Abstract
Gene expression in mammals is regulated by noncoding elements that can impact physiology and disease, yet the functions and target genes of most noncoding elements remain unknown. We present a high-throughput approach that uses CRISPR interference (CRISPRi) to discover regulatory elements and identify their target genes. We assess >1 megabase (Mb) of sequence in the vicinity of 2 essential transcription factors, MYC and GATA1, and identify 9 distal enhancers that control gene expression and cellular proliferation. Quantitative features of chromatin state and chromosome conformation distinguish the 7 enhancers that regulate MYC from other elements that do not, suggesting a strategy for predicting enhancer-promoter connectivity. This CRISPRi-based approach can be applied to dissect transcriptional networks and interpret the contributions of noncoding genetic variation to human disease.
A fundamental goal in modern biology is to identify and characterize the noncoding regulatory elements that control gene expression in development and disease, yet we have lacked systematic approaches to do so. Studies of individual regulatory elements have revealed principles of their function, such as the ability of enhancers to recruit activating transcription factors, modify chromatin state, and physically interact with target genes (1, 2). From these insights, systematic mapping of chromatin state and chromosome conformation across cell types has been used to identify putative regulatory elements (3–6). However, these measurements do not determine which (if any) genes are regulated or assess the quantitative effects on gene expression. Indeed, the rules that connect regulatory elements with their target genes in the genome appear to be complex. Regulatory elements do not necessarily affect the closest gene, but instead may act across long distances (7, 8). It remains unclear how many regulatory elements control any given gene, or how many genes are regulated by any given element (2, 3, 8).
We developed a high-throughput approach that utilizes the programmable properties of CRISPR/Cas9 to characterize the regulatory functions of noncoding elements in their native contexts. We use pooled CRISPR screens in combination with CRISPR interference (CRISPRi) — which alters chromatin state at targeted loci through recruitment of a KRAB effector domain fused to catalytically dead Cas9 (dCas9) (9–12) — to simultaneously characterize the regulatory effects of up to 1 Mb of sequence on a gene of interest (Fig. 1A) (13).
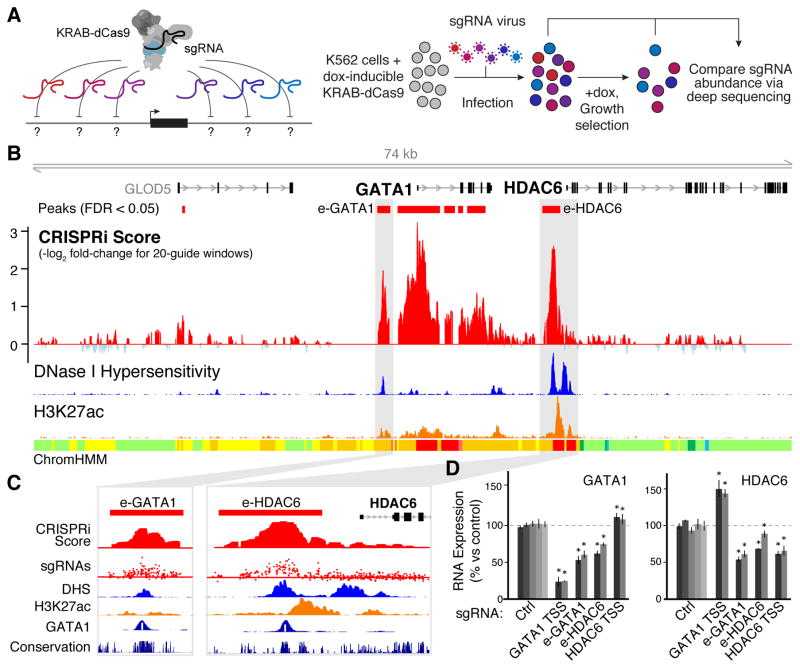
Fig. 1. Systematic mapping of noncoding elements that regulate GATA1.
(A) CRISPRi method for identifying gene regulatory elements. Cells expressing KRAB-dCas9 from a dox-inducible promoter are infected with a pool of single guide RNAs (sgRNAs) targeting every possible site across a region of interest. In a proliferation-based screen, cells expressing sgRNAs that target essential regulatory elements will be depleted in the final population. (B) CRISPRi screen results in the GATA1 locus. A high CRISPRi score indicates strong depletion over the course of the screen. Red boxes: Windows showing significant depletion compared to negative control sgRNAs (13). DNase I hypersensitivity, H3K27ac ChIP-Seq, and histone modification annotations (ChromHMM) in K562 cells are from ENCODE (4). (C) Close-up of e-GATA1 and e-HDAC6. sgRNA track shows CRISPRi scores for each individual sgRNA in the region. White bar in GATA1 ChIP-seq track represents the GATA1 motif. (D) qPCR for GATA1 and HDAC6 mRNA in cells expressing individual sgRNAs. KRAB-dCas9 expression was activated for 24 hours before measurement. Gray bars: different sgRNAs for each target. Ctrl: negative control sgRNAs without a genomic target. Error bars: 95% confidence intervals (CI) for the mean of 3 biological replicates (13). *: p < 0.05 in T-test versus Ctrl.
We studied two gene loci, GATA1 and MYC, that affect proliferation of K562 erythroleukemia cells in a dose-dependent manner (Fig. S1). This allowed us to search for regulatory elements that quantitatively tune GATA1 or MYC expression using a proliferation-based pooled assay (Fig. 1A). Importantly, GATA1 and MYC are not located near other strongly essential genes (Fig. S1); thus, proliferation defects caused by sgRNAs targeted to sequences near these genes can be attributed to elements regulating GATA1 or MYC. We designed a library containing 98,000 sgRNAs tiling across a total of 1.29 Mb of genomic sequence around GATA1 and MYC as well as 85 kb of control noncoding regions (13). We infected K562 cells expressing KRAB-dCas9 under a doxycycline-inducible promoter with a lentiviral sgRNA library and sequenced the representation of sgRNAs before and after growing cells in doxycycline for 14 population doublings (Fig. 1A). As expected, internal control sgRNAs targeting the promoters of known essential genes (10) were depleted (Fig. S2A) and correlated across biological replicates (R = 0.91, Fig. S2B).
We examined the quantitative depletion of sgRNAs in a 74 kb region surrounding GATA1, which encodes a key erythroid transcription factor (Fig. 1B). Because the efficiency of different sgRNAs for CRISPRi can vary dramatically (10), we used a sliding window approach, averaging the scores of 20 consecutive sgRNAs and assessing the false discovery rate (FDR) of this metric through comparison to negative control, non-essential regions (13) (Fig. S3). Because the average spacing between consecutive sgRNAs was 16 bp, the regions targeted by 20 consecutive sgRNA spanned an average of 314 bp (Fig. S3C,D). With this approach, the window with the highest score (strongest depletion) overlapped the GATA1 TSS itself (Fig. 1B, Fig. S3F). In addition, we identified 3 distal elements that significantly affected cellular proliferation (FDR < 0.05, Fig. 1B) (13). One such element (e-GATA1) is located ~3.6 kb upstream of GATA1 and corresponds to a DNase I hypersensitive site (DHS) marked by H3K27ac (Fig. 1C); notably, this element shows high sequence conservation among vertebrates, and the syntenic sequence in mouse is required for proper Gata1 expression in murine erythroid progenitor cells (14). The second distal element (e-HDAC6) corresponds to a conserved DHS located ~1.5 kb upstream of HDAC6 (Fig. 1C). A third significant element is located at a DHS near the promoter of GLOD5, which itself is not essential and only weakly expressed in K562 cells. The first two elements overlap GATA1 ChIP-Seq peaks and sequence motifs (Fig. 1C), consistent with known auto-regulatory loops in which GATA1 activates its own expression (15). All three elements reside in close linear and spatial proximity to GATA1 (Fig. S4A). Finally, multiple regions in the gene body of GATA1 scored as significantly depleted in the screen (Fig. 1B), but, because recruitment of KRAB-dCas9 to these sites may directly interfere with transcription (9), we focused on distal regulatory elements in subsequent analysis.
To characterize these elements, we measured GATA1 expression using quantitative PCR in cell lines stably expressing individual sgRNAs (13). As expected, targeting KRAB-dCas9 to the GATA1 TSS reduced GATA1 expression (76% reduction, Fig. 1D). sgRNAs targeting e-GATA1 or e-HDAC6 reduced GATA1 expression by 44% and 33%, respectively (Fig. 1D), and affected the expression of genes known to be regulated by the GATA1 transcription factor (Fig. S4B), confirming that these enhancers regulate GATA1. In contrast, sgRNAs targeting the HDAC6 TSS did not reduce GATA1 expression despite reducing HDAC6 expression (Fig. 1D), indicating that (i) the pooled screen accurately predicted that this region does not reduce GATA1 expression and (ii) the effects seen for the e-GATA1 and e-HDAC6 sgRNAs are not due to general effects of targeting KRAB-dCas9 to the gene neighborhood. Additionally, both e-GATA1 and e-HDAC6 can activate the expression of a plasmid-based reporter gene (Fig. S4C) (13). Together, these results support the specificity of this CRISPRi-based approach and demonstrate that e-GATA1 and e-HDAC6 quantitatively control GATA1 expression in K562 cells.
Considering the close proximity of GATA1 to HDAC6 (Fig. 1B, S4A), we tested whether this pair of enhancers also regulates HDAC6. sgRNAs targeting e-GATA1 and e-HDAC6 reduced HDAC6 expression by 42% and 22%, respectively, comparable to their effects on GATA1 (Fig. 1D). Intriguingly, inhibition of the GATA1 promoter led to an increase in HDAC6 expression (+47%, Fig. 1D), and inhibition of the HDAC6 promoter modestly activated GATA1 (+9%, Fig. 1D); this suggests that GATA1 and HDAC6 may compete for these shared enhancers, similar to observations for other pairs of neighboring genes (16, 17). Interestingly, histone deacetylases are required for erythropoiesis (18) and HDAC6 has been implicated in cellular proliferation in multiple cancers (19). Thus, although HDAC6 does not score as essential in proliferation assays in K562 cells, it is possible that proliferative defects observed upon inhibition of e-GATA1 or e-HDAC6 result from the combined effects on both GATA1 and HDAC6 expression (13), and the genomic proximity of these genes may be important for coordinating their expression in vivo. These observations indicate a complex connectivity between enhancers and promoters in their native genomic contexts (Fig. S4D).
We next investigated the cis regulatory architecture of MYC, a critical transcription factor encoded within a 3-Mb topological domain that contains hundreds of putative enhancers. Several enhancers in this domain regulate MYC in other cell types (13), but chromatin state varies dramatically across cell types and it is unclear which of these elements regulate MYC in a given cell type. Notably, the domain contains over 60 genetic haplotypes associated (through genome-wide association studies) with human phenotypes, including cancer susceptibility (20).
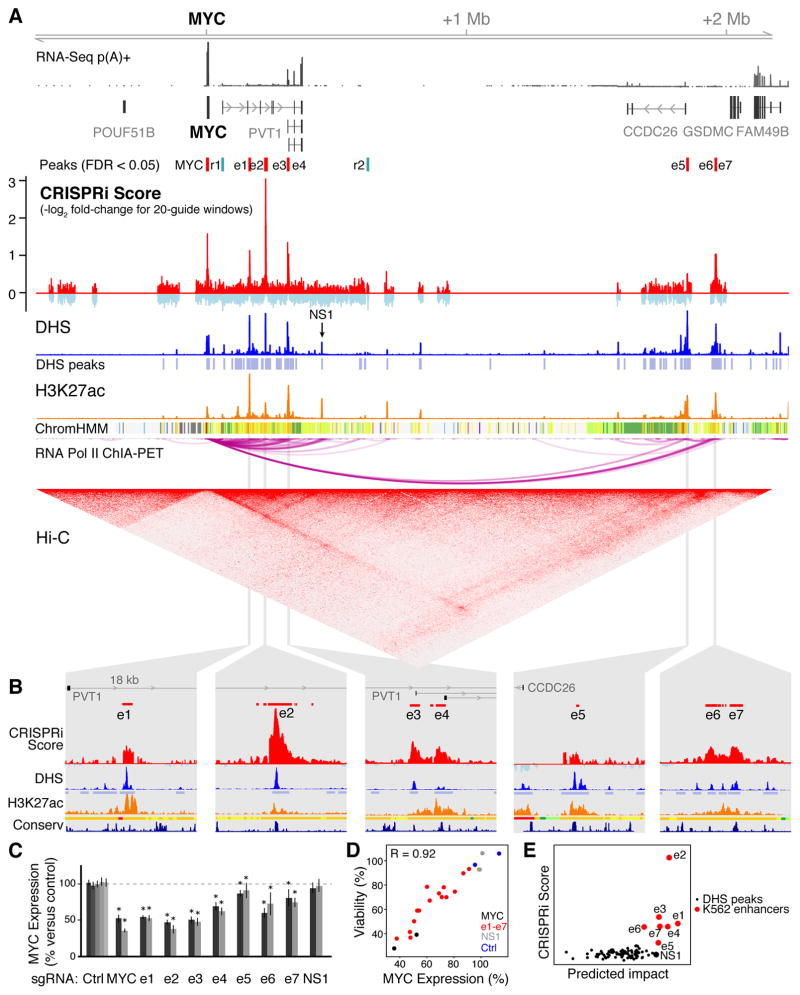
To identify elements that regulate MYC in K562 cells, we tiled sgRNAs across ~1.2 Mb of sequence in this topological domain (Fig. 2A). A sliding window analysis identified several regions whose inhibition reproducibly reduced cellular proliferation, including a known promoter-proximal element located 2 kb upstream of the MYC TSS (Fig. S5A) (21), the transcribed region of the MYC gene body (Fig. S5A), and seven distal regions (labeled e1 through e7) located between 0.16 and 1.9 Mb downstream of MYC (Fig. 2A, S5B,C). We also identified two regions that significantly increased cell proliferation (r1 and r2), and thus may repress MYC expression (Fig. 2A, Fig. S5D,E) (13).
Fig. 2. Identification and prediction of elements that regulate MYC.
(A) CRISPRi screening identifies 7 distal enhancers (e1–e7) that activate MYC and two repressive elements (r1, r2) that may act to repress MYC. NS1: an element that does not score in the screen. (B) 18-kb windows around each of the 7 distal enhancers. Y-axis scales are equivalent between panels. (C) qPCR for MYC mRNA in cells expressing individual sgRNAs 24 hours after KRAB-dCas9 activation. Gray bars: 2 different sgRNAs per target, or 5 for non-targeting controls (Ctrl). Error bars: 95% CI for the mean of 12 biological replicates (13). *: p < 0.05 in T-test versus negative controls. (D) Correlation between MYC expression and relative cell viability for e1–e7, MYC TSS, NS1, and Ctrl sgRNAs (13). Pearson’s R = 0.92 includes e1–e7 sgRNAs only; with the others, R = 0.95. (E) Predicted impact of DHS elements on MYC expression (a function of quantitative DHS, H3K27ac, and Hi-C signal) versus their experimentally derived CRISPRi scores (13).
Each of the seven putative activating elements is marked by high levels of DNase I hypersensitivity (Fig. 2A); is bound by multiple transcription factors (Fig. S6A); and shows patches of sequence conservation across mammals (Fig. 2B). Each enhancer frequently contacts the MYC promoter in three dimensions as assayed by Hi-C and ChIA-PET in K562 cells (Fig. 2A) (3, 6); elements e5 and e6/7 form very long-range (>1.8 Mb) loops to the MYC promoter and are located within 10 kb of CTCF ChIP-Seq peaks with motifs oriented toward MYC (Fig. S5B,C), consistent with the convergent rule for CTCF-mediated chromatin loops (6). Two elements (e3 and e4) correspond to alternative TSSs for the long noncoding RNA PVT1 (Fig. 2A); knockdown experiments indicate that the mature PVT1 RNA transcript itself is likely not essential in K562 cells (Fig. S1) and so e3 and e4 likely affect cellular proliferation through direct regulation of MYC (13).
We experimentally characterized these seven activating elements to test whether they regulate MYC. CRISPRi inhibition of each of these elements with individual sgRNAs led to proliferation defects in a competitive growth assay (Fig. S6B) and led to a 9–62% reduction in MYC expression (Fig. 2C). The magnitude of the change in gene expression correlated with the proliferation defect, consistent with a quantitative relationship between cell growth and precise MYC expression levels (Pearson R = 0.92, Fig. 2D). In a plasmid-based reporter assay, each putative regulatory element led to >5-fold up-regulation of a reporter gene relative to a control sequence (Fig. S6C) (13). For a subset of the elements (e2, e3, and e4), we generated clonal cell lines containing genetic deletions on one or two of the three chromosome 8 alleles (K562 cells are triploid) and measured the expression of MYC from each allele (13). For each element, we found that genetic deletions reduced MYC expression from the corresponding allele(s), confirming our CRISPRi results (Fig. S7). Together, these data support the hypothesis that these seven elements, spanning 1.6 Mb of noncoding sequence, act as enhancers to control MYC expression and cellular proliferation.
In addition to e1–e7, we characterized one noncoding element (NS1) that did not score in the screen (Fig. 2A). In K562 cells, NS1 displays strong DHS and H3K27ac occupancy, binds to multiple transcription factors (Fig. S6A), and participates in a long-range chromatin loop to the MYC promoter (Fig. 2A). In a lung adenocarcinoma cell line, NS1 regulates MYC as assayed by CRISPRi inhibition with individual sgRNAs (22). Accordingly, we wondered whether NS1 regulates MYC in K562 cells despite not being detected as such in our CRISPRi screen. To explore this possibility, we targeted KRAB-dCas9 to NS1 with individual sgRNAs in K562 cells and found that CRISPRi successfully reduced H3K27ac occupancy to an extent similar to that observed when targeting other MYC enhancers (Fig. S6D). Despite affecting chromatin state at NS1 in K562 cells, these sgRNAs did not substantially impact cellular proliferation or MYC expression (Fig. 2C,D), consistent with the results from the pooled screen. These observations support the ability of the CRISPRi screening approach to distinguish elements that do and do not regulate a given gene. However, we note that some regulatory elements, such as those that act redundantly with others in the locus, may not be discoverable by this method (13).
The ability to systematically test gene regulatory elements will help to train predictive models of functional enhancer-promoter connectivity. Notably, existing annotations and catalogs of enhancer-promoter predictions performed poorly at distinguishing e1–e7 from enhancers that do not impact MYC expression (13). For example, ENCODE annotates 185 Kb of sequence in this domain as putative “strong enhancer” in K562 cells (Fig. 2A), but only 8% of this sequence, corresponding to e1–e7, appears to regulate MYC. We sought to improve the ability to predict enhancers and connect them with genes that they regulate. When we examined chromatin state maps (including DHS, H3K27ac and Hi-C), we found that quantitative DHS or H3K27ac signal could distinguish most of the seven MYC enhancers but ranked them in the wrong order (Fig. S8A): for example, e5 shows the strongest DHS signal yet has the weakest effect on MYC expression (Fig. 2). Accordingly, we considered a framework (Fig. S8B) wherein the impact of an enhancer on gene expression is determined both by its intrinsic activity level (for which we use quantitative DHS and H3K27ac levels as a proxy) and the frequency at which the enhancer contacts its target promoter (for which we use Hi-C data as a proxy) (13). This metric correctly ranked 6 of the 7 distal enhancers as the most important of 93 DHS elements in K562 cells (Fig. 2E) and provided a reasonable ordering of their relative effects (Spearman correlation = 0.79). We note that this approach did not perfectly distinguish between enhancers that do and do not regulate MYC: NS1 was ranked 7 and e6 was ranked 11. Nonetheless, quantitative measures of chromatin state and chromosome conformation are strongly predictive of enhancers that regulate MYC in K562 cells.
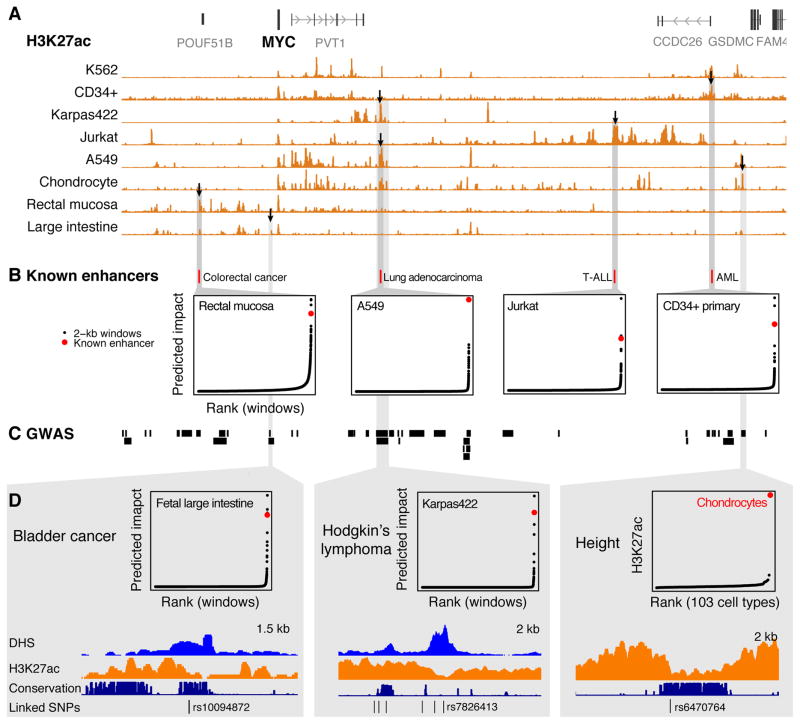
To determine whether this approach might be applicable in other cellular contexts, we examined 4 MYC enhancers identified in other cell types (Fig. 3A,B) (13). In each case our metric ranked these known elements among the 3 most important in the corresponding cell type (Fig. 3B). We also identified multiple instances where elements predicted to regulate MYC in one or more cell types harbor single nucleotide polymorphisms (SNPs) associated with human traits including cancer susceptibility and height (Fig. 3C,D, Table S1). Additional CRISPRi-based functional mapping in other cell types and gene loci might allow the derivation of general models to predict functional enhancer-promoter connections and help to understand noncoding genetic variation.
Fig. 3. A heuristic model predicts disease-associated MYC enhancers across cell types.
(A) H3K27ac occupancy around MYC varies among 8 cell types and primary tissues. Black arrows: elements highlighted in panels below. (B) Locations of 4 enhancers previously shown to regulate MYC expression in other cell types and their predicted impact in a corresponding cell type. Points show predicted impact of 2-kb windows tiled in 100-bp increments across the MYC locus (13). T-ALL: T-cell acute lymphoblastic leukemia. AML: Acute myeloid leukemia. For each cell type, predicted impact is calculated based on available data (13). (C) Haplotype blocks of SNPs linked to human diseases and phenotypes (R2 > 0.8 with index SNP in genome-wide association study). (D) SNPs associated with bladder cancer and Hodgkin’s lymphoma overlap regulatory elements predicted by our metric to regulate MYC in a corresponding cell type or tissue. A SNP associated with height overlaps a conserved element that is active only in chondrocytes. Karpas422: diffuse large B cell lymphoma cell line.
In summary, CRISPRi screens can accurately identify and characterize the regulatory functions and connectivity of noncoding elements. In the MYC and GATA1 loci, CRISPRi reveals complex and non-obvious dependencies between multiple genes and enhancers, including relationships that suggest regulation of multiple genes by the same enhancer, coordinated activity of multiple enhancers to control a single gene, and competition between neighboring promoters. Thus, learning the principles and connectivity of transcriptional networks requires dissecting putative regulatory elements in their native genomic contexts.
While we used cellular proliferation as a readout to investigate 2 essential genes, this CRISPRi approach can be applied to identify regulatory elements that control an arbitrary gene or phenotype of interest through alternative assays, for example by tagging an endogenous gene locus with green fluorescent protein (GFP) and sorting cells by GFP expression (23).
Together with complementary methods using catalytically active Cas9 (13, 23–25), CRISPRi-based functional mapping provides a broadly applicable approach (13) to dissect transcriptional networks and interpret the contributions of noncoding genetic variation in gene regulatory elements to human disease.
Supplementary Material
Acknowledgments
We thank T. Wang and R. Issner for technical advice and reagents; and R. Ryan, B. Bernstein, N. Sanjana, J. Wright, and F. Zhang for discussions. This work was supported by funds from the Broad Institute (E.S.L.). C.P.F. is supported by the National Defense Science and Engineering Graduate Fellowship. J.M.E. is supported by the Fannie and John Hertz Foundation. M.M. is supported by a DFG Research Fellowship. S.R.G. is supported by NIGMS T32GM007753. The Broad Institute, which E.S.L. directs, holds patents and has filed patent applications on technologies related to other aspects of CRISPR. Data presented in this paper can be found in the supplemental materials.
Footnotes
Author Contributions: J.M.E. conceived the study. J.M.E., C.P.F., M.M., and S.R.G. designed experiments. C.P.F., M.M., R.A., G.M., E.M.P., M.K., and J.M.E. performed experiments. C.P.F., J.M.E., and B.C. analyzed data. C.P.F., J.M.E., and E.S.L. wrote the manuscript with input from all authors.
Materials and Methods
Supplemental references 26–83:
References and Notes
- 1.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 3.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Project Consortium ENCODE. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao SSP, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 8.van Arensbergen J, van Steensel B, Bussemaker HJ. In search of the determinants of enhancer-promoter interaction specificity. 2014;24:695–702. doi: 10.1016/j.tcb.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kearns NA, et al. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thakore PI, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015 doi: 10.1038/nmeth.3630.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.See supplementary materials on Science online.
- 14.Suzuki M, Moriguchi T, Ohneda K, Yamamoto M. Differential contribution of the Gata1 gene hematopoietic enhancer to erythroid differentiation. Mol Cell Biol. 2009;29:1163–1175. doi: 10.1128/MCB.01572-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimura S, et al. A GATA box in the GATA-1 gene hematopoietic enhancer is a critical element in the network of GATA factors and sites that regulate this gene. Mol Cell Biol. 2000;20:713–723. doi: 10.1128/mcb.20.2.713-723.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi OR, Engel JD. Developmental regulation of beta-globin gene switching. Cell. 1988;55:17–26. doi: 10.1016/0092-8674(88)90005-0. [DOI] [PubMed] [Google Scholar]
- 17.Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujieda A, et al. A putative role for histone deacetylase in the differentiation of human erythroid cells. Int J Oncol. 2005;27:743–748. [PubMed] [Google Scholar]
- 19.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2015;14:219–219. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 20.Burdett T, et al. The NHGRI-EBI Catalog of published genome-wide association studies. doi: 10.1093/nar/gkw1133. (available at http://www.ebi.ac.uk/gwas) [DOI] [PMC free article] [PubMed]
- 21.Gombert WM, Krumm A. Targeted deletion of multiple CTCF-binding elements in the human C-MYC gene reveals a requirement for CTCF in C-MYC expression. PLoS One. 2009;4:e6109. doi: 10.1371/journal.pone.0006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2015;48:176–182. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajagopal N, et al. High-throughput mapping of regulatory DNA. Nat Biotechnol. 2016;34:167–174. doi: 10.1038/nbt.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015 doi: 10.1038/nature15521.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korkmaz G, et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol. 2016 doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.