Abstract
Climate change causes permafrost thawing, and we are confronted with the unpredictable risk of newly discovered permafrost microbes that have disease-causing capabilities. Here, we first characterized the detailed chemical structure of the lipid A moiety from a Pseudomonas species that was isolated from thawing arctic permafrost using MALDI-based mass spectrometric approaches (i.e., MALDI-TOF MS and MALDI-QIT-TOF MSn). The MALDI multi-stage mass spectrometry (MS) analysis of lipid A extracted from the Pseudomonas sp. strain PAMC 28618 demonstrated that the hexaacyl lipid A ([M−H]− at m/z 1616.5) contains a glucosamine (GlcN) disaccharide backbone, two phosphates, four main acyl chains and two branched acyl chains. Moreover, the lipid A molecule–based structural activity relationship with other terrestrial Gram-negative bacteria indicated that strain PAMC 28618 has an identical lipid A structure with the mesophilic Pseudomonas cichorii which can cause rot disease in endive (Cichorium endivia) and that their bacterial toxicities were equivalent. Therefore, the overall lipid A validation process provides a general strategy for characterizing bacteria that have been isolated from arctic permafrost and analyzing their respective pathogenicities.
Introduction
The Arctic has experienced permafrost thawing due to a rapid temperature increase1, 2. Consequently, a number of native and undiscovered microorganisms in Arctic permafrost may be exposed and activated. The diversity of the microbial community across permafrost sites directly reflects the distinct conditions of the permafrost environment2. Such microorganisms could also utilize the ancient organic carbon in permafrost soils, which is thought to be mainly responsible for carbon dioxide and methane emissions3. Therefore, the characterizing the community structure and functional diversity of permafrost microbes is highly important for understanding the permafrost ecosystem and the emissions of greenhouse gasses in the permafrost. The isolation of permafrost bacteria and studies on permafrost microbes have increased recently4–6. The researchers who handle and study the unknown microbes, however, should be concerned about the pathogenicity of bacteria exposed from permafrost because some strains may cause unpredictable infectious diseases.
To screen and identify the endotoxins (i.e., lipopolysaccharide (LPS)) of unknown microbes, the most common approach is the Limulus Amebocyte Lysate (LAL) test7. In this assay, a proenzyme in LAL that is activated by the bacterial endotoxins catalyzes the cleavage of p-Nitroaniline (pNA) from a colorless peptide substrate. The quantity of released pNA increases proportionally with the endotoxin concentration. The LAL assay has generally been used to detect the endotoxin contamination in food, pharmaceutical and clinical specimens8, 9. Unfortunately, however, the results of the LAL test cannot directly link the presence of LPS to toxicity of lipid A from LPS10, 11. Therefore, more meticulous structural analysis of lipid A is required to determine the toxicity of newly discovered extremophile bacteria12, 13.
Lipopolysaccharides (LPS) are embedded in the external membranes of Gram-negative bacteria. These lipoglycans are virulence factors in the pathogenesis of host animal or plant cells14, 15. They have three major components: (1) the hydrophilic O-antigen, which is constitutes three to five repeating oligosaccharide subunits and serves as a major surface antigen; (2) the core oligosaccharide region, which contains a non-repeating short chain of sugar (e.g., 2-kedo-3-deoxy-octulonic acid) and maintains outer membrane stability; and (3) lipid A, which is the conserved amphipathic molecule anchored in the outer membrane. Importantly, the toxic activities of the Gram-negative bacteria that are associated with septic shock are mainly associated with the lipid A molecule16. However, there are multiple structural requirements for the toxicity of lipid A17–19. For example, lipid A that is toxic to animals or plants should contain all of the following structural components: (1) a glucosamine disaccharide, (2) diphosphorylation and (3) normal fatty acids20. Therefore, the lipid A from the newly discovered bacteria should be validated by a detailed structural analysis to understand the contribution of chemical changes to toxicity. To determine the detailed structure of lipid A, mass spectrometry (MS) has become a crucial technique because it allows the precise determination of the chemical structure and the molecular mass21–30. After the structural characterization of lipid A, the phenotypic toxicity of an invasive microorganism should be validated.
In this study, we characterized the chemical structure of lipid A from an unknown artic bacterial isolate that was originally found in thawing permafrost and validated its phenotypical toxicity (Fig. 1). First, the newly screened arctic bacterium was identified using the 16S rRNA sequencing method as a Pseudomonas sp. (named Pseudomonas sp. strain PAMC 28618) and detected by the LAL assay. Then, MALDI-TOF MS and MALDI-QIT-TOF MSn were applied to determine the partial and complete structures of the lipid A molecule. For this purpose, tandem MS which permits multiple stages of fragmentation (up to MS5) is essential for identifying more detailed structural information about the lipid A from Gram-negative bacteria. Through the multiple stages of fragmentation, we could precisely characterize the number of glucosamine (GlcN) and phosphate moieties, as well as the position of the ester-linked fatty acyl chains of lipid A. Interestingly, the chemical structure of lipid A from Pseudomonas sp. strain PAMC 28618 was identical to that of lipid A from Pseudomonas cichorii (ATCC 10857) which can cause rot disease in endive (Cichorium endivia). To verify the phenotypical toxicity of the Pseudomonas sp. strain PAMC 28618, the structure-activity relationship assay was performed with P. cichorii. This overall analytical process will be valuable for further polar microorganism research, and for studying the infectious diseases associated with newly discovered bacteria.
Figure 1.
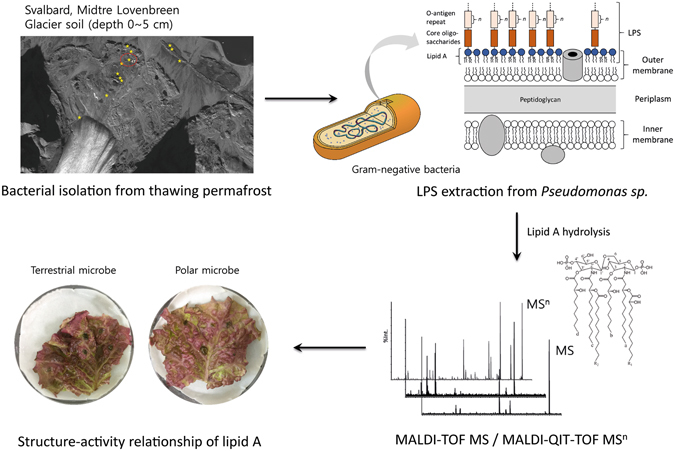
Overall workflow of the characterization of the lipid A molecule from polar Gram-negative bacteria by MALDI-based mass spectrometric approaches and the determination of pathogenical activity via structure-activity relationship.
Results and Discussion
Taxonomic analysis and LAL assay
Taxonomic analysis (BLASTn) of the five representative Arctic strains (PAMC 28615, 28616, 28617, 28618, and 28619) had shown a maximum similarity index for the Pseudomonas isolated from different environmental niches. Strains PAMC 28615, 28616, 28617, 28618, and 28619 had 99.9% sequence matches with Pseudomonas sp. 3–17 (KX378955), Pseudomonas sp. HP7R (KM187527), Pseudomonas sp. MDT3-38 (JX949569), Pseudomonas frederiksbergensis IHB B 10366 (KR233782), and Pseudomonas sp. JSZCNM3 (KU643204), respectively. The taxonomic relationship was confirmed with the classifier program of Ribosomal Database Project II release 11.4 (http://rdp.cme.msu.edu/). Therefore, 16S rRNA gene sequences and taxonomic relatedness have revealed that the isolated strains belong to genus Pseudomonas, and a similar group of organisms has been reported in Arctic and Antarctic glacier environments31–33. Typical homologous (98–99%) reference sequences were obtained from the seqmatch program of RDPII and used to construct the NJ phylogenetic tree (Fig. 2). Phylogenetic analysis showed that our Arctic pseudomonads sequences of 16S rRNA genes were matched to conserved regions of previously characterized 16S rRNA sequences of Arctic and Antarctic pseudomonads.
Figure 2.
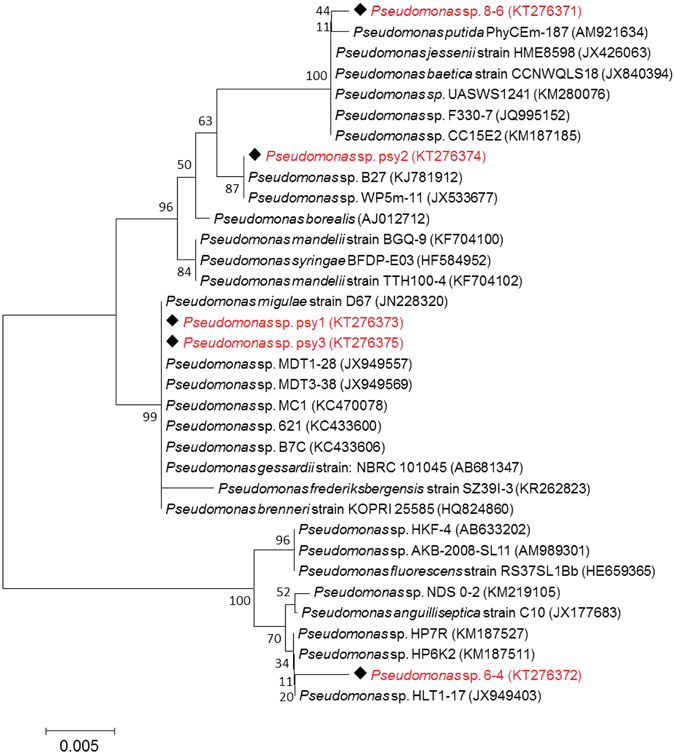
Neighbor-Joining (NJ) bootstrapping (100) phylogenetic tree of arctic glacier soil Pseudomonas spp. and their closest NCBI (BLASTn) strains based on the 16S rRNA gene sequences. Phylogenetic trees were developed based on the maximum composite likelihood method using MEGA 6.06 version.
Previously, several studies have examined isolated bacteria in the Arctic and Antarctic soil to discover the microbial diversity34–37. However, the pathogenicity assay has not been actively performed with Arctic soil bacteria. As a model system, we focused on the toxicity of strain PAMC 28618 among the newly discovered bacteria in arctic soil, and an LAL assay was performed to investigate the presence of LPSs as endotoxins embedded in the external membrane of Gram-negative bacteria. The activity of endotoxins in the sonicated whole cell (strain PAMC 28618) was calculated using the standard calibration curve of endotoxin, which showed a correlation between the optical densities (OD) measured at 405 nm and endotoxin unit (EU) per mL (Fig. S3). As a result, the activity of LPS in Pseudomonas sp. strain PAMC 28618 (12 CFU) was calculated as approximatively 0.38 EU/mL. However, Takayama et al. reported that the positive LAL test does not directly correlate with its toxicity and there should be multiple structural requirements for the lipid A moiety: (1) a glucosamine disaccharide, (2) diphosphorylation, and (3) normal fatty acids20. To evaluate the toxicity of the newly discovered strain PAMC 28618, required a more detailed mass spectrometry-based structural analysis of lipid A, which is known to play a major role in the activity of endotoxins38, 39.
MS analysis of lipid A from Pseudomonas sp. strain PAMC 28618 by MALDI-TOF MS
The lipid A sample was prepared by hydrolysis of LPS that had been extracted from Pseudomonas sp. strain PAMC 28618 with phenol/water extraction40. First, to determine the parent ion mass of lipid A from strain PAMC 28618, the negative ion MALDI-TOF MS was applied. The MALDI-based analytical condition can generate more fragment ions of the lipid A molecule, which can provide structural information of the parent ion. The major peak at m/z 1616.5 is attributed to hexaacyl diphosphoryl lipid A which may contain two C10:0 (3-OH), two C12:0 (3-OH), one C12:0 (2-OH) and one C12:0 (Fig. 3C). The other peaks in the mass spectrum arise from fragmentation products of hexaacyl lipid A, where m/z 1536.5, 1446.3, 1418.3 and 1366.4 correspond to the elimination of the phosphate group at C-1, the acyl chain ‘d’ (C10:0 (3-OH)) at C-3′, the acyl chain ‘R1’ (C12:0 (2-OH)) at C-2 and both the acyl chain ‘d’ at C-3 and the phosphate group at C-1, respectively (Fig. 3A,B and Table S1). The intense peak at m/z 1536.5 is a product ion that is generated by the removal of the phosphate group at C-1 because the phosphate group at C-4′ may be more stable than that of C-1 due to the oxygen atom next to C-141. With only the full mass spectrum of MALDI-TOF MS, we can sufficiently deduce the number of glucosamine (GlcN), phosphate groups and acyl chains of the lipid A molecule of strain PAMC 28618. However, to determine the structure of lipid A in more detail (i.e., the position and linkage of acyl chains and phosphates) from the tandem mass spectrometric analysis (MSn), MALDI-QIT TOF MS/MS was performed.
Figure 3.
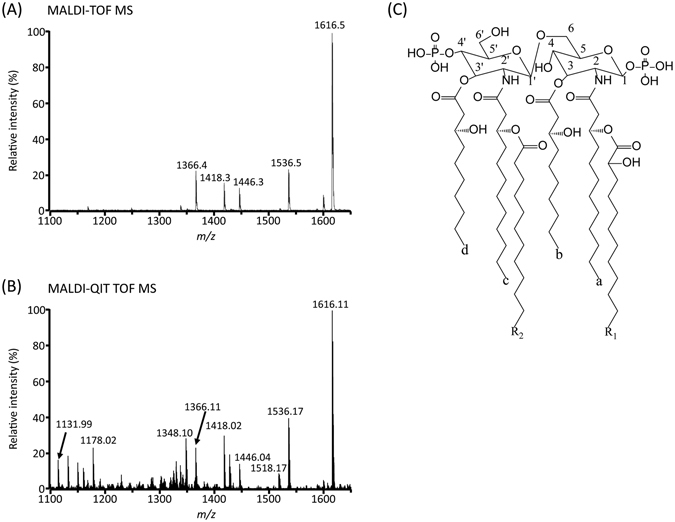
Negative-ion MALDI-TOF MS (A), MALDI-QIT TOF MS mass spectra of lipid A (B) and the lipid A molecule structure of Pseudomonas sp. strain PAMC 28618 (C).
MS analysis of the De-O-acylated lipid A from Pseudomonas sp. strain PAMC 28618 and Pseudomonas cichorii by MALDI-TOF MS
MALDI analysis of de-O-acylated lipid A can offer more detailed structural information about the fatty acid species and position (Fig. S4). To remove ester-linked fatty acids, mainly at C-3 and C-3′, lipid A was treated to mild alkaline condition as previously described21, 42. Silipo et al. reported O-acyl hydrolysis rate order that primary fatty acid is hydrolyzed first21. Thus, a peak at m/z 1276.0 is readily explained by the absence of the ester-linked primary fatty acids (C10:0 (3-OH) ‘b’ and ‘d’) at the C-3 and C-3′ position. Additionally, a peak at m/z 1247.8 and 1013.8 indicates the presence of secondary fatty acid R1 and R2 linked N-acyl residue, respectively (Fig. S4). However, the position of the secondary fatty acid was not identified. Thus, tandem mass spectrometric analysis via MALDI-QIT TOF MS was performed to determine the location of the secondary fatty acid.
MSn analysis of lipid A from Pseudomonas sp. strain PAMC 28618 by MALDI-QIT TOF MS
First, the MALDI-QIT TOF mass spectrum exhibited the prominent fragmental ions of lipid A of Pseudomonas sp. strain PAMC 28618, which were detected by MALDI-TOF MS analysis. With the full mass MALDI-TOF and MALDI-QIT TOF mass spectra, we can confirm that the parent ion of lipid A from strain PAMC 28618 is at 1616.11 [M−H]−. To characterize the detailed chemical structure of lipid A, we conducted a multiple stages of fragmentation (up to MS5) study and selected the parental ion at m/z 1616.27 in the first MS/MS stage. In the MS2 spectrum, according to Luisa Sturiale et al., a peak at m/z 824.95 was identified as a Y-type fragment ion of GlcN I via in-source decay25. Thus, the peak can be unequivocally explained by the presence of the secondary fatty acid C12:0 (2-OH) on N-linked to GlcN I. Additionally, the ion at m/z 850.95 indicated the presence of the secondary fatty acid C12:0 linked amide primary fatty acid C12:0 of GlcN II. The major fragment peaks were observed at m/z 1518.32, 1428.18, 1330.24 and 1114.10 corresponding to the cleavage of the phosphate group at C-1, ‘b’ acyl chain at C-3, the phosphate group and ‘b’ acyl chain at C-3, and the phosphate and two acyl groups ‘b’ at C-3 and ‘R1’ at C-2, respectively (Fig. 4 and Table S2). Interestingly, in the intense peak at m/z 1428.18, we deduced that the fragment peak was generated by the elimination of the acyl chain ‘b’, but not ‘d’. This is because the ion at m/z 808.96 can only be detected when the ‘d’ acyl chain remained at C-3′ during the MS3 stage of m/z 1428.19 ions (Fig. S5 and Table S6). The QIT MS2 fragmentation patterns were quite different from the MALDI-MS fragments, which means that each MSn stage will provide more detailed structural information for characterizing the lipid A of strain PAMC 28618. The highly abundant ions at m/z 1518.32 and 1428.18 were isolated for the MS3 stage for further structural characterization.
Figure 4.
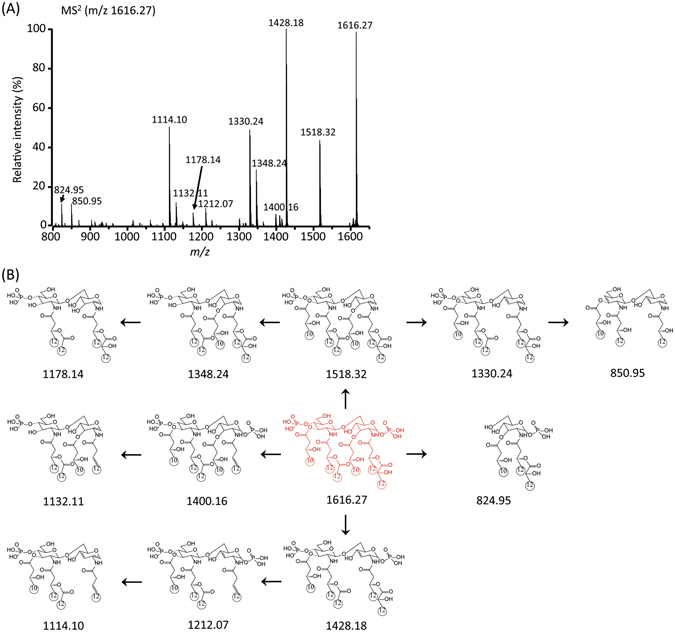
Negative-ion MALDI-QIT-TOF MS2 spectrum of parent peak at m/z 1616.27 (A) and fragment structures corresponding to each fragment ion peak were drawn as neutral molecules (B).
In the MS3 stage, the product ions of m/z 1348.24, 1330.24 and 1302.20 were generated by eliminating a ‘d’ acyl chain at C-3′, ‘b’ acyl chain at C-3 and ‘R1’ at C-2 from m/z 1518.32 as shown in Fig. 5A and Table S3. Two fragmentation mechanisms with CID (collision-induced dissociation) might be applied to the product ions at m/z 1348.24 and 1330.24. First, the charge-driven process resulted in the neutral loss of an acyl chain as a ketone derivative from the parent ion at C-3′. Next, the charge-remote process led to the neutral loss of an acyl chain as a free fatty acid at C-3. When we compare the product ion peak intensities, the results suggest that the charge-driven and charge-remote fragmentation processes compete with each other in the CID experiments using MALDI-QIT-TOF MS (Figs 5B and S2).
Figure 5.
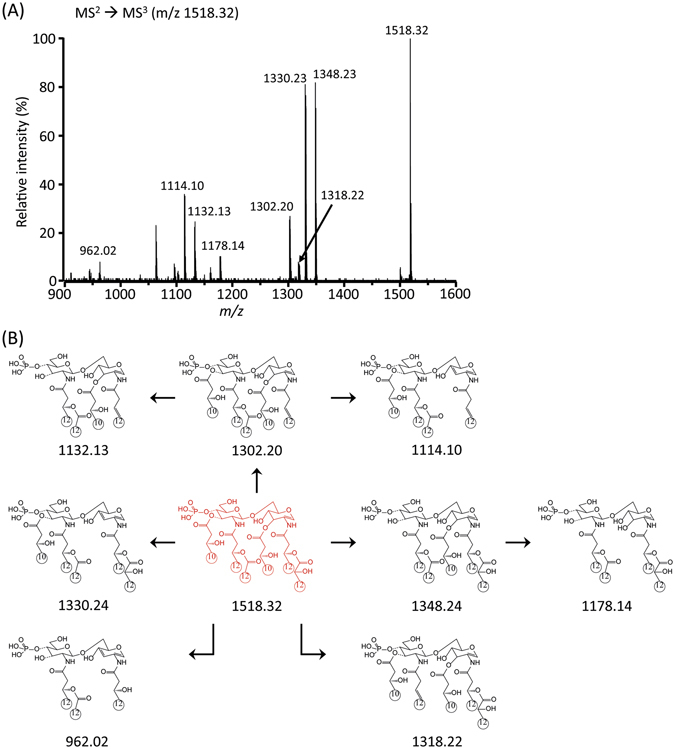
Negative-ion MALDI-QIT-TOF MS3 spectrum of monophosphoryl lipid A at m/z 1518.32 (A) and fragment structures (B).
The abundant product ions at m/z 1348.23 (Fig. 5A) and 1330.24 (Fig. S5A) from the MS3 stage were isolated for the MS4 stage (Figs 6 and S6; Tables S4 and S). The highest fragment ion at m/z 1132.07 corresponded to the elimination of ‘R1’ at C-2 from m/z 1348.16. The other fragment ions clearly supported the chemical structure of the parent ion at m/z 1348.24. Finally, the ion at m/z 1132.07 from the MS4 stage was selected for the subsequent MS5 analysis. The existence of the free fatty acid (C10:0 (3-OH)) at C-3 was confirmed by the formation of an ion at m/z 943.96 and 962.01 by MS5 (Figs 7 and S7; Tables S5 and S8). Taken together, the tandem mass analysis of lipid A from strain PAMC 28618 demonstrated the exact chemical structure of hexaacyl diphosphoryl lipid A. The two phosphate groups are located at the C-1 and C-4′ positions of the glucosamine dimer. In addition, two C10:0 (3-OH) acyl chains are at the C-3 and C-3′ positions via ester bonds and two C12:0 (3-OH) acyl chains are at C-2 and C-2′ via amide bonds. The two branched chains (i.e., C12:0 and C12:0 (2-OH)) are substituted at the main chains of C-2 and C-2′, respectively.
Figure 6.
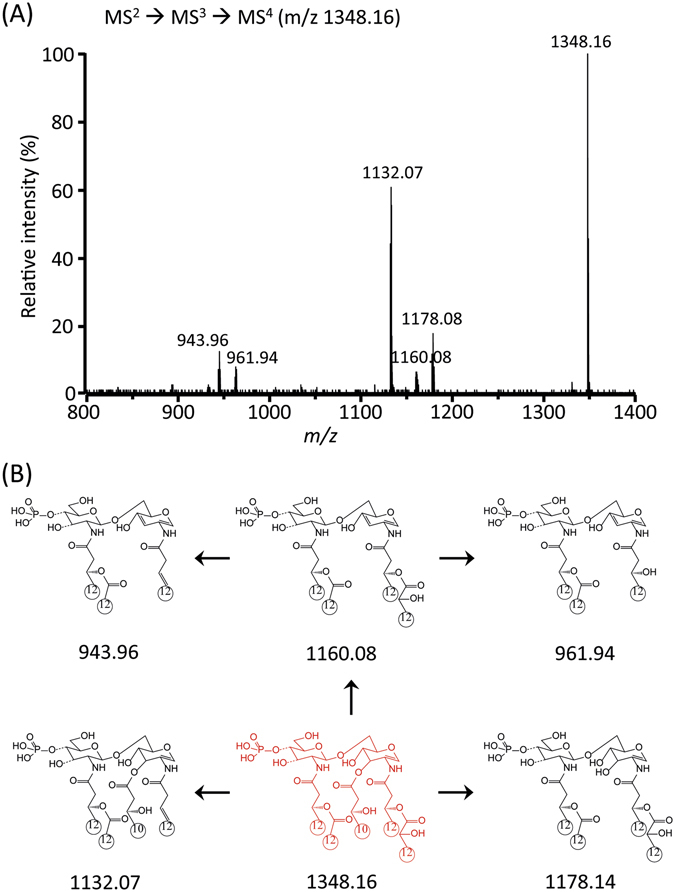
Negative-ion MALDI-QIT-TOF MS4 spectrum of m/z 1348.16 corresponding to removal of acyl group at C-3′ (A) and fragment structures (B).
Figure 7.
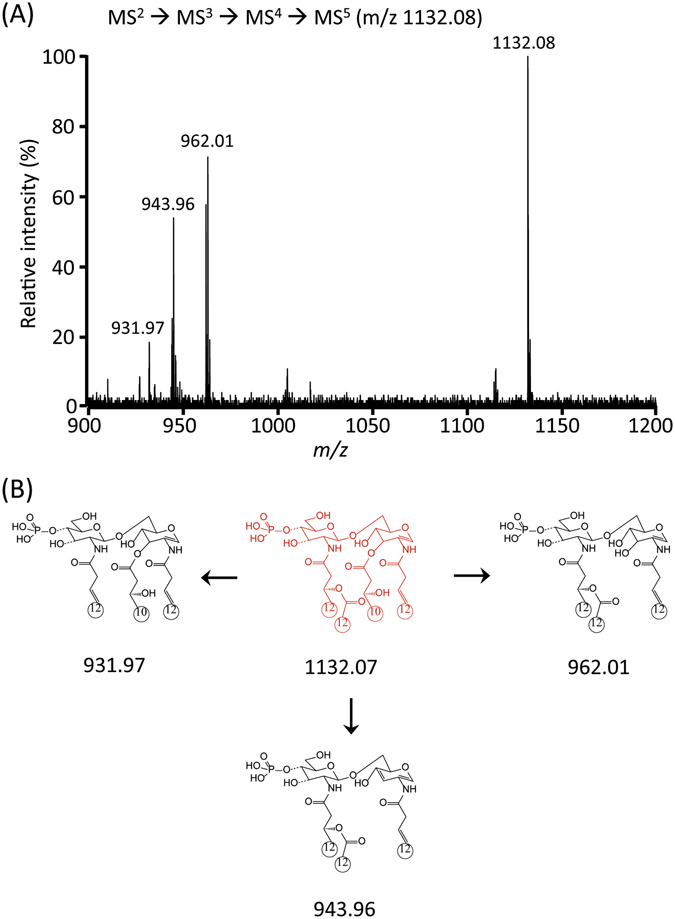
Negative-ion MALDI-MS5 spectrum of m/z 1132.08 (A) and fragment structure (B).
Structure-activity relationship of lipid A from Pseudomonas sp. strain PAMC 28618 with Pseudomonas cichorii
After we had characterized the chemical structure of lipid A from Pseudomonas sp. strain PAMC 28618, we were curious whether any mesophilic Pseudomonas sp. has the same chemical structure of lipid A. Interestingly, Molinaro et al. reported the structure of the lipid A fraction from LPS of the Pseudomonas cichorii using 2D-NMR and MALDI-TOF MS43. According to our taxonomic analysis (BLASTn), P. cichorii had 97% sequence matches with strain PAMC 28618. The intact lipid A from P. cichorii also consisted of glucosamine disaccharides, two C12:0 (3-OH) at C-2 and C-2′, and two C10:0 (3-OH) at C-3 and C-3′. C12:0 (2-OH) and C12:0 are linked to both C12:0 (3-OH) at C-2 and C-2′. Finally, each phosphate group is linked to C-1 and C-4′. Taken together, the overall structure of lipid A from P. cichorii is identical to that from Pseudomonas sp. strain PAMC 28618. To validate the chemical structure of lipid A from P. cichorii which was determined by 2D-NMR and MALDI-TOF MS43, we conducted multiple-stage MS analysis of lipid A from P. cichorii (Figs S8–11, Tables S9–12). With the full MALDI-TOF and MALDI-QIT TOF mass spectra, we can confirm that the parent ion of lipid A of P. cichorii is at 1616.10 [M−H]−. Moreover, the MSn analysis of lipid A from P. cichorii demonstrated that the lipid A of both P. cichorii and strain PAMC 28618 has an identical fine structure. The Gram-negative bacterium Pseudomonas cichorii has been identified to cause a destructive disease known as “varnish spot” on lettuce, cabbage, celery and chrysanthemum plants44, 45. Previous studies have revealed that LPS plays an important role in the pathogenesis of Gram-negative bacterial infection, and that its endotoxic effects such as cytokine production, inflammation and shock are largely determined by its lipid A46. Therefore, based on the structural identity between P. cichorii and strain PAMC 28618, we investigated the bacterial toxicity of the newly discovered polar microorganism.
Lettuce that had been cultured at regulated conditions (temperature = 20 °C, humidity = 50% and pH = 6.0 to 7.0) was inoculated with a high population density of P. cichorii, Pseudomonas sp. strain PAMC 28618 and P. oleovorans (94 × 104, 54 × 104 and 44 × 105 CFU) to verify proof-of-concept with real plant sample. Pseudomonas oleovorans was used as a control (Fig. 8). Inoculation was conducted using pipette tips to prick a hole in the leaves on Whatman chromatography paper in a petri dish. This inoculation process was slightly modified from the work of Dong et al.47.
Figure 8.
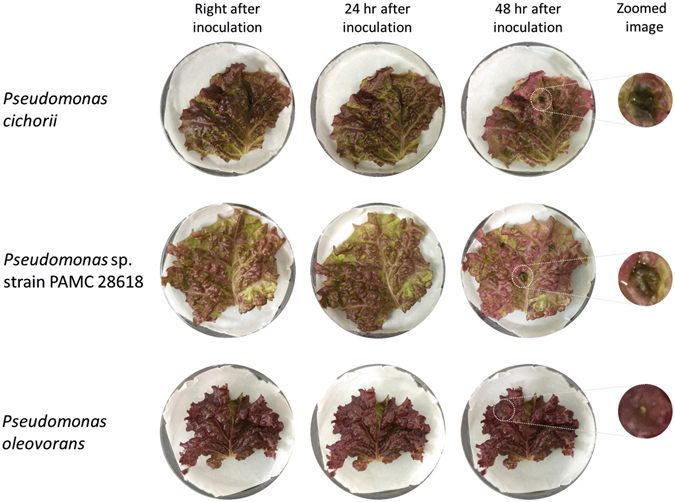
Phenotypical toxicity test was performed by inoculating lettuce leaves with Pseudo- monas cichorii, strain PAMC 28618 or P. oleovorans as a control and leaves were observed every 24 hr for 48 hr. After 48 h, the spot that had been inoculated with P. cichorii and strain PAMC 28618 were showed rot disease, but lettuce inoculated with P. oleovorans did not develop rot disease.
After 24 hours, we could identify symptoms in the early stage of the disease at inoculated spots with strain PAMC 28618 and P. cichorii. After 48 hours, necrotic dark-brown areas were clearly observed and were larger than the brown areas recorded in the same spots at 24 hours. As expected, there were no symptoms at the spot inoculated with Pseudomonas oleovorans. Therefore, the newly screened polar microorganism has an identical lipid A structure as the mesophilic P. cichorii and their bacterial toxicity was equivalent. To the best of our knowledge, this is the first report to identify the relationship between the lipid A molecule structure of a polar microbe and its toxicity. However, further studies may be needed to demonstrate that all Gram-negative bacteria with the same lipid A structure show similar toxicity.
Conclusions
In this study, we characterized the chemical structure of the lipid A moiety from Pseudomonas sp. strain PAMC 28618 which was newly discovered in thawing arctic soils using mass spectrometric approaches with MALDI-TOF and MALDI-QIT-TOF MS. Based on the structural relationship of lipid A, we directly compared the phenotypical toxicity of strain PAMC 28618 with P. cichorii. During the multi-stage tandem mass spectrometry (MS5), the diagnostic fragment ions were used to characterize the hexaacyl diphosphoryl lipid A from strain PAMC 28618. Although the genotypical (16S rRNA sequencing) characterization of strain PAMC 28618 was not completely identical to that of other terrestrial Gram-negative bacteria, the structural relationships of lipid A may be significantly correlated with the bacteria’s phenotypic toxicities. It is believed that the overall lipid A analytical process would be valuable for studying the newly discovered Arctic bacteria which are not free of diseases, and for tackling critical and largely unmet challenges in polar microbiology.
Materials and Methods
Site description and sample collection
The sampling site is located in the glacier foreland of Midtre Lovenbreen, Svalbard (78.9°N, 12.0°E). The field campaign was permitted by the Svalbard Science Forum (RIS ID: 6752) in July, 2014. The glacier foreland soil samples were collected from different sites (Fig. S1) at 0~5 cm depth, placed in sterile 50 mL Falcon tubes and the samples were stored at 5 °C in the Arctic. Then, the samples were transported to the laboratory in ice buckets, containing ice packs that had been precooled to −70 °C and were stored at 4 °C until they were processed for bacterial isolation.
Isolation and culturing of Arctic pseudomonads
Approximately 1 g of glacier foreland soil sample was inoculated into 10 mL of Nutrient Broth (Acumedia, Lansing, MI, USA) and 10 mL of lysogeny broth (LB) (Merck KGaA, Darmstadt, Germany)48. The inoculated media were incubated at 20 °C for 24 hr under aerobic condition. The enriched culture was then serially diluted and 100 μl was plated on Pseudomonas Isolation Agar (17208, Sigma-Aldrich, St. Louis, MO, USA) plates (Peptic digest of animal tissue, 20 g/L; Potassium sulfate, 10 g/L; Magnesium chloride, 1.4 g/L; Triclosan (Irgasan), 0.025 g/L; Agar, 13.6 g/L; pH, 7.0 ± 0.2). Agar plates were incubated at 4, 15, 20 and 25 °C for two weeks under aerobic condition. Single pseudomonads colonies were obtained and each colony was repeatedly plated on Pseudomonas Agar (P2102, Sigma-Aldrich, St. Louis, MO, USA) to obtain pure isolated aerobic pseudomonads. Pure cultures were stored at 4 °C and cryopreserved in 20% (v/v) glycerol at −70 °C. Subsequently, Arctic pseudomonad strains (8–6, 6–4, psy1, psy2, and psy3) were deposited into the Korea Polar Research Institute (KPRI) based Polar and Alpine Microbial Collection (PAMC) center under the general deposit category with accession numbers PAMC 28615 to PAMC 28619, respectively.
DNA extraction, 16S rRNA gene amplification, and phylogenetic analysis
Genomic DNA was extracted from the five Arctic pseudomonads, and 16S rRNA gene was amplified by polymerase chain reaction (PCR) using universal degenerate primers 27 F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492 R (5′-GGT TAC CTT GTT ACG ACT T-3′) as previously described49. The PCR amplicon was cloned using TOPO TA cloning kit (Invitrogen, CA, USA) and then sequenced. 16S rRNA gene sequences obtained from the strains PAMC 28615 to PAMC 28619 were compared with those of other pseudomonads using NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) for their pair-wise identities. Multiple sequence alignment was carried out using the ClustalW2 version of EBI (http://www.ebi.ac.uk/Tools/msa/clustalw2/) with 0.5 transition weight. Phylogenetic trees were constructed using the MEGA software version 6.06 (http://www.megasoftware.net) by neighbor joining (NJ) with the Kimura two-parameter model. Moreover, the Ribosomal database project (RDP-11) (https://rdp.cme.msu.edu/) was also used to determine the strain identity with a seqmatch and classifier program. The 16S rRNA gene sequences of the Arctic pseudomonads were deposited in the GenBank nucleotide sequence database under the following accession numbers: PAMC 28615 (KT276371), PAMC 28616 (KT276372), PAMC 28617 (KT276373), PAMC 28618 (KT276374), and PAMC 28619 (KT276375).
Extraction of LPS from Pseudomonas sp. strain PAMC 28618 and Pseudomonas cichorii
Pseudomonas sp. strain PAMC 28618 and Pseudomonas cichorii was grown at 30 °C in lysogeny broth (LB) and Nutrient broth (NB), respectively, with incubation for 24 hr at 250 rpm in a shaking incubator. A 2% (v/v) of bacterial culture was used as the inoculum for culturing in a 100 mL Erlenmeyer flask.
LPS extraction from the cultured cells was performed using an LPS extraction kit (Intron Biotechnology Inc., Sungnam, Korea). First, 10 mL of lysis buffer containing phenol from the kit was added to 25 mg of the cell and vigorously vortexed to improve the lysis of the bacterial cell. After 2 mL of chloroform was added to the lysis buffer, it was vortexed for 30 sec. The mixture was then centrifuged at 13000 rpm and 4 °C for 10 min. Subsequently, 4 mL of supernatant was transferred to a new 50 mL Falcon tube and 8 mL of purification buffer was added. The mixture was incubated for 30 min at −20 °C to purify the LPS of other cell extract components, such as protein, nucleic acid, and lipids, and it was then centrifuged at 13,000 rpm and 4 °C for 20 min. After 8 mL of supernatant had been transferred to a new 50 mL Falcon tube, the LPS pellet was obtained by centrifugation under previously described conditions. The extracted LPS was washed with 10 mL of 70% EtOH and completely dried. Through this extraction process, 2.1 mg and 2.2 mg of LPS were extracted from 25 mg of strain PAMC 28618 and P. cichorii, respectively.
Lipid A extraction
The LPS pellet was suspended in 200 μl of 1% acetic acid. It was denatured for 2 hr at 100 °C and lipid A was extracted using the chloroform methanol extraction method. The chloroform methanol extraction process was performed by mixing 200 μl of the solution with 400 μl of chloroform and 200 μl of distilled water. The mixed solution was centrifuged at 8000 rpm for 15 min at 15 °C. The bottom layer was transferred to a new tube and dried by nitrogen gas.
De-O-acylation of lipid A
Extracted lipid A was de-O-acylated with 200 μl of 1:3 diluted ammonium hydroxide hydrolysis for 16 h at room temperature and it was dried under nitrogen gas as previously reported21.
MALDI-TOF MS and MALDI-QIT-TOF MSn analysis
Dried lipid A samples were dissolved with 10 μl of 50% (v/v) methanol/chloroform. Then, 1 μl of the lipid A sample was mixed with 1 μl of 50 mg 2,5-dihydroxybenzoic acid (DHB) solution in 1 mL 70% (v/v) acetonitrile/water. Subsequently, 2 μl of the mixture was loaded on a MSP 96 target polished steel MALDI plate and dried at room temperature. The lipid A samples were analyzed via a Bruker Daltonics Microflex LRF MALDI-TOF MS instrument and controlled by FlexControl 3.0 software (Bruker, Bremen, Germany). The operating conditions were as follows: negative-ion and reflectron mode, detector gain = 6.9, laser frequency = 60.0 Hz and laser power = 70%. Data acquisition and processing were performed by Flexanalysis 3.3 software (Bruker, Bremen, Germany). MS/MS analysis was performed using an Axima Resonance MALDI-quadrupole ion trap-TOF instrument (Shimadzu, Manchester, UK). The instrument was operated in negative ion and reflectron mode. Fragment ions were analyzed by collision-induced dissociation (CID) of the precursor ions and argon gas was used as a collision gas. Operating conditions were as follows: intro endcap = −5.5 kV, extr endcap = 10.0 kV, flight tube = 10.0 kV, reflectron center = −0.2 kV, reflectron back = −0.2 kV, and Detector = 2.2 kV. The data acquisition and processing were performed using the Launchpad 2.9.3 software (Kratos Analytical, Manchester, UK).
LAL (Limulus amebocyte lysate) assay
The LAL assay was performed using LAL Chromogenic Endotoxin Quantitation Kit (Thermo Scientific Pierce, Rockford, IL, USA) to confirm the existence of LPS in Pseudomonas strain PAMC 28618. The assay kit was verified to be working using standard endotoxin (0.1 to 1.0 EU/mL). Then, the LAL assay was performed using serially diluted cell lysate (12 CFU) of strain PAMC 28618 and according to the manufacturer’s protocol.
Inoculation of bacteria in lettuce for pathological test
Pseudomonas sp. strain PAMC 28618 and Pseudomonas oleovorans were cultured in lysogeny broth (LB) and the P. cichorii strain was cultured in Nutrient broth (NB). All three strains were cultured at 30 °C for 24 hr. The lettuce was grown for 2~3 weeks under controlled conditions. Regulated conditions were as follows: temperature = 20 °C, humidity = 50% and pH = 6.0 to 7.0. The lettuce was placed in Petri dishes containing Whatman Chromatography paper drenched with sterile water (3 mL) to create a damp environment. A volume of 20 μl of the cultured bacteria was used to inoculate the punched lettuce leaves with the end of the pipette tip beforehand. Then, the Petri dishes were placed in a 30 °C incubator and evaluated every 24 hr for 48 hr.
Electronic supplementary material
Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF-2015M1A5A1037196, NRF-2015R1A2A2A04006014, NRF-2016M1A5A1901797, NFR-2016M1A3A3A02018239, NRF-2015R1C1A1A01053048). We also deeply appreciate Prof. Dominique Laffly (Université de Toulouse 2, Toulouse, France) who kindly provided the map for the moraine soil at the Midtre Lovenbreen, Svalbard (Fig. S1).
Author Contributions
H.G.P., G.S., C.H.H., J.H.K., G.B., Y.G.K. were involved in the design and execution of the experiments. H.G.P., D.H.A., H.W.R., Y.K.L., Y.H.Y., Y.G.K. were involved in the research design, data interpretation and supervision of the studies. All authors contributed to the writing of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02145-w
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yoo Kyung Lee, Email: yklee@kopri.re.kr.
Yung-Hun Yang, Email: seokor@konkuk.ac.kr.
Yun-Gon Kim, Email: ygkim@ssu.ac.kr.
References
- 1.Schuur EA, Abbott B. Climate change: High risk of permafrost thaw. Nature. 2011;480:32–33. doi: 10.1038/480032a. [DOI] [PubMed] [Google Scholar]
- 2.Hultman J, et al. Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature. 2015;521:208–212. doi: 10.1038/nature14238. [DOI] [PubMed] [Google Scholar]
- 3.Zimov SA, Schuur EA, Chapin FS., III Permafrost and the global carbon budget. Science(Washington) 2006;312:1612–1613. doi: 10.1126/science.1128908. [DOI] [PubMed] [Google Scholar]
- 4.McCalley CK, et al. Methane dynamics regulated by microbial community response to permafrost thaw. Nature. 2014;514:478–481. doi: 10.1038/nature13798. [DOI] [PubMed] [Google Scholar]
- 5.Gyeong HR, et al. Domibacillus tundrae sp. nov., isolated from active layer soil of tussock tundra in Alaska, and emended description of the genus Domibacillus. Int. J. Syst. Evol. Microbiol. 2015;65:3407–3412. doi: 10.1099/ijsem.0.000429. [DOI] [PubMed] [Google Scholar]
- 6.Xue K, et al. Tundra soil carbon is vulnerable to rapid microbial decomposition under climate warming. Nature Climate Change. 2016;6:595–600. doi: 10.1038/nclimate2940. [DOI] [Google Scholar]
- 7.Webster CJ. Principles of a quantitative assay for bacterial endotoxins in blood that uses Limulus lysate and a chromogenic substrate. J. Clin. Microbiol. 1980;12:644–650. doi: 10.1128/jcm.12.5.644-650.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Townsend S, Barron JC, Loc-Carrillo C, Forsythe S. The presence of endotoxin in powdered infant formula milk and the influence of endotoxin and Enterobacter sakazakii on bacterial translocation in the infant rat. Food Microbiol. 2007;24:67–74. doi: 10.1016/j.fm.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Hurley JC. Endotoxemia: methods of detection and clinical correlates. Clin. Microbiol. Rev. 1995;8:268–292. doi: 10.1128/cmr.8.2.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henricson B, Benjamin W, Vogel S. Differential cytokine induction by doses of lipopolysaccharide and monophosphoryl lipid A that result in equivalent early endotoxin tolerance. Infect. Immun. 1990;58:2429–2437. doi: 10.1128/iai.58.8.2429-2437.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribi E. Beneficial modification of the endotoxin molecule. J. Immunother. 1984;3:1–9. [PubMed] [Google Scholar]
- 12.Di Lorenzo F, et al. Thermophiles as Potential Source of Novel Endotoxin Antagonists: the Full Structure and Bioactivity of theLipo‐oligosaccharide from Thermomonas hydrothermalis. ChemBioChem. 2014;15:2146–2155. doi: 10.1002/cbic.201402233. [DOI] [PubMed] [Google Scholar]
- 13.Silipo A, et al. Structure and immunological activity of the lipopolysaccharide isolated from the species Alkalimonas delamerensis. Eur. J. Org. Chem. 2013;2013:2653–2665. doi: 10.1002/ejoc.201201702. [DOI] [Google Scholar]
- 14.Cryz S, Pitt T, Fürer E, Germanier R. Role of lipopolysaccharide in virulence of Pseudomonas aeruginosa. Infect. Immun. 1984;44:508–513. doi: 10.1128/iai.44.2.508-513.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaper JB, Nataro JP, Mobley HL. Pathogenic escherichia coli. Nature Reviews Microbiology. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 16.Wilson J, et al. Mechanisms of bacterial pathogenicity. Postgrad. Med. J. 2002;78:216–224. doi: 10.1136/pmj.78.918.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schletter J, Heine H, Ulmer AJ, Rietschel ET. Molecular mechanisms of endotoxin activity. Arch. Microbiol. 1995;164:383–389. doi: 10.1007/BF02529735. [DOI] [PubMed] [Google Scholar]
- 18.Takayama K, et al. Influence of fine structure of lipid A on Limulus amebocyte lysate clotting and toxic activities. Infect. Immun. 1984;45:350–355. doi: 10.1128/iai.45.2.350-355.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rietschel ET, et al. Bacterial endotoxin: molecular relationships of structure to activity and function. The FASEB Journal. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 20.Takayama K, Qureshi N, Ribi E, Cantrell JL. Separation and characterization of toxic and nontoxic forms of lipid A. Review of Infectious Diseases. 1984;6:439–443. doi: 10.1093/clinids/6.4.439. [DOI] [PubMed] [Google Scholar]
- 21.Silipo A, Lanzetta R, Amoresano A, Parrilli M, Molinaro A. Ammonium hydroxide hydrolysis a valuable support in the MALDI-TOF mass spectrometry analysis of Lipid A fatty acid distribution. J. Lipid Res. 2002;43:2188–2195. doi: 10.1194/jlr.D200021-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Sforza S, et al. Determination of fatty acid positions in native lipid A by positive and negative electrospray ionization mass spectrometry. J. Mass Spectrom. 2004;39:378–383. doi: 10.1002/jms.598. [DOI] [PubMed] [Google Scholar]
- 23.Silipo A, et al. Structural characterizations of lipids A by MS/MS of doubly charged ions on a hybrid linear ion trap/orbitrap mass spectrometer. J. Mass Spectrom. 2008;43:478–484. doi: 10.1002/jms.1333. [DOI] [PubMed] [Google Scholar]
- 24.Sturiale L, et al. New conditions for matrix‐assisted laser desorption/ionization mass spectrometry of native bacterial R‐type lipopolysaccharides. Rapid Commun. Mass Spectrom. 2005;19:1829–1834. doi: 10.1002/rcm.1994. [DOI] [PubMed] [Google Scholar]
- 25.Sturiale L, et al. Reflectron MALDI TOF and MALDI TOF/TOF mass spectrometry reveal novel structural details of native lipooligosaccharides. J. Mass Spectrom. 2011;46:1135–1142. doi: 10.1002/jms.2000. [DOI] [PubMed] [Google Scholar]
- 26.Aussel L, et al. Novel variation of lipid A structures in strains of different Yersinia species. FEBS Lett. 2000;465:87–92. doi: 10.1016/S0014-5793(99)01722-6. [DOI] [PubMed] [Google Scholar]
- 27.Therisod H, Labas V, Caroff M. Direct microextraction and analysis of rough-type lipopolysaccharides by combined thin-layer chromatography and MALDI mass spectrometry. Anal. Chem. 2001;73:3804–3807. doi: 10.1021/ac010313s. [DOI] [PubMed] [Google Scholar]
- 28.Boué SM, Cole RB. Confirmation of the structure of lipid A from Enterobacter agglomerans by electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 2000;35:361–368. doi: 10.1002/(SICI)1096-9888(200003)35:3<361::AID-JMS943>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 29.Qureshi N, Takayama K, Heller D, Fenselau C. Position of ester groups in the lipid A backbone of lipopolysaccharides obtained from Salmonella typhimurium. J. Biol. Chem. 1983;258:12947–12951. [PubMed] [Google Scholar]
- 30.Chan S, Reinhold VN. Detailed structural characterization of lipid A: electrospray ionization coupled with tandem mass-spectrometry. Anal. Biochem. 1994;218:63–73. doi: 10.1006/abio.1994.1141. [DOI] [PubMed] [Google Scholar]
- 31.Janek T, Łukaszewicz M, Krasowska A. Identification and characterization of biosurfactants produced by the Arctic bacterium Pseudomonas putida BD2. Colloids Surf. B. Biointerfaces. 2013;110:379–386. doi: 10.1016/j.colsurfb.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Janek T, Łukaszewicz M, Rezanka T, Krasowska A. Isolation and characterization of two new lipopeptide biosurfactants produced by Pseudomonas fluorescens BD5 isolated from water from the Arctic Archipelago of Svalbard. Bioresour. Technol. 2010;101:6118–6123. doi: 10.1016/j.biortech.2010.02.109. [DOI] [PubMed] [Google Scholar]
- 33.Abraham WP, Thomas S. Draft genome sequence of Pseudomonas psychrophila MTCC 12324, isolated from the Arctic at 79 N. Genome announcements. 2015;3:e00578–00515. doi: 10.1128/genomeA.00578-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steven B, Pollard WH, Greer CW, Whyte LG. Microbial diversity and activity through a permafrost/ground ice core profile from the Canadian high Arctic. Environ. Microbiol. 2008;10:3388–3403. doi: 10.1111/j.1462-2920.2008.01746.x. [DOI] [PubMed] [Google Scholar]
- 35.Teixeira LC, et al. Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. The ISME journal. 2010;4:989–1001. doi: 10.1038/ismej.2010.35. [DOI] [PubMed] [Google Scholar]
- 36.Yergeau E, et al. Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. The ISME journal. 2012;6:692–702. doi: 10.1038/ismej.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yergeau E, Newsham KK, Pearce DA, Kowalchuk GA. Patterns of bacterial diversity across a range of Antarctic terrestrial habitats. Environ. Microbiol. 2007;9:2670–2682. doi: 10.1111/j.1462-2920.2007.01379.x. [DOI] [PubMed] [Google Scholar]
- 38.Rietschel ET, et al. Lipid A, the endotoxic center of bacterial lipopolysaccharides: relation of chemical structure to biological activity. Prog. Clin. Biol. Res. 1986;231:25–53. [PubMed] [Google Scholar]
- 39.Morrison DC. Bacterial endotoxins and pathogenesis. Review of Infectious Diseases. 1983;5:S733–S747. doi: 10.1093/clinids/5.Supplement_4.S733. [DOI] [PubMed] [Google Scholar]
- 40.Takayama S, Saitoh E, Kimizuka R, Yamada S, Kato T. Effect of eel galectin AJL-1 on periodontopathic bacterial biofilm formation and their lipopolysaccharide-mediated inflammatory cytokine induction. Int. J. Antimicrob. Agents. 2009;34:355–359. doi: 10.1016/j.ijantimicag.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Lee CS, Kim YG, Joo HS, Kim BG. Structural analysis of lipid A from Escherichia coli O157: H7: K− using thin‐layer chromatography and ion‐trap mass spectrometry. J. Mass Spectrom. 2004;39:514–525. doi: 10.1002/jms.614. [DOI] [PubMed] [Google Scholar]
- 42.Bhat R, Marx A, Galanos C, Conrad R. Structural studies of lipid A from Pseudomonas aeruginosa PAO1: occurrence of 4-amino-4-deoxyarabinose. J. Bacteriol. 1990;172:6631–6636. doi: 10.1128/jb.172.12.6631-6636.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molinaro A, et al. Determination of the Structure of the Lipid A Fraction from the Lipopolysaccharide of Pseudomonas Cichorii by Means of NMR and MALDI‐TOF Mass Spectrometry. Eur. J. Org. Chem. 2002;2002:3119–3125. doi: 10.1002/1099-0690(200209)2002:18<3119::AID-EJOC3119>3.0.CO;2-M. [DOI] [Google Scholar]
- 44.Grogan R, et al. Varnish spot, destructive disease of lettuce in California caused by Pseudomonas cichorii. Phytopathology. 1977;67:957–960. doi: 10.1094/Phyto-67-957. [DOI] [Google Scholar]
- 45.Paula Wilkie J, Dye D. Pseudomonas cichorii causing tomato and celery diseases in New Zealand. N. Z. J. Agric. Res. 1974;17:123–130. doi: 10.1080/00288233.1974.10420990. [DOI] [Google Scholar]
- 46.Loppnow H, et al. Cytokine induction by lipopolysaccharide (LPS) corresponds to lethal toxicity and is inhibited by nontoxic Rhodobacter capsulatus LPS. Infect. Immun. 1990;58:3743–3750. doi: 10.1128/iai.58.11.3743-3750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong Y-H, et al. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature. 2001;411:813–817. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- 48.Sathiyanarayanan G, et al. Exopolysaccharide from psychrotrophic Arctic glacier soil bacterium Flavobacterium sp. ASB 3-3 and its potential applications. RSC Adv. 2015;5:84492–84502. doi: 10.1039/C5RA14978A. [DOI] [Google Scholar]
- 49.Jiang H, et al. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl. Environ. Microbiol. 2006;72:3832–3845. doi: 10.1128/AEM.02869-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


