Abstract
The gut microbiota is an important contributor to human health. Vegetable/fruit juices provide polyphenols, oligosaccharides, fiber and nitrate (beet juice), which may induce a prebiotic-like effect. Juice-based diets are becoming popular. However, there is a lack of scientific evidence of their health benefits. It was our hypothesis that changes in the intestinal microbiota induced by a juice-based diet play an important role in their health benefits. Twenty healthy adults consumed only vegetable/fruit juices for 3 days followed by 14 days of customary diet. On day 4 we observed a significant decrease in weight and body mass index (p = 2.0E−05), which was maintained until day 17 (p = 3.0E−04). On day 4 the proportion of the phylum Firmicutes and Proteobacteria in stool was significantly decreased and Bacteroidetes and Cyanobacteria was increased compared to baseline and was partially reversed on day 17. On day 4 plasma and urine nitric oxide was increased by 244 ± 89% and 450 ± 360%, respectively, and urinary lipid peroxidation marker malondialdehyde was decreased by 32 ± 21% compared to baseline. General well-being score was increased at the end of the study. In summary a 3-day juice-based diet altered the intestinal microbiota associated with weight loss, increase in the vasodilator NO, and decrease in lipid oxidation.
Introduction
Vegetable/fruit juice-based diets have been very popular recently. However, well designed controlled research studies with clinical outcome measures providing scientific evidence of potential health benefits of juice only diets are limited1. The consumption of vegetable/fruit juice during the abstinence from food provides essential nutrients and improves compliance.
Fruit and vegetables are rich sources of several biologically active components that contribute to general health and decrease the risk of chronic diseases such as cardiovascular disease2. They are the most ubiquitous source of phenolic compounds3. Polyphenols exert a variety of physiological effects in vitro including antioxidative, immunomodulatory and antimicrobial activities4.
The absorption of polyphenols in the small intestine is limited and considerable amounts of these polyphenols can be found in the colon. There the colonic bacteria metabolize polyphenols to smaller compounds, which in turn alter the abundance of bacteria in the intestinal microbiome. In addition, fruit and vegetable are rich in fermentable fiber with prebiotic activity. High fiber intake is associated with decreased risk of cardiovascular disease, type 2 diabetes and some forms of cancer5. Fiber is composed of oligosaccharides, which resist digestion in the small intestine and are transported to the colon where they provide energy for gut bacteria6. Growing evidence is demonstrating the role of the microbiota in the health benefits of dietary fiber consumption6.
In the human intestine the gut microbiota is an important contributor to human health and has been implicated in the development of obesity and obesity-related diseases such as diabetes and cardiovascular disease7–9. The two most abundant bacterial phyla in humans and in mice are Firmicutes (40–60%) and Bacteroidetes (20–40%) with lower abundance of Actinobacteria, Fusobacteria, Proteobacteria and Verrucomicrobia10. Recent studies show that dietary interventions with polyphenol rich extracts and foods, including dealcoholized red wine polyphenols, cocoa-derived flavanols, quercetin and grape anthocyanins, modulate the human gut microbiota by decreasing the abundance of Firmicutes and increasing Bifidobacteria, Lactobacillus and Verrucomicrobia11–13, which is also a key difference in the gut microbiota found in obese and lean individuals14, 15.
We therefore investigated whether the consumption of fruit and vegetable juices (6 bottles daily of mixtures of greens, roots, citrus, lemon, cayenne and vanilla almond) as part of a 3-day juice only program alters the intestinal microbiota in twenty healthy participants. Secondary outcomes of the study were to determine the effect of the 3-day juice based diet on change in weight and body composition and biomarkers of oxidation (urine malondialdehyde) and vasodilation (plasma and urine nitric oxide).
Results
Thirty participants were screened. Twenty-five participants met enrollment criteria and were randomized and completed the 31-day study. Five participants were not able to provide stool samples at either day 4 or 17 and were excluded. All data included in this manuscript includes the twenty participants only (Table 1).
Table 1.
Baseline characteristics of the study participants.
| Variable | |
|---|---|
| Study Participants | |
| Screened (N) | 30 |
| Enrolled/randomized (N) | 25 |
| Participants with all stool samples (N) | 20 |
| Age, years | 32 ± 8 |
| Gender | |
| Male (N) | 5 |
| Female (N) | 15 |
| Race | |
| African American (N) | 1 |
| Caucasian (N) | 9 |
| Asian (N) | 8 |
| Bi-Racial (N) | 2 |
| Height, cm | 167 ± 5 |
| Weight, kg | 71 ± 18 |
| BMI, kg/m2 | 25.5 ± 5 |
Body weight and composition
According to the calorie content provided by the manufacturer the total calorie intake per person per day was 1310 kcal. During the 3-day juice intervention a significant weight loss of 1.7 ± 1.2 kg was observed (p = 2.0E−05) (Fig. 1a). After the 2-week follow up period body weight remained decreased (0.91 ± 0.9 kg) compared to baseline weight. Body mass index (BMI) was decreased by 0.6 ± 0.4 after the 3-day juice fast and remained decreased by 0.33 ± 0.3 after the 2 week follow up period (Fig. 1b).
Figure 1.
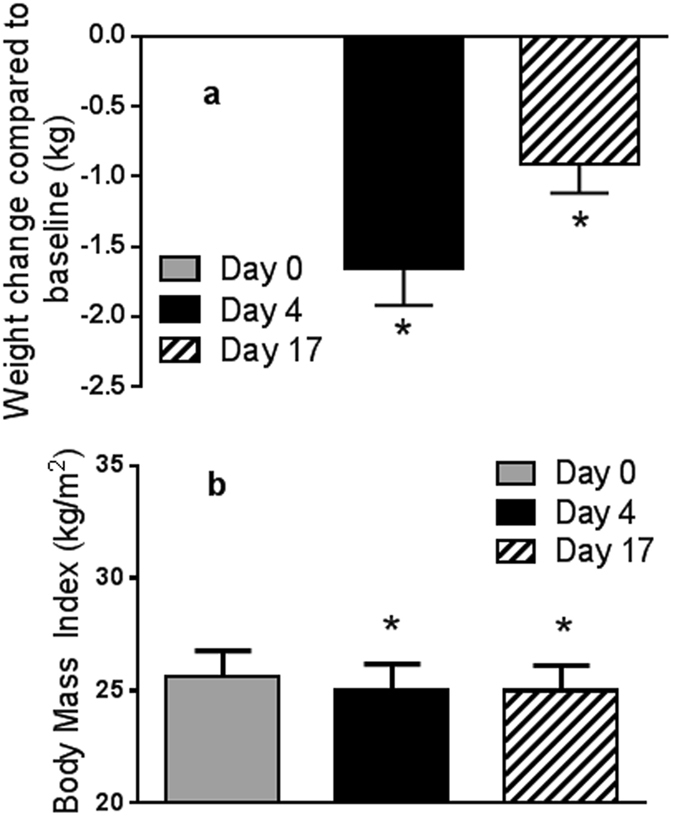
Effect of juice based diet on body weight (a) and body mass index (b). Data are mean ± SEM, n = 20. One way repeat measures ANOVA was performed. Difference to baseline is indicated; *p < 0.05.
Intestinal microbiome
Fecal microbiota composition of 20 participants collected at day 0, 4 and 17 was analyzed by sequencing the V4 region of the 16S rDNA gene. Rarefaction curves for bacterial DNA sequences for each stool sample approached a plateau, indicating that sequence coverage was sufficient to encompass the majority of diversity contained within each sample (results not shown). The distribution of 16S rDNA genes at the phylum level was composed of Firmicutes > Bacteroidetes > Proteobacteria = Verrucomicrobia > Actinobacteria (52 ± 4, 41 ± 4, 2.3 ± 1, 2.3 ± 1 and 5 ± 4%, respectively). The juice consumption, however, was associated with a significant decrease in the proportion of the bacterial phylum Firmicutes (p = 0.014) and increase in Bacteroidetes (p = 0.026) and Cyanobacteria (0.003) at day 4 compared to baseline and was partially reversed to baseline proportions at day 17 (Fig. 2). The proportions of Verrucomicrobia, Proteobacteria, Actinobateria and Fusobacteria were not changed significantly (Fig. 2). Linear regression analysis of data from this study showed a significant positive correlation with day 4 body weight and Firmicutes proportion (p = 0.006), and a negative correlation with Bacteroidetes (p = 0.011) (Supplementary Fig. S1). There was no difference in indices of fecal community richness and diversity (α-diversity indices) throughout the intervention period (Supplementary Fig. 2; p = 0.49).
Figure 2.
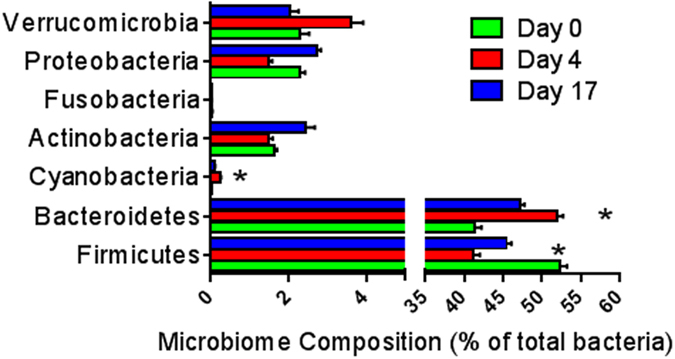
Effect of juice based diet on fecal microbiota (phylum) comparing baseline (day 0), after the juice intervention (day 4) and after two weeks of customary diet (day 17). Values are mean ± SEM (n = 20). Difference to baseline is indicated by *p < 0.05.
On the genus level the most prominent bacterium was Bacteroides, which was increased significantly at day 4 compared to baseline (Table 2 and Supplementary Fig. S3). Twenty different Bacteroides species were identified and the abundance of eight of them (B. acidifaciens, B. caccae, B. fragilis, B. massiliensis, B. nordii, B. salyersiae, B. thetaiotaomicron, B. uniformis) was increased on day 4 compared to baseline (Supplementary Fig. S3).
Table 2.
Effect of juice based diet on fecal microbiota (genus).
| Genus | Day 0 | Day 7 | Day 17 |
|---|---|---|---|
| Anaeroarcus | 0.2 ± 0.03b | 0.3 ± 0.05a | 0.1 ± 0.02b |
| Bacteroides | 25.6 ± 3.4b | 37.0 ± 3.5a | 30.2 ± 2.7a,b |
| Barnesiella | 0.4 ± 0.15b | 0.8 ± 0.3a | 0.6 ± 0.2a,b |
| Bilophila | 0.6 ± 0.3a,b | 0.5 ± 0.2b | 1.2 ± 0.4a |
| Butyricimonas | 0.1 ± 0.05a,b | 0.2 ± 0.07a | 0.1 ± 0.03b |
| Dialister | 3.6 ± 1.19a | 2.4 ± 0.9b | 3.6 ± 1.5a,b |
| Eisenbergiella | 0.2 ± 0.06a | 0.1 ± 0.01b | 0.2 ± 0.05a |
| Erysipelatoclostridium | 0.2 ± 0.04a,b | 0.3 ± 0.11a | 0.1 ± 0.02b |
| Faecalibacterium | 6.7 ± 1.9a,b | 4.2 ± 1.02b | 7.5 ± 1.4a |
| Haemophilus | 0.02 ± 0.008a | 0.002 ± 0.001b | 0.007 ± 0.004a,b |
| Halospirulina | 0.02 ± 0.007b | 0.2 ± 0.07a | 0.09 ± 0.07b |
| Odoribacter | 0.3 ± 0.11b | 0.6 ± 0.1a | 0.4 ± 0.09b |
| Oscillospira | 2.3 ± 1.0a | 2.1 ± 0.5b | 2.6 ± 0.84a,b |
| Parabacteroides | 2.5 ± 0.66a | 2.4 ± 0.52a,b | 3.3 ± 0.6b |
| Paraprevotella | 0.2 ± 0.12b | 0.8 ± 0.4a | 0.8 ± 0.38a,b |
| Ralstonia | 0.002 ± 0.000a | 0.001 + 0.000b | 0.001 ± 0.000a,b |
| Ruminiclostridium | 1.0 ± 0.28a | 0.5 ± 0.15b | 0.5 ± 0.12a,b |
| Streptococcus | 1.5 + 0.63a | 0.1 + 0.03b | 0.3 + 0.09b |
| Subdoligranulum | 4.1 ± 1.16a | 1.5 ± 0.34b | 2.5 ± 0.7a,b |
This table includes only significantly altered bacteria. Supplementary Table S2 includes bacteria that did not change significantly. Data presents proportions of bacteria as percent of total count. Values are mean ± SEMs (n = 20). a,bLabeled means without a common superscript letter differ, p < 0.05.
On the genus level the following bacterial populations were also significantly increased at day 4 compared to baseline (percent of baseline): Halospirulina (1467%), Paraprevotella (348%), Barnesiella (200%), Odoribacter (200%) and Bacteroides (144%) (Table 2). On the other hand, proportions of the following bacterial genera were decreased significantly at day 4 compared to baseline (percent of baseline): Streptococcus (8%), Subdoligranulum (30%), Eisenbergiella (40%), Ruminiclostridium (50%), and Dialister (67%) (Table 2). Except for Streptococcus, which was still significantly decreased to 19% of baseline at day 17, the proportions of all other bacterial genera were changed back to baseline values (Table 2).
The functional analysis of the microbial metagenome using PICRUST showed no difference in bacterial nitrogen metabolism (Supplementary Fig. S4).
Plasma antioxidant capacity and urine lipid peroxidation
The effect of juice based diet on plasma antioxidant capacity was determined by analyzing the trolox equivalents (TE) using the TEAC method. There was no change in plasma TE comparing samples collected at day 4 and 17 with baseline samples (data not presented). Lipid peroxidation was determined by the analysis of urine malondialdehyde (MDA). Urine MDA was significantly decreased by 40% in 6-hour urine collected at day 4 compared to baseline urine (p = 0.01) and returned to baseline levels at day 17 (Fig. 3).
Figure 3.
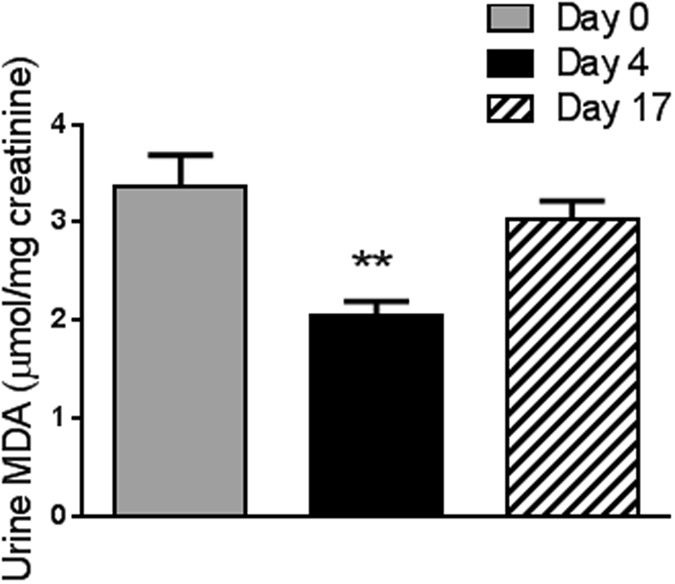
Effect of juice based diet on urinary malondialdehyde concentration. Data are mean ± SEM. One way repeat measures ANOVA was performed. The difference to baseline is indicated **p < 0.001.
Plasma and urine nitric oxide
To evaluate potential effects of vegetable and fruit nitrate content on vasodilation we determined the effect of juice based diet on plasma and urine nitric oxide (NO) concentrations. Both plasma and urine NO were significantly increased 3-fold for plasma and 5-fold for urine at 4 days compared to baseline (p = 1.0E−06) (Fig. 4). All NO values returned to baseline concentrations at 17 days (Fig. 4).
Figure 4.
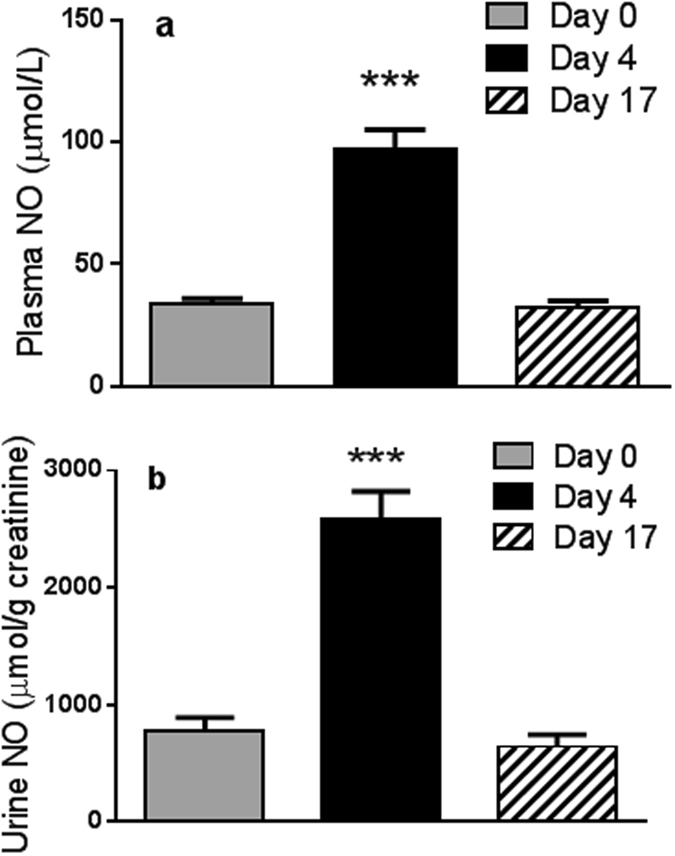
The effect of juice based diet on (a). Plasma nitric oxide and (b). Urinary nitric oxide. Data are mean ± SEM (n = 20). One way repeat measures ANOVA was performed. The difference to baseline is indicated ***p < 1.0E−08.
General well-being score
All participants completed a general well-being questionnaire at baseline, day 4 and at the end of study. There was no difference in well-being score after the 3-day juice period (82.4 ± 14) compared to baseline (82.2 ± 10). However, at the end of the study the well-being score (87 ± 11) was significantly increased (p = 0.006) (Fig. 5).
Figure 5.
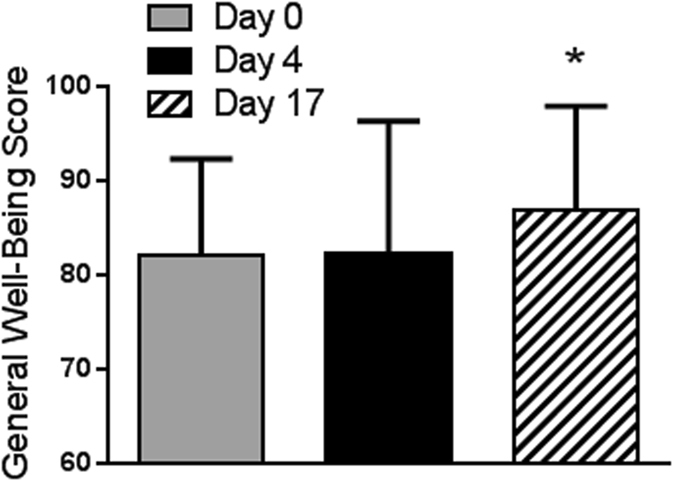
Effect of juice based diet on Wellness Score. Data are mean ± SEM (n = 20). One way repeat measures ANOVA was performed. The difference to baseline is indicated *p < 0.05.
Discussion
The scientific base for the popularity of juice based diets has not been explored. Here we demonstrated that a 3-day juice-based diet of drinking 6 bottles of fruit/vegetable juice blends (16 oz ea) resulted in a significant decrease in body weight (p = 2.0E−05). The observed weight loss remained significant and persisted over the following 2 weeks and may be related to changes in the microbiome (p = 0.003).
The two most abundant bacterial phyla in humans are Firmicutes (40–60%) and Bacteroidetes (20–40%)10. Relative proportional abundance of Firmicutes has been associated with increased body weight and that of Bacteroidetes with low body weight. The 3-day juice based diet of only drinking 6 bottles of fruit/vegetable juice blends induced significant changes in the intestinal microbiota. The proportional abundance of the phylum Firmicutes was significantly decreased, while Bacteroidetes was significantly increased when comparing end of juice based diet (day 4) to baseline (Table 1). Human Bacteroides species are able to degrade diverse plant fiber and complex polysaccharides, including pectin and xylans from fruit and vegetable16, 17. Bacteroides ovatus, B. thetaiotaomicron and B. uniformis ferment a particularly wide range of complex polysaccharides16. In the present study eight Bacteroides species were significantly increased after the juice based diet. A significant increase in the proportion of these Bacteroides species (B. ovatus, B. acidifaciens and B. xylanisolvens) was also observed in another human intervention study in individuals with metabolic syndrome who included the prebiotic resistant starch in their diet18. In that study the increase in Bacteroides was associated with a decrease in markers of metabolic syndrome (cholesterol, blood pressure, etc.). In another study B. thetaiotaomicron was used as probiotic in combination with a prebiotic in rats fed a high fat diet, resulting in a significant decrease in weight and postprandial plasma triglyceride levels19. Products of fiber/complex carbohydrate metabolism are short chain fatty acids, which have been shown to play an important role in the cardiovascular health benefits of fiber consumption20.
The only other human study investigating the effect of juice based diet on changes in the gut microbiota was published by Remely et al.21. They observed changes in the stool microbiome in participants following a traditional program in an Austrian Monastery including a small amount of cereal, vegetable, fruit and herbal tea21. After one week of the program an increase in microbial diversity was observed as well as the abundance of Akkermansia and Bifidobacteria was increased and the abundance of Enterobacteria and Lactobacilli was decreased as determined by quantitative PCR of 16S rDNA of the bacteria of interest21.
In the present study the consumption of the juices was associated with a significant increase in plasma and urine nitric oxide. Nitric oxide (NO), a known vasodilator, is an important factor in cardiovascular disease. Impaired eNOS activity and lack of bioavailable NO play a role in the development and progression of endothelial dysfunction that leads to arterial stiffness and an increase in blood pressure22. A study by Velmurugan et al. demonstrated that the consumption of dietary nitrate from beetroot juice resulted in significant increase in ultrasound flow-mediated dilatation in hypercholesterolemic patients23. Dietary interventions leading to increased blood NO concentrations may be beneficial in the prevention of heart disease.
Sources of nitric oxide (NO) are either the endogenous formation through the endothelial arginine nitric oxide synthase (eNOS) pathway or from dietary sources in form of nitrate24. One of the juices consumed during the juice fast was prepared from green leafy vegetable and one from beetroot, which are both good dietary sources of nitrate24. Polyphenols have been shown to enhance eNOS activity22, 24. Due to the high polyphenol content of the fruit/vegetable juices stimulation of endogenous eNOS activity may have contributed to the increase in NO. Endothelial dysfunction is one of the hallmark warning signs of cardiovascular disease and is involved in disease states such as atherosclerosis and hypertension, in which the normal function of endothelium is severely affected25. Dietary nitrate is mostly reduced to nitrite by nitrate-reducing bacteria in the oral cavity but also by bacteria in the lower intestinal tract24, 26, 27. In the present study, however, our functional analysis of the microbial metagenome using PICRUST showed no difference in the nitrogen metabolism (p = 0.24).
We observed a decrease in the concentration of urinary lipid peroxidation product malondialdehyde (MDA) after the 3-day juice fast. This may have been based on the high polyphenol content providing antioxidant protection for lipids in the intestine during the digestion. Similar effects on postprandial lipid peroxidation were observed by our laboratory and others utilizing other polyphenol sources. For example the addition of a spice mix or red wine to a high fat meal dramatically decreased postprandial urinary MDA formation28, 29. Another study by Bub et al. also supports the antioxidant effect of polyphenol rich fruit juices30. In this randomized, crossover-design study in healthy men, a daily consumption of polyphenol-rich juices (330 ml/d) consumed for 2 weeks decreased plasma malondialdehyde compared to baseline30. In addition, urine MDA may have been decreased based on the low fat content of the juice fast (15% calories from fat).
The vegetable/fruit juice based diet consumption showed a delayed response to improved well-being after participants returned to their regular diet for two weeks. Since the microbiota changes mostly returned to baseline levels, it appears unlikely that changes in the well-being were related to the gut microbiota. As mediator of mood, appetite and sleep, serum levels of the monoamine neurotransmitter serotonin could play a role in well-being. However, no significant change in serum serotonin levels were found (data not shown). Possibly participants felt good about the weight loss that was maintained through the two weeks after returning to the customary diet.
In summary the 3-day vegetable/fruit juice-based diet induced significant changes in the intestinal microbiota which were associated with weight loss. Further mechanistic studies are required to confirm that changes in the microbiota are directly linked to weight loss. The juice- based diet also, significantly increased serum and urine NO and decreased a marker of lipid oxidation. Future studies will need to be performed to determine if these biochemical changes are associated with vasodilation and improved cardiovascular health.
Methods
Study design and juice intervention
The study was carried out in accordance with the guidelines of the Office for Protection of Research Subjects of the University of California, Los Angeles. The clinical protocol was approved by the internal review board (IRB) of the University of California, Los Angeles. The study was registered at the NIH Clinical Trial Registry: NCT02377063 on 2/18/2015.
All subjects provided written informed consent before the study began. The study was divided into three periods: 2-week run-in period, 3-day juice fast and 2-week follow-up period. Healthy subjects who consume <3 servings of fruits/vegetables were recruited to the study. During the run-in period the participants were asked to continue their usual diet and refrain from consuming vitamins and antibiotics. During the intervention period all participants only consumed 6 bottles of vegetable/fruit juice daily for 3 days. The 6 different types of vegetable/fruit blends were prepared from the following fruits and vegetables: Green mixes were blended from apple, cucumber, celery, romaine lettuce, lemon and limited amount (<2%) of spinach, kale and parsley. One of the green mixes also contained ginger. The root mix was a juice blend of apple, lemon, ginger and beet. The citrus mix contained apple, pineapple, and very limited amount (<1%) of lemon and mint. Lemon cayenne water consisted of filtered water with cayenne and lemon and “vanilla almond (VA)” was a blend of almond, dates, sea salt and vanilla bean. The nutritional facts are listed in Supplementary Table S1. Each bottle contained 16 oz. or 2 servings. The total caloric content of all 6 bottles providing 1310 kcal per day. The juices provided 38 g of total fat per day (266 kcal, 20% calories from fat) and 28 g of fiber.
During the follow-up period participants resumed their usual diet consumed during the run-in period following the same fruit and vegetable serving, vitamin and antibiotic restrictions. Juices were obtained from Pressed Juicery (Santa Monica, CA). Nutrition facts are provided in Supplementary Table S1. On day 0, 4 and 17 participants completed a Psychological General Well Being Questionnaire (http://www.opapc.com).
Subjects
Twenty-five healthy women and men 18–50 years of age with a custom diet including <3 servings of fruits/vegetables per day were recruited through local advertisement. Subjects with a history of diabetes mellitus on medications, hyperlipidemia on medications, or other serious medical condition within 6 months prior to screening were excluded. In addition, individuals using antibiotics or laxatives during 2 months prior to the study were excluded.
Blood and urine collection
Fasting
EDTA blood samples and 24 hr urine samples were collected at baseline, day 4 and day 17. Blood was centrifuged 1500 × g for 10 min at 10 °C and plasma stored at 80 °C until analysis.
Stool Collection
A total of three stool samples were collected from each subject: at baseline, day 4 and day 17. Each time an aliquot of the stool specimen was collected by the participant and delivered to the UCLA Center for Human Nutrition in a cooler within a few hours of collection. At the laboratory stool was aliquoted into smaller vials and frozen immediately and stored at −80 °C.
Bacterial DNA Sequencing
DNA from stool was extracted using the MoBio power soil DNA isolation kit (MoBio Laboratories, Inc., Carlsbad, CA). The quality and quantity of the DNA was confirmed using a Nanodrop 1000 (Thermo Fisher Scientific, Wilmington, DE). The 16S rRNA gene V4 variable region PCR primers 530/926 with barcode on the forward primer were used in a 30 cycle PCR using the HotStarTaq Plus Master Mix Kit (Qiagen, USA) under the following conditions: 94 °C for 3 min, followed by 28 cycles of 94 °C for 30 s, 53 °C for 40 s and 72 °C for 1 min, after which a final elongation step at 72 °C for 5 min was performed. After amplification, PCR products are checked in 2% agarose gel to determine the success of amplification and the relative intensity of bands. Sequencing was performed at MR DNA (www.mrdnalab.com, Shallowater, TX, USA) on a MiSeq (Illumina, San Diego, CA) following the manufacturer’s guidelines. Sequence data were processed using a proprietary analysis pipeline (MR DNA, Shallowater, TX, USA). Operational taxonomic units (OTUs) were defined by clustering at 3% divergence (97% similarity). Final OTUs were taxonomically classified using BLASTn against a curated GreenGenes database31. Within community diversity (α-diversity) was calculated using Quantitative Insights Into Microbial Ecology (QIIME) software package32. Analysis of α-diversity (Shannon index) was performed by a one-way ANOVA. β-diversity was measured by calculating the weighted UniFrac distances33 using QIIME default scripts, and weighted UniFrac PCoA biplot was visualized using EMPeror. Statistical difference between different time points was analyzed by PERMDISP. Microbiome data was further analyzed using PICRUST (phylogenetic investigation of communities by reconstruction of unobserved states) to predict the functional composition of the microbial community’s metagenome from its 16S profile by using marker gene data and a database of reference genomes34.
Body Composition
Body composition was measured using the Tanita-BC418 bioelectrical impedance analyzer (Tanita Corp., Japan).
Malondialdehyde
Urine malondialdehyde concentration was determined in form of thiobarbituric acid (TBA) reactive substances by deproteinization and derivatization with TBA according to the method of Korchazhkina et al.35. In brief, 0.6 mL urine was mixed with 0.4 mL of water and 3 mL of H3PO4 (1%V/V) and vortexed. One ml of TBA solution was added and incubated in a 100 °C water bath for 60 min. Samples were cooled on ice and centrifuged at 18,000 × g for 10 min. An aliquot of 50 µL supernatant was injected into the HPLC system for analysis. Chromatographic determinations were performed on a Waters 2690 HPLC (Waters, Milford, MA) equipped with a Waters 474 fluorescence detector (Waters, Milford, MA) at Ex (ex-citation) 515 nm and Em (emission) 550. An Agilent Zorbax SB C18 reversed-phase column (150 mm, 4.6-mm inside diameter; 3.5-µm particles; Waters, Milford, MA) was used for separation at an ambient temperature. A linear gradient from 10% A and 90% B to 60% A and 40% B over 20 min (A: acetonitrile and B: 0.1% phosphoric acid in water) at a flow rate of 0.8 mL/min was used for the elution. The reagent 1,1,3,3-tetramethoxypropane was used to prepare a standard curve.
Nitric oxide
Nitric oxide (NO) is oxidized in the body to the metabolites nitrate (NO3−) and nitrite (NO2−), which are excreted in the urine. Nitrate/nitrite concentrations were measured in the form of NO in urine and serum samples by an ozone chemiluminescence method (Sievers Instruments, Boulder, CO). Serum samples were mixed with ethanol in a ratio of 1:2 and stored at 4 °C overnight. After centrifugation for 20 min at 17 000 × g an aliquot of the supernatant was injected into the reaction chamber of the NO analyzer. Urine was centrifuged at the same speed and injected. Total nitrate and nitrite were analyzed by injecting 5 μL of each microdialysate sample into a purge vessel containing a solution of vanadium (III) chloride (50 mmol/L) in hydrochloric acid (1 mol/L) at 95 °C, continuously purged with a stream of nitrogen gas, connected to a Sievers 280i Nitric Oxide Analyser (GE Analytical Instruments, Boulder, CO, USA). The concentration was determined in comparison to a sodium nitrate standard calibration curve.
Urine creatinine
The urinary creatinine concentration was measured by using a Stanbio Direct Creatinine LiquiColor Kit (Stanbio LaboratoryBoerne, TX, USA) according to manufacturer’s instructions. A creatinine standard calibration curve was generated by plotting the absorbance measured at 510 nm of a series concentration of creatinine standard solutions reacting with working reagent provided by the kit. The urinary creatinine level was measured in the same manner after a proper dilution with distilled water.
Statistics
Statistical analyses were performed using IBM SPSS Statistics version 22 (IBM Corporation, Armonk, NY, USA). Data were evaluated with One-way repeated measures ANOVA with a Bonferroni’s post-test if the assumption are met, and those without were analyzed using non-parametric test (Friedman test with a Dunn’s post-test). P-values < 0.05 were considered statistically significant. Correlation was evaluated using GraphPad Prism 6 (La Jolla, CA).
Electronic supplementary material
Acknowledgements
Supported by departmental funds from the Center for Human Nutrition, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles.
Author Contributions
S.M.H. and Z.L. wrote the main manuscript text, J.Y. and A.L. performed QIIME analysis, R.P.L. and J.H. performed chemical analyses, P.S., M.H. and G.T. coordinated clinical study, Q.Y.L. evaluated juices, D.H. participated in manuscript preparation. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02200-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Horne BD, Muhlestein JB, Anderson JL. Health effects of intermittent fasting: hormesis or harm? A systematic review. The American journal of clinical nutrition. 2015;102:464–470. doi: 10.3945/ajcn.115.109553. [DOI] [PubMed] [Google Scholar]
- 2.Tome-Carneiro, J. & Visioli, F. Polyphenol-based nutraceuticals for the prevention and treatment of cardiovascular disease: Review of human evidence. Phytomedicine: international journal of phytotherapy and phytopharmacology, doi:10.1016/j.phymed.2015.10.018 (2015). [DOI] [PubMed]
- 3.Abuajah CI, Ogbonna AC, Osuji CM. Functional components and medicinal properties of food: a review. Journal of food science and technology. 2015;52:2522–2529. doi: 10.1007/s13197-014-1396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li AN, et al. Resources and biological activities of natural polyphenols. Nutrients. 2014;6:6020–6047. doi: 10.3390/nu6126020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahl, W. J. et al. Health Benefits of Fiber Fermentation. Journal of the American College of Nutrition 1–10, doi:10.1080/07315724.2016.1188737 (2017). [DOI] [PubMed]
- 6.Simpson HL, Campbell BJ. Review article: dietary fibre-microbiota interactions. Alimentary pharmacology & therapeutics. 2015;42:158–179. doi: 10.1111/apt.13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korpela K, et al. Gut microbiota signatures predict host and microbiota responses to dietary interventions in obese individuals. PloS one. 2014;9:e90702. doi: 10.1371/journal.pone.0090702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut. 2014;63:1513–1521. doi: 10.1136/gutjnl-2014-306928. [DOI] [PubMed] [Google Scholar]
- 9.Tuohy KM, Fava F, Viola R. ‘The way to a man’s heart is through his gut microbiota’–dietary pro- and prebiotics for the management of cardiovascular risk. The Proceedings of the Nutrition Society. 2014;73:172–185. doi: 10.1017/S0029665113003911. [DOI] [PubMed] [Google Scholar]
- 10.Million M, Lagier JC, Yahav D, Paul M. Gut bacterial microbiota and obesity. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2013;19:305–313. doi: 10.1111/1469-0691.12172. [DOI] [PubMed] [Google Scholar]
- 11.Etxeberria U, et al. Reshaping faecal gut microbiota composition by the intake of trans-resveratrol and quercetin in high-fat sucrose diet-fed rats. The Journal of nutritional biochemistry. 2015;26:651–660. doi: 10.1016/j.jnutbio.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Roopchand DE, et al. Dietary Polyphenols Promote Growth of the Gut Bacterium Akkermansia muciniphila and Attenuate High-Fat Diet-Induced Metabolic Syndrome. Diabetes. 2015;64:2847–2858. doi: 10.2337/db14-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duenas M, et al. A survey of modulation of gut microbiota by dietary polyphenols. BioMed research international. 2015;2015:850902–15. doi: 10.1155/2015/850902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 15.Stenman, L. K., Burcelin, R. & Lahtinen, S. Establishing a causal link between gut microbes, body weight gain and glucose metabolism in humans - towards treatment with probiotics. Beneficial microbes 1–12, doi:10.3920/BM2015.0069 (2015). [DOI] [PubMed]
- 16.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravcheev DA, Godzik A, Osterman AL, Rodionov DA. Polysaccharides utilization in human gut bacterium Bacteroides thetaiotaomicron: comparative genomics reconstruction of metabolic and regulatory networks. BMC genomics. 2013;14:873. doi: 10.1186/1471-2164-14-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Upadhyaya B, et al. Impact of dietary resistant starch type 4 on human gut microbiota and immunometabolic functions. Scientific reports. 2016;6:28797. doi: 10.1038/srep28797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olli K, et al. Independent and Combined Effects of Lactitol, Polydextrose, and Bacteroides thetaiotaomicron on Postprandial Metabolism and Body Weight in Rats Fed a High-Fat Diet. Frontiers in nutrition. 2016;3:15. doi: 10.3389/fnut.2016.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marques, F. Z. et al. High Fibre Diet and Acetate Supplementation Change the Gut Microbiota and Prevent the Development of Hypertension and Heart Failure in DOCA-Salt Hypertensive Mice. Circulation, doi:10.1161/CIRCULATIONAHA.116.024545 (2016). [DOI] [PubMed]
- 21.Remely M, et al. Increased gut microbiota diversity and abundance of Faecalibacterium prausnitzii and Akkermansia after fasting: a pilot study. Wiener klinische Wochenschrift. 2015;127:394–398. doi: 10.1007/s00508-015-0755-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cutler BR, Petersen C, Anandh Babu PV. Mechanistic insights into the vascular effects of blueberries: Evidence from recent studies. Molecular nutrition & food research. 2016 doi: 10.1002/mnfr.201600271. [DOI] [PubMed] [Google Scholar]
- 23.Velmurugan S, et al. Dietary nitrate improves vascular function in patients with hypercholesterolemia: a randomized, double-blind, placebo-controlled study. The American journal of clinical nutrition. 2016;103:25–38. doi: 10.3945/ajcn.115.116244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bondonno CP, Croft KD, Ward N, Considine MJ, Hodgson JM. Dietary flavonoids and nitrate: effects on nitric oxide and vascular function. Nutrition reviews. 2015;73:216–235. doi: 10.1093/nutrit/nuu014. [DOI] [PubMed] [Google Scholar]
- 25.Chistiakov DA, Orekhov AN, Bobryshev YV. Endothelial Barrier and Its Abnormalities in Cardiovascular Disease. Frontiers in physiology. 2015;6:365. doi: 10.3389/fphys.2015.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weitzberg E, Lundberg JO. Novel aspects of dietary nitrate and human health. Annual review of nutrition. 2013;33:129–159. doi: 10.1146/annurev-nutr-071812-161159. [DOI] [PubMed] [Google Scholar]
- 27.Tiso M, Schechter AN. Nitrate reduction to nitrite, nitric oxide and ammonia by gut bacteria under physiological conditions. PloS one. 2015;10:e0119712. doi: 10.1371/journal.pone.0119712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorelik S, Ligumsky M, Kohen R, Kanner J. A novel function of red wine polyphenols in humans: prevention of absorption of cytotoxic lipid peroxidation products. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2008;22:41–46. doi: 10.1096/fj.07-9041com. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, et al. Antioxidant-rich spice added to hamburger meat during cooking results in reduced meat, plasma, and urine malondialdehyde concentrations. The American journal of clinical nutrition. 2010;91:1180–1184. doi: 10.3945/ajcn.2009.28526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bub A, et al. Fruit juice consumption modulates antioxidative status, immune status and DNA damage. The Journal of nutritional biochemistry. 2003;14:90–98. doi: 10.1016/S0955-2863(02)00255-3. [DOI] [PubMed] [Google Scholar]
- 31.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature biotechnology. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korchazhkina O, Exley C, Andrew Spencer S. Measurement by reversed-phase high-performance liquid chromatography of malondialdehyde in normal human urine following derivatisation with 2,4-dinitrophenylhydrazine. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2003;794:353–362. doi: 10.1016/S1570-0232(03)00495-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


