Abstract
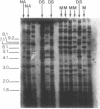
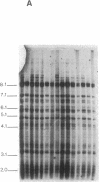
The measurement of clonal heterogeneity is central to understanding the evolutionary and population genetics of the roughly 50 species of vertebrates that lack effective genetic recombination. Simple-sequence DNA fingerprinting with oligonucleotide probes (CAC)5 and (GACA)4 is a sensitive and efficient means of detecting this heterogeneity in natural populations of two clonal fishes, Poecilia formosa, an apomictic unisexual, and Rivulus marmoratus, a selfing hermaphrodite. The fingerprints are clonally stable for at least three generations. The technique clearly differentiates allozymically identical laboratory lines of R. marmoratus that were previously distinguishable only by histocompatibility analysis. The technique also reveals apparent cases of shifts in clonal composition of a natural population of each species. Clonal variation in most natural populations is quite high. For example, a sample of 19 specimens of P. formosa from one station on the Rio Soto la Marina contained 16 clones (average clonal frequency = 0.07). This level of clonal diversity implies that mutation, subsequent to the founding of clonal lineages, is an important source of variation in these populations. It also suggests that chance (sampling error) has a previously unappreciated role in determining the clonal composition of populations even though some of the clones may be divergent in biologically significant features.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Hancock J. M., Dover G. A. Molecular coevolution among cryptically simple expansion segments of eukaryotic 26S/28S rRNAs. Mol Biol Evol. 1988 Jul;5(4):377–391. doi: 10.1093/oxfordjournals.molbev.a040505. [DOI] [PubMed] [Google Scholar]
- Harrington R. W., Jr Oviparous Hermaphroditic Fish with Internal Self-Fertilization. Science. 1961 Dec 1;134(3492):1749–1750. doi: 10.1126/science.134.3492.1749. [DOI] [PubMed] [Google Scholar]
- Schäfer R., Zischler H., Epplen J. T. DNA fingerprinting using non-radioactive oligonucleotide probes specific for simple repeats. Nucleic Acids Res. 1988 Oct 11;16(19):9344–9344. doi: 10.1093/nar/16.19.9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989 Aug 25;17(16):6463–6471. doi: 10.1093/nar/17.16.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]