no evidence for complex formation as INO80 is essential for histone eviction during non-homologous end joining prior to nucleosome reassembly mediated by the HIRA or CAF-1 complexes independent of INO80 79. While the possibility remains open, more comprehensive studies are needed to identify how the HIRA complex functions, whether alone or in complex with an ATP-dependent chromatin remodeler.
Conclusion
The HIRA complex is an evolutionarily conserved histone chaperone complex that operates to mediate H3.3-specific deposition in a wide variety of chromatin environments. While this review is not meant to be an exhaustive summary of HIRA complex biological function, a more comprehensive review is available57 that features detailed analysis of the biological functions of the S. cerevisiae Hir complex and the HIRA complex in a number of species. Through summarizing the existing biochemical and structural data, this review has aimed to improve the current understanding of the architecture of the HIRA complex and to provide a testable model for H3.3-specific histone deposition by the complex. Nonetheless, it is clear that our model would benefit from more structural information, and significant work still needs to be done to uncover the detailed molecular mechanism for the H3.3-specific histone chaperone function of the human HIRA complex.
Research Highlights.
Molecular basis for assembly of the H3.3-specific HIRA histone chaperone complex
Structural analysis of the mechanism underlying H3.3-specific binding by the HIRA complex
Proposed model for (H3.3/H4)2 tetramer formation by the HIRA complex
Acknowledgments
This work was supported by National Institutes of Health grants R01 GM060293, R35 GM118090 and P01 AG031862 to R.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 3.Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 4.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. Journal of molecular biology. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 5.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 6.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 7.Maze I, Noh KM, Soshnev AA, Allis CD. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nature reviews. Genetics. 2014;15:259–271. doi: 10.1038/nrg3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21 doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laskey RA, Honda BM, Mills AD, Finch JT. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- 10.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20 doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattiroli F, D’Arcy S, Luger K. The right place at the right time: chaperoning core histone variants. EMBO reports. 2015;16:1454–1466. doi: 10.15252/embr.201540840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winkler DD, Zhou H, Dar MA, Zhang Z, Luger K. Yeast CAF-1 assembles histone (H3-H4)2 tetramers prior to DNA deposition. Nucleic acids research. 2012;40:10139–10149. doi: 10.1093/nar/gks812. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Tsunaka Y, Fujiwara Y, Oyama T, Hirose S, Morikawa K. Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes & development. 2016;30 doi: 10.1101/gad.274183.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 15.Tang Y, et al. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nature structural & molecular biology. 2006;13 doi: 10.1038/nsmb1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaillard PH, et al. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 17.Banumathy G, et al. Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells. Mol Cell Biol. 2009;29:758–770. doi: 10.1128/MCB.01047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rai TS, et al. Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex. Mol Cell Biol. 2011;31:4107–4118. doi: 10.1128/MCB.05546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pchelintsev NA, et al. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep. 2013;3:1012–1019. doi: 10.1016/j.celrep.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin C, et al. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banaszynski LA, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orsi GA, et al. Drosophila Yemanuclein and HIRA cooperate for de novo assembly of H3.3-containing nucleosomes in the male pronucleus. PLoS Genet. 2013;9:e1003285. doi: 10.1371/journal.pgen.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adam S, Polo SE, Almouzni G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 2013;155:94–106. doi: 10.1016/j.cell.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 25.Zhang R, et al. HP1 proteins are essential for a dynamic nuclear response that rescues the function of perturbed heterochromatin in primary human cells. Mol Cell Biol. 2007;27:949–962. doi: 10.1128/MCB.01639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray-Gallet D, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Molecular cell. 2011;44:928–941. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Schneiderman JI, Orsi GA, Hughes KT, Loppin B, Ahmad K. Nucleosome-depleted chromatin gaps recruit assembly factors for the H3.3 histone variant. Proc Natl Acad Sci U S A. 2012;109:19721–19726. doi: 10.1073/pnas.1206629109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elsasser SJ, et al. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature. 2012;491:560–565. doi: 10.1038/nature11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elsasser SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522:240–244. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elsaesser SJ, Allis CD. HIRA and Daxx constitute two independent histone H3.3-containing predeposition complexes. Cold Spring Harb Symp Quant Biol. 2010;75:27–34. doi: 10.1101/sqb.2010.75.008. [DOI] [PubMed] [Google Scholar]
- 31.Corpet A, Olbrich T, Gwerder M, Fink D, Stucki M. Dynamics of histone H3.3 deposition in proliferating and senescent cells reveals a DAXX-dependent targeting to PML-NBs important for pericentromeric heterochromatin organization. Cell Cycle. 2014;13:249–267. doi: 10.4161/cc.26988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ricketts MD, et al. Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nature communications. 2015;6:7711. doi: 10.1038/ncomms8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye X, et al. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol Cell Biol. 2007;27:2452–2465. doi: 10.1128/MCB.01592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang R, et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Developmental cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 35.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 37.Tyler JK, et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 38.Lamour V, et al. A human homolog of the S. cerevisiae HIR1 and HIR2 transcriptional repressors cloned from the DiGeorge syndrome critical region. Hum Mol Genet. 1995;4:791–799. doi: 10.1093/hmg/4.5.791. [DOI] [PubMed] [Google Scholar]
- 39.Lindsay EA, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 40.Sherwood PW, Tsang SV, Osley MA. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Molecular and cellular biology. 1993;13:28–38. doi: 10.1128/mcb.13.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spector MS, Osley MA. The HIR4–1 mutation defines a new class of histone regulatory genes in Saccharomyces cerevisiae. Genetics. 1993;135:25–34. doi: 10.1093/genetics/135.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dutta D, et al. Regulation of angiogenesis by histone chaperone HIRA-mediated incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. The Journal of biological chemistry. 2010;285:41567–41577. doi: 10.1074/jbc.M110.190025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dilg D, et al. HIRA Is Required for Heart Development and Directly Regulates Tnni2 and Tnnt3. PloS one. 2016;11:e0161096. doi: 10.1371/journal.pone.0161096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prochasson P, Florens L, Swanson SK, Washburn MP, Workman JL. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes & development. 2005;19:2534–2539. doi: 10.1101/gad.1341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green EM, et al. Replication-independent histone deposition by the HIR complex and Asf1. Current biology : CB. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aho S, et al. Ubinuclein, a novel nuclear protein interacting with cellular and viral transcription factors. J Cell Biol. 2000;148:1165–1176. doi: 10.1083/jcb.148.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aho S, et al. Characterization of the ubinuclein protein as a new member of the nuclear and adhesion complex components (NACos) Biology of the cell / under the auspices of the European Cell Biology Organization. 2009;101:319–334. doi: 10.1042/BC20080072. [DOI] [PubMed] [Google Scholar]
- 48.Youn HD, Liu JO. Cabin1 represses MEF2-dependent Nur77 expression and T cell apoptosis by controlling association of histone deacetylases and acetylases with MEF2. Immunity. 2000;13:85–94. doi: 10.1016/s1074-7613(00)00010-8. [DOI] [PubMed] [Google Scholar]
- 49.Liu W, Youn HD, Liu JO. Thapsigargin-induced apoptosis involves Cabin1-MEF2-mediated induction of Nur77. European journal of immunology. 2001;31:1757–1764. doi: 10.1002/1521-4141(200106)31:6<1757::aid-immu1757>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 50.Han A, et al. Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2. Nature. 2003;422:730–734. doi: 10.1038/nature01555. [DOI] [PubMed] [Google Scholar]
- 51.Jang H, Choi SY, Cho EJ, Youn HD. Cabin1 restrains p53 activity on chromatin. Nature structural & molecular biology. 2009;16:910–915. doi: 10.1038/nsmb.1657. [DOI] [PubMed] [Google Scholar]
- 52.Hammond DR, Udvadia AJ. Cabin1 expression suggests roles in neuronal development. Developmental dynamics : an official publication of the American Association of Anatomists. 2010;239:2443–2451. doi: 10.1002/dvdy.22367. [DOI] [PubMed] [Google Scholar]
- 53.Ray-Gallet D, Quivy JP, Sillje HW, Nigg EA, Almouzni G. The histone chaperone Asf1 is dispensable for direct de novo histone deposition in Xenopus egg extracts. Chromosoma. 2007;116:487–496. doi: 10.1007/s00412-007-0112-x. [DOI] [PubMed] [Google Scholar]
- 54.Bonnefoy E, Orsi GA, Couble P, Loppin B. The essential role of Drosophila HIRA for de novo assembly of paternal chromatin at fertilization. PLoS genetics. 2007;3:1991–2006. doi: 10.1371/journal.pgen.0030182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song Y, et al. Dissecting the roles of the histone chaperones reveals the evolutionary conserved mechanism of transcription-coupled deposition of H3.3. Nucleic Acids Res. 2013;41:5199–5209. doi: 10.1093/nar/gkt220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balaji S, Iyer LM, Aravind L. HPC2 and ubinuclein define a novel family of histone chaperones conserved throughout eukaryotes. Mol Biosyst. 2009;5:269–275. doi: 10.1039/b816424j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amin AD, Vishnoi N, Prochasson P. A global requirement for the HIR complex in the assembly of chromatin. Biochimica et biophysica acta. 2013;1819:264–276. doi: 10.1016/j.bbagrm.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 58.Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 59.Esau C, et al. Deletion of calcineurin and myocyte enhancer factor 2 (MEF2) binding domain of Cabin1 results in enhanced cytokine gene expression in T cells. The Journal of experimental medicine. 2001;194:1449–1459. doi: 10.1084/jem.194.10.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang JH, et al. Histone chaperones cooperate to mediate Mef2-targeted transcriptional regulation during skeletal myogenesis. Biochemical and biophysical research communications. 2011;407:541–547. doi: 10.1016/j.bbrc.2011.03.055. [DOI] [PubMed] [Google Scholar]
- 61.Ray-Gallet D, et al. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- 62.Loppin B, et al. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- 63.Kang B, et al. Phosphorylation of H4 Ser 47 promotes HIRA-mediated nucleosome assembly. Genes Dev. 2011;25:1359–1364. doi: 10.1101/gad.2055511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agez M, et al. Structure of the histone chaperone ASF1 bound to the histone H3 C-terminal helix and functional insights. Structure. 2007;15:191–199. doi: 10.1016/j.str.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 65.Adkins MW, Tyler JK. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. The Journal of biological chemistry. 2004;279:52069–52074. doi: 10.1074/jbc.M406113200. [DOI] [PubMed] [Google Scholar]
- 66.English CM, Maluf NK, Tripet B, Churchill ME, Tyler JK. ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of the H3-H4 heterotetramer on DNA. Biochemistry. 2005;44:13673–13682. doi: 10.1021/bi051333h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Natsume R, et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 69.Banks DD, Gloss LM. Folding mechanism of the (H3-H4)2 histone tetramer of the core nucleosome. Protein science : a publication of the Protein Society. 2004;13:1304–1316. doi: 10.1110/ps.03535504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang Y, et al. Identification of an Ubinuclein 1 Region Required for Stability and Function of the Human HIRA/UBN1/CABIN1/ASF1a Histone H3.3 Chaperone Complex. Biochemistry. 2012;51:2366–2377. doi: 10.1021/bi300050b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bowman A, Ward R, El-Mkami H, Owen-Hughes T, Norman DG. Probing the (H3-H4)2 histone tetramer structure using pulsed EPR spectroscopy combined with site-directed spin labelling. Nucleic acids research. 2010;38:695–707. doi: 10.1093/nar/gkp1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu M, et al. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 73.Huang H, et al. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nature structural & molecular biology. 2015;22:618–626. doi: 10.1038/nsmb.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Richet N, et al. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic acids research. 2015;43:1905–1917. doi: 10.1093/nar/gkv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen S, et al. Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2. Genes & development. 2015;29:1326–1340. doi: 10.1101/gad.261115.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fazly A, et al. Histone chaperone Rtt106 promotes nucleosome formation using (H3-H4)2 tetramers. The Journal of biological chemistry. 2012;287:10753–10760. doi: 10.1074/jbc.M112.347450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bowman A, et al. The histone chaperones Nap1 and Vps75 bind histones H3 and H4 in a tetrameric conformation. Mol Cell. 2011;41:398–408. doi: 10.1016/j.molcel.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley MA. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol Cell. 1999;4:75–83. doi: 10.1016/s1097-2765(00)80189-6. [DOI] [PubMed] [Google Scholar]
- 79.Li X, Tyler JK. Nucleosome disassembly during human non-homologous end joining followed by concerted HIRA- and CAF-1-dependent reassembly. eLife. 2016;5 doi: 10.7554/eLife.15129. [DOI] [PMC free article] [PubMed] [Google Scholar]