Abstract
Tissue macrophages provide immune defense and contribute to establishment and maintenance of tissue homeostasis. Here we used constitutive and inducible mutagenesis to delete the nuclear transcription regulator methyl-CpG binding protein 2 (Mecp2) in defined tissue macrophages. Animals lacking the Rett syndrome-associated gene in macrophages did not show signs of neurodevelopmental disorder, but displayed spontaneous obesity, which could be linked to impaired brown adipose tissue (BAT) function. Specifically, mutagenesis of a BAT-resident CX3CR1+ macrophage subpopulation compromised homeostatic, though not acute cold-induced thermogenesis. Mechanistically, BAT malfunction of pre-obese mice harboring mutant macrophages was associated with decreased sympathetic innervation and local norepinephrine titers, resulting in reduced adipocyte expression of thermogenic factors. Mutant macrophages over-expressed PlexinA4, which might contribute to the phenotype by repulsion of Sema6A-expressing sympathetic axons. Collectively, we report a previously unappreciated homeostatic role of macrophages in the control of tissue innervation, disruption of which in BAT results in metabolic imbalance.
Macrophages are myeloid immune cells that are strategically positioned throughout the organism. As professional phagocytes, they ingest and degrade debris and foreign material, including pathogens, and orchestrate inflammatory processes 1. Macrophages can be generated from two sources: an early, currently considered transient hematogenic wave initiated in the yolk sac that seeds tissue macrophages prenatally, and a pathway involving hematopoietic stem cells and monocytes, that persists throughout adult life 2. Most macrophage compartments are established prenatally, and develop independently from each other in their respective host tissue under the influence of the local microenvironment. Studies revealed critical contributions of tissue macrophages to organ development and homeostasis 3. However, specific involvement of macrophages in the maintenance of the healthy balanced steady state remains for most tissues incompletely defined.
Here we probed the role of mutant macrophages in the development of Rett syndrome, a neurodevelopmental disorder characterized by developmental stagnation and regression in neurological function 4 caused by mutations in the gene encoding methyl-CpG binding protein 2 (Mecp2). Using two complementary conditional mutagenesis approaches we establish that a macrophage-restricted Mecp2 deficiency alone was insufficient to trigger the neurodevelopmental disorder. Instead, macrophage mutagenesis resulted in spontaneous obesity that could be linked to altered homeostatic energy expenditure and impaired brown adipose tissue (BAT) thermogenesis. Specifically, mice harboring a Mecp2 deficiency in a Cx3cr1+ BAT macrophage subset, displayed reduced expression of uncoupling protein 1 (Ucp1) in BAT that resulted from compromised sympathetic innervation. Finally, comparative translatome profiling of mutant and wild type BAT macrophages revealed a potential role of the Plexin-Semaphorin axis in macrophage-axon cross talk. Taken together, we report a novel homeostatic role of macrophages in the control of tissue innervation.
Results
Macrophage-restricted Mecp2 deficiency causes obesity
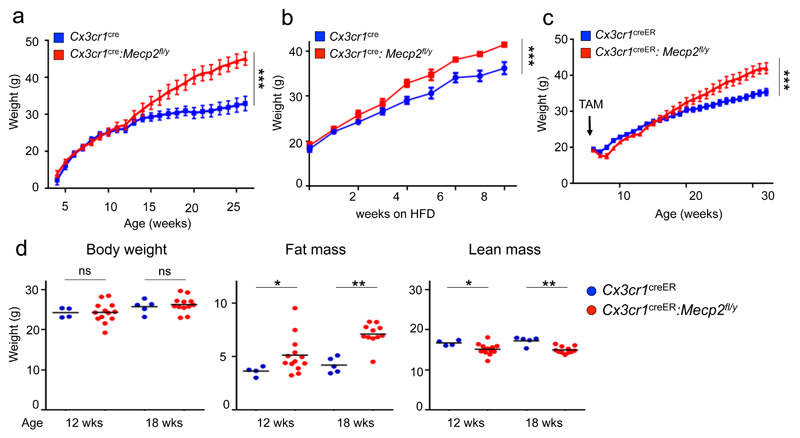
To probe for potential contributions of mutant macrophages in Rett syndrome 4 we generated mice bearing a largely macrophage-restricted mutation of Mecp2. Specifically, we crossed Cx3cr1cre animals 5 with mice that harbor a ‘floxed’ allele of the X chromosome-linked Mecp2 gene (Mecp2fl/y) 6 (Supplementary Fig. 1a). Deletion of the Mecp2 gene in microglia of Cx3cr1cre:Mecp2fl/y mice was confirmed by genomic PCR (Supplementary Fig. 1b). Unlike Mecp2fl animals crossed to ubiquitous or neuron-restricted Cre deleter strains 6, Cx3cr1cre:Mecp2fl/y mice did not exhibit overt Rett-like symptoms, such as impaired hind leg clasping or survival defects (Supplementary Fig. 1c, d), but were indistinguishable from co-housed Cx3cr1cre littermates until the age of 3-4 months. At that time, mutant mice unexpectedly started to gain weight and by the age of 6 months, all Cx3cr1cre:Mecp2fl/y mice were overtly obese, weighing ~30% more than controls (Fig. 1a, Supplementary Fig. 1e). Six month old obese Cx3cr1cre:Mecp2fl/y mice exhibited fatty and enlarged livers, as well as larger visceral, subcutaneous adipose tissue, but no signs of splenomegaly (Supplementary Fig. 1f, g). Histology of obese Cx3cr1cre:Mecp2fl/y animals revealed enlarged adipocytes and the formation of characteristic 'crown-like structure', as well as fat-laden hepatocytes (Supplementary Fig. 1h). The phenotype developed in mutant mice kept on standard chow, but high fat diet (HFD) given from the age of 6 weeks accelerated obesity (Fig. 1b).
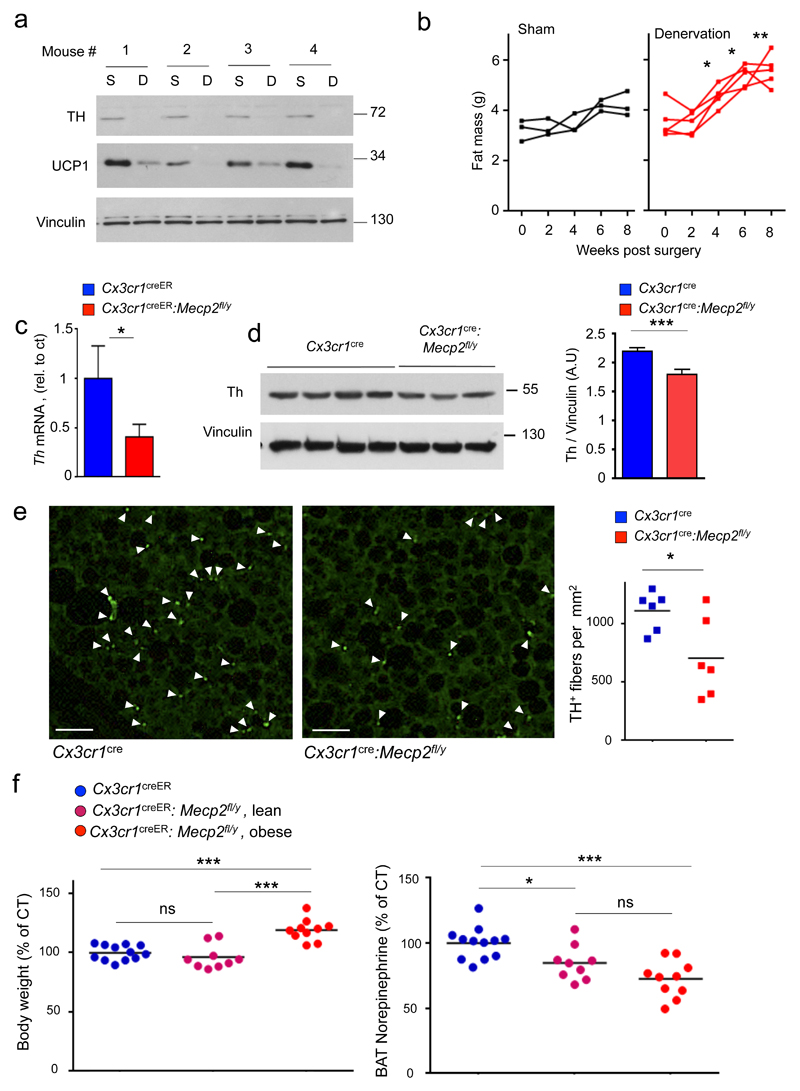
Fig. 1. Macrophage-restricted Mecp2 deletion causes spontaneous obesity in adulthood.
(a) Weight of Cx3cr1cre (n=9) and Cx3cr1cre:Mecp2fl/y mice (n=14) over 6 months. *** p<0.001, F20,360=22.24, two- way ANOVA, interaction effect.
(b) Weight of Cx3cr1cre (n=4) and Cx3cr1cre:Mecp2fl/y mice (n=7) kept on high-fat diet (HFD) for 8 weeks starting from 6 weeks of age. *** p<0.001, F8,72=5.871, two- way ANOVA, interaction effect.
(c) Weight of Cx3cr1creER:Mecp2fl/y (n=13) and Cx3cr1creER (n=14) mice over 6 months, all administered TAM at the age of 6 weeks. *** p<0.001, F26, 624=15.57, two- way ANOVA, interaction effect.
(d) Body composition of pre-obese Cx3cr1creER:Mecp2fl/y mice (n=13) and littermate controls (n=4) 6 weeks after TAM treatment (age 12 weeks), and Cx3cr1creER:Mecp2fl/y mice (n=12) and littermate controls (n=5) 12 weeks after TAM treatment (age 18 weeks). *p<0.05, **p<0.01, Mann-Whitney’s U-test.
In Cx3cr1cre mice the Cre transgene is expressed in macrophage precursors and thus results in mutagenesis of all tissue macrophages 5. To further restrict the mutation, we generated Cx3cr1creER:Mecp2fl/y animals. Rearrangements in this mouse strain require tamoxifen (TAM) treatment to induce nuclear translocation of the latent cytosolic CreER fusion protein and are restricted to cells expressing Cx3cr1 at the time of TAM application 5. Mecp2 was efficiently deleted in Cx3cr1+ macrophages of TAM-treated Cx3cr1creER:Mecp2fl/y mice and also these animals did not show signs of Rett syndrome, and consistent with a previous study7. When administered TAM at 6 weeks of age, only Cx3cr1creER:Mecp2fl/y mice, but not controls exhibited a significant weight gain by 6 months post TAM (Fig. 1c), similar to Cx3cr1cre:Mecp2fl/y mice. Control littermates were co-housed and TAM-treated to exclude potential confounding effects of microbiota 8 or the estrogen receptor (ER) modulator itself 9. The observed obesity is hence likely due to the Mecp2 deficiency in Cx3cr1+ macrophages. Moreover, as the metabolic alteration and obesity developed when the mutation was introduced in adulthood, the phenomenon can be dissociated from developmental impairment.
To investigate the cause of the obesity, we next focused on pre-obese animals, to minimize potential secondary effects. Indeed, TAM-treated, but not untreated Cx3cr1creER:Mecp2fl/y animals or TAM-treated Cx3cr1creER littermate controls displayed altered body compositions well before weight changes, as reflected in increased fat mass and decreased lean mass measured by Nuclear Magnetic Resonance (NMR) spectroscopy (Fig. 1d, Supplementary Fig. 2a).
Mecp2 deficiency causes brown adipose tissue impairment
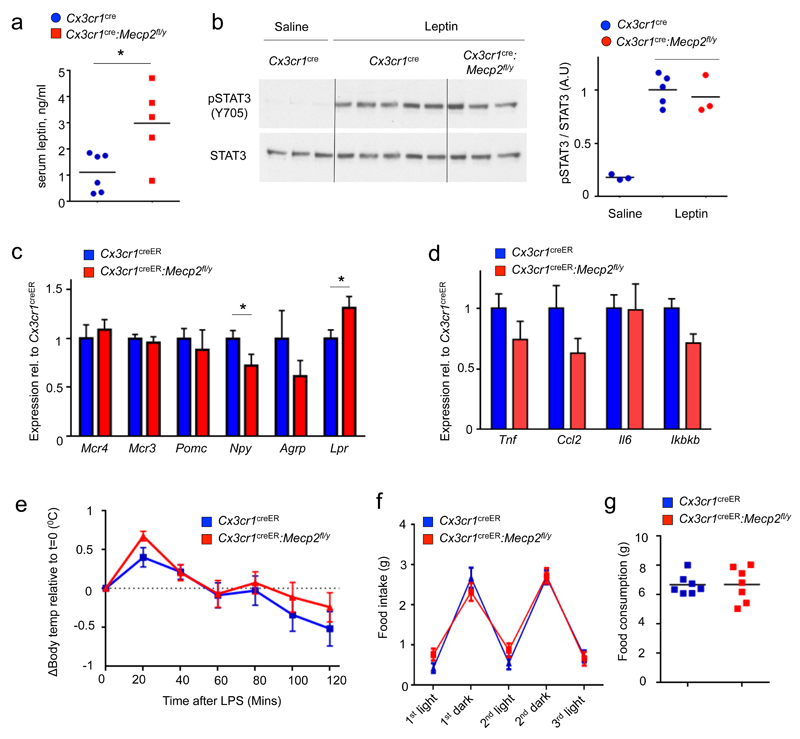
Energy balance is centrally regulated in the hypothalamus, where interactions between hypothalamic nuclei maintain homeostasis through regulation of food intake and energy expenditure 10. Pre-obese mutant mice exhibited higher serum leptin levels (Fig. 2a), likely resulting from enlarged fat mass. Specifically, leptin resistance could be ruled out since the hypothalamus of the mutant animals responded normally to challenge with this hormone, as indicated by Stat3 phosphorylation 11 (Fig. 2b). Also expression of hypothalamic central feeding regulators, such as Pomc and Agrp, was unchanged in pre-obese Mecp2fl/yCx3cr1-creER mice, whereas Npy and leptin receptor (Lpr) expression were down- and up-regulated, respectively (Fig. 2c). Hypothalamic expression of inflammatory genes, such as Tnf, Ccl2, Il6 and Ikbkb, which is elevated under high fat diet 12, was unaltered (Fig. 2d). Moreover, LPS-injected Cx3cr1creER:Mecp2fl/y developed a hypothalamus-controlled fever response resembling that of control littermates, including rapid hyperthermia followed by a prolonged hypothermia (Fig. 2e). Finally, Cx3cr1creER:Mecp2fl/y mice presented normal circadian oscillations in food intake and did not consume more diet compared to littermate controls (Fig. 2f, g). Collectively, these findings argue against a functional impairment of the hypothalamus or inflammation as causes for the observed metabolic disorder.
Fig. 2. Macrophage-restricted Mecp2 deletion does not impair central functions.
(a) Serum leptin levels in pre-obese starved Cx3cr1cre:Mecp2fl/y (n=5) and littermate controls (n=6), age 10 weeks. *p<0.05, Mann-Whitney’s U-Test.
(b) Western blot of phosphorylated and total Stat3 in hypothalami isolated from pre-obese starved Cx3cr1cre:Mecp2fl/y mice and littermate controls 45 minutes after leptin or saline administration.
(c) qRT-PCR analysis of hypothalami obtained from TAM-treated Cx3cr1creER and Cx3cr1creER:Mecp2fl/y mice (all n=6) for central feeding regulators. *p<0.05,Student's T test, for Npy T9=1.865, p=0.0476, for Lpr T11=2.111, p=0.0292.
(d) qRT-PCR analysis of hypothalami obtained from TAM-treated Cx3cr1creER and Cx3cr1creER:Mecp2fl/y mice for inflammation-associated genes (same mice as in Fig 2C).
(e) Body temperature differences (Body temperature(t)-body temperature(0)) of TAM-treated Cx3cr1creER (n=10) and Cx3cr1creER:Mecp2fl/y mice mice (n=8) after LPS treatment. Data are pooled from two independent experiments.
(f) Daily food consumption of TAM-treated Cx3cr1creER (n=8) and Cx3cr1creER:Mecp2fl/y (n=7) mice divided by dark and light phases.
(g) Accumulated food intake of 96 hours of (f)
Pre-obese Cx3cr1creER:Mecp2fl/y mice 3 months after TAM administration exhibited similar glucose and insulin tolerance, as indicated by intra-peritoneal glucose tolerance and insulin tolerance tests (GTT; ITT) (Supplementary Fig. 2b, c). Moreover, fasting insulin levels of mutants and controls were comparable; hence the observed obesity did not stem from primary hyper-insulinemia (Supplementary Fig. 2d). In contrast, once obese, Cx3cr1cre:Mecp2fl/y mice developed glucose intolerance and insulin resistance, indicating that secondary diabetes is a result of obesity but does not precede it (Supplementary Fig. 2e-f). The observed body composition alterations hence do not result from primary impairment in glucose metabolism.
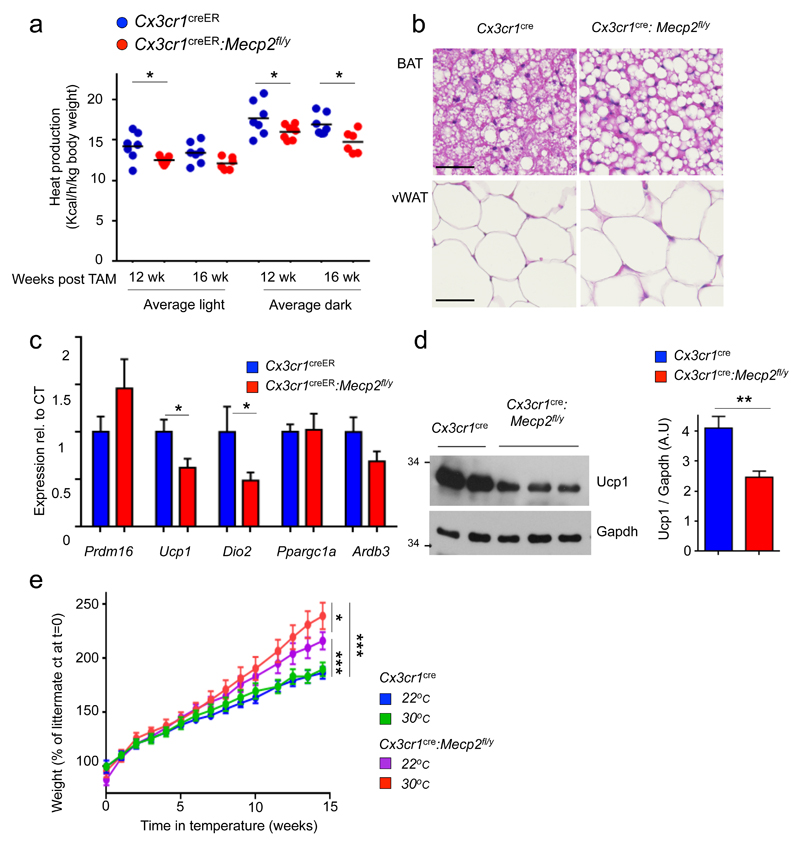
Metabolic cage analysis revealed unaltered respiratory exchange ratio (RER) in the mutants (Supplementary Fig. 3a, b). However, calculations by indirect calorimetric measurement showed that some groups of pre-obese TAM-treated Cx3cr1creER:Mecp2fl/y mice displayed significantly reduced heat production, as compared to controls. (Fig. 3a, Supplementary Fig. 3c). This suggested that the mutation affects energy expenditure.
Fig. 3. Macrophage-restricted Mecp2 deletion impairs BAT steady state function.
(a) Graphical summary of heat production of Cx3cr1creER mice (n=8) and Cx3cr1creER:Mecp2fl/y littermates (n=7) at 3 and 4 months after TAM, calculated by indirect calorimetry measurements. *p<0.05, 2-way ANNOVA followed by multiple comparisons, Taverage light after 3 months= 3.295, Taverage dark after 3 months= 3.194, Taverage dark after 4 months= 3.154, all df=22.
(b) H & E histology of iBAT and visceral WAT (vWAT) of Cx3cr1cre:Mecp2fl/y mice and control littermates at the age of 9 weeks. Pictures are representative of 3 independent mice with similar results. Scale bars, 40 μm.
(c) qRT-PCR analysis of BAT obtained from Cx3cr1creER:Mecp2fl/y mice (n=7) and littermates (n=8), 4 months after TAM treatment. Threshold Cycle (CT) for Ucp1- average for Cre mice 15.5 *p<0.05; for Ucp1 T11=2.419, p=0.017, for Dio2 T11=2.082, p=0.0297, Student’s T-test.
(d) Representative western blot analysis for Ucp1 and Gapdh in tissues obtained from 10 week old pre-obese Cx3cr1cre:Mecp2fl/y mice and littermates controls (left). Bar graph of densitometry analysis of Ucp1 normalized to Gapdh bands from three independent experiments Cx3cr1cre:Mecp2fl/y (n=10) mice and littermates controls (n=11) (right) **p=0.0028, T18=3.451, Student’s T-test.
(e) Weight gain of Cx3cr1cre:Mecp2fl/y (n=5, 5) mice and littermate controls (n=5, 6) under either 22°C or 30°C (thermoneutrality). Mice were examined starting from the age of 5 weeks onwards. *p<0.05, *** p<0.001, two-way ANOVA followed by Tuckey’s post-hoc test, Genotype effect F3,255<0.001.
The prime organ responsible for homeostatic non-shivering heat production in mammals is the brown adipose tissue (BAT) 13. BAT thermogenesis is critical for temperature balance of newborns and contributes in the adult to heat production following cold stress 14, as well as steady state energy expenditure termed 'diet-induced thermogenesis' 15. The impaired homeostatic energy expenditure we observed in mice harboring Mecp2-deficient macrophages might result from BAT impairment. Indeed, histological analysis of pre-obese Cx3cr1cre:Mecp2fl/y mice revealed alterations of their intrascapular BAT (iBAT), the main murine BAT depot 16. Specifically, brown adipocytes showed fat accumulation and large lipid droplets, whereas the morphology of white adipose tissue (WAT) appeared unaltered (Fig. 3b). Brown adipocyte development seemed unaffected in TAM-treated Cx3cr1creER:Mecp2fl/y mice, as indicated by unimpaired expression of Prdm16, a zinc finger protein required for brown adipocyte fate determination 17 (Fig. 3c).
BAT thermogenesis is linked to expression of the thermogenic factors Ucp1 18 and type II deiodinase (Dio2) 19. The transporter Ucp1 disrupts the mitochondrial proton gradient and thereby shunts energy generated by mitochondrial fatty acid (FA) oxidation from ATP formation to thermogenesis 13,14,20. Comparative analysis of iBAT isolated from TAM-treated pre-obese Cx3cr1creER:Mecp2fl/y and control mice revealed unaltered expression of peroxisome proliferator-activated receptor co-activator (Pgc-1α; Ppargc1a) that regulates energy metabolism 21 (Fig. 3c). Expression of the adrenergic β3 receptor was reduced in mutant iBAT, albeit without reaching significance. Macrophage mutagenesis resulted however in significantly reduced dio2 and ucp1 mRNA levels in iBAT (Fig. 3c), which was also reflected in Ucp1 protein decrease (Fig. 3d). Of note, and highlighting the role of Ucp1 in thermogenesis, Ucp1–/– mice are cold sensitive 22; however these animals only become obese when kept at thermo-neutrality (30°C) 15. Likewise, also Cx3cr1creER:Mecp2fl/y mice gained more weight, when housed at thermo-neutrality (Fig. 3e), supporting a role of Ucp1 in the phenotype. Collectively, these data suggest reduced Ucp1 expression in Cx3cr1cre- and Cx3cr1creER:Mecp2fl/y mice and thus altered energy expenditure, as the cause of obesity development in mutant animals.
Mutant mice respond to an acute cold challenge
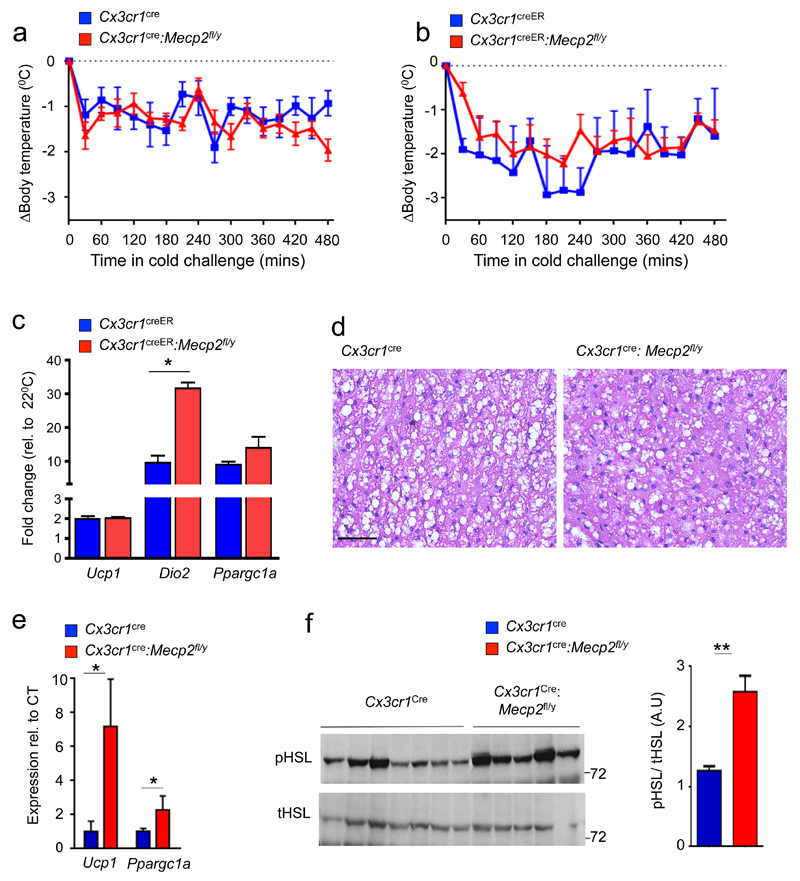
Following cold shock, body temperature is restored through shivering thermogenesis, BAT hyper-activation 14 and subcutaneous WAT (scWAT) 'beiging' 20. BAT responses to cold shock and WAT beiging have been reported to involve immune cells, including macrophages 23,24. To test, if the acute cold shock response of iBAT is affected in animals harboring MeCP2 null macrophages, we subjected pre-obese Cx3cr1cre:Mecp2fl/y and TAM-treated Cx3cr1creER:Mecp2fl/y mice to a temperature change (4°C, for 8 hours). Both strains were able to maintain their body temperature like control littermates (Fig. 4a, b). BAT of cold-challenged TAM-treated Cx3cr1creER:Mecp2fl/y and control mice displayed a significant induction of ucp1, ppargc1a and dio2 mRNA following the 8 hour cold exposure (Fig. 4c). Moreover, histological assessment showed depletion of fat stores from mutant iBAT (Fig. 4d). In scWAT of challenged Cx3cr1cre:Mecp2fl/y mice, Ucp1 and Ppargc1a mRNAs were found significantly increased compared to controls (Fig. 4e), albeit Ucp1 protein was not yet detected by this time point (data not shown). Hyper-activation of scWAT was also reflected in increased phosphorylation of hormone-sensitive lipase (Hsl), a key enzyme in fat mobilization (Fig. 4f). Increased lipolysis in scWAT of the mutant mice might indicate FA mobilization to the muscles to compensate for non-optimal BAT activity. Collectively, Cx3cr1cre:Mecp2fl/y mice display at facility room temperature (22°C) reduced Ucp1 and lower lipid combustion; however following acute cold challenge, iBAT of the mutant animals responded like that of littermate controls by inducing Ucp1 and Dio2 transcription and fat depot consumption.
Fig. 4. Normal acute cold acclimation and scWAT hyper activation in absence of macrophage Mecp2.
(a) Body temperature of pre-obese Cx3cr1cre:Mecp2fl/y (n=7) mice and control littermates (n=4) at 4°C for a period of 8h.
(b) Body temperature of pre-obese Cx3cr1creER:Mecp2fl/y mice and control littermates (all n=4) at 4°C for a period of 8h.
(c) qRT-PCR of iBAT taken from TAM-treated Cx3cr1creER:Mecp2fl/y and controls in (b) after 8 hours of cold challenge compared to animals housed at room temperature. *p<0.05,Mann-Whitney’s U test.
(d) Representative H&E stain of the iBAT of mice in (a). Scale bar, 50 μm.
(e) qRT-PCR of scWAT taken from Cx3cr1cre:Mecp2fl/y (n=7) mice and control littermates (n=5) at 4°C for a period of 8h. Threshold Cycle (CT) for ucp1 average for Cx3cr1cre:Mecp2fl/y mice 19.5 *p<0.05, Mann-Whitney’s U test.
(f) Western blot analysis for total and phosphorylated Hsl in scWAT of mice in (e). Bar graph of densitometry analysis of pHsl normalized to total Hsl ** p<0.01, Mann-Whitney’s U test. Data are representative of two independent experiments with similar results.
Macrophage Mecp2 deficiency causes impaired BAT innervation
BAT thermogenesis requires noradrenergic stimulation by the sympathetic autonomous nerve system 14,25,26. Specifically, tyrosine hydroxylase (Th)-expressing axons release norepinephrine (NE) that stimulates lipolysis and induction of the thermogenic factors in brown adipocytes. Animals deficient of beta adrenergic receptors display metabolic imbalance 27 and steady state thermogenesis is thought to require a tonic NE signal 28. Corroborating this notion, unilateral iBAT denervation in WT mice led to a reduction of Th in iBAT extracts, concomitant with a significant decrease of Ucp1, as compared to the sham treated control pad (Fig. 5a). Moreover as also shown earlier 29,30, following bilateral BAT denervation the ensuing impairment of BAT thermogenesis sufficed to alter body composition, as reflected in increased fat mass (Fig. 5b). We hence hypothesized that the obesity we observed in the mutant mice might result from impaired BAT innervation. Indeed, analysis of iBAT of pre-obese TAM-treated Cx3cr1creER:Mecp2fl/y mice revealed reduced axonal th transcription, as compared to littermate controls (Fig. 5c). Moreover, mutant iBAT displayed also a significant reduction of axonal Th protein (Fig. 5d). Histological staining for Th on iBAT of Cx3cr1cre:Mecp2fl/y mice revealed a significant reduction of sympathetic axons compared to control mice (Fig. 5e). To probe for a functional outcome of the reduced innervation, we performed Liquid chromatography-tandem mass spectrometry (LC-MS/MS) to directly measure local NE tissue levels. Analysis of iBAT of pre-obese Cx3cr1creER:Mecp2fl/y mice revealed a significant reduction (~20-30%) of the neurotransmitter, as compared to littermate controls (Fig. 5f, Supplementary Fig. 3d). Moreover, NE reduction was observed in TAM-treated Cx3cr1creER:Mecp2fl/y mice independent of their body mass alteration, suggesting that it precedes the latter. Collectively, these data establish that macrophage-restricted Mecp2 mutagenesis results in reduced sympathetic innervation of iBAT compromising thermogenesis, altering body composition and culminating in obesity.
Fig. 5. Impact of experimental BAT denervation and reduced BAT innervation in the absence of macrophage Mecp2.
(a) Western blot analysis of Th, Ucp1 and Vinculin in iBAT obtained from WT mice 10 days after unilateral iBAT denervation. From each mouse, both denervated BAT (designated D) and sham treated control BAT (designated S) are presented. n=4.
(b) Individual fat mass measurements of sham lesioned (n=3) and bilaterally denervated C57BL/6 mice (n=5), 0-8 weeks after surgery. *p<0.05, **p<0.01, 2 way ANNOVA followed by Bonferoni’s multiple comparison test, surgery effect F1,6=14.28.
(c) qRT-PCR for Th mRNA expression in whole BAT of Cx3cr1creER:Mecp2fl/y mice (n=7) and control littermates (n=8) 4 months after TAM administration. T11=1.904, p=0.0411.
(d) Western blot analysis for Th and Vinculin in tissues obtained from 10 week old pre-obese Cx3cr1creER:Mecp2fl/y mice (n=7) and control littermates (n=10). Bar graph of densitometry analysis of Th normalized to Vinculin bands. T15=4.175, ***p=0.008, Student’s T-test.
(e) Immunofluorescent stain for Th in BAT of Cx3cr1cre:Mecp2fl/ymice and control littermates (Each group n=6; 6 sections/ mouse; 36 sections/ genotype). Bar graph of iBAT Th+ fibers per mm2.*p=0.0259, T10=2.613, Student’s T test. Scale bar, 25μm.
(f) Body weight (left panel, control mean 28gr) and total BAT norepinephrine (right panel, control mean 19.5 ng) in age-matched, Cx3cr1creER (n=12), lean Cx3cr1creER:Mecp2fl/y (n=9) and obese Cx3cr1creER:Mecp2fl/y mice (n=10) 3 months after TAM., *p<0.05, ***p<0.001, one-way ANNOVA followed by Tukey’s multiple comparison test, for body weight F2,28=20.58, for BAT norepinephrine F2,28=11.99.
Characterization of BAT resident macrophage populations
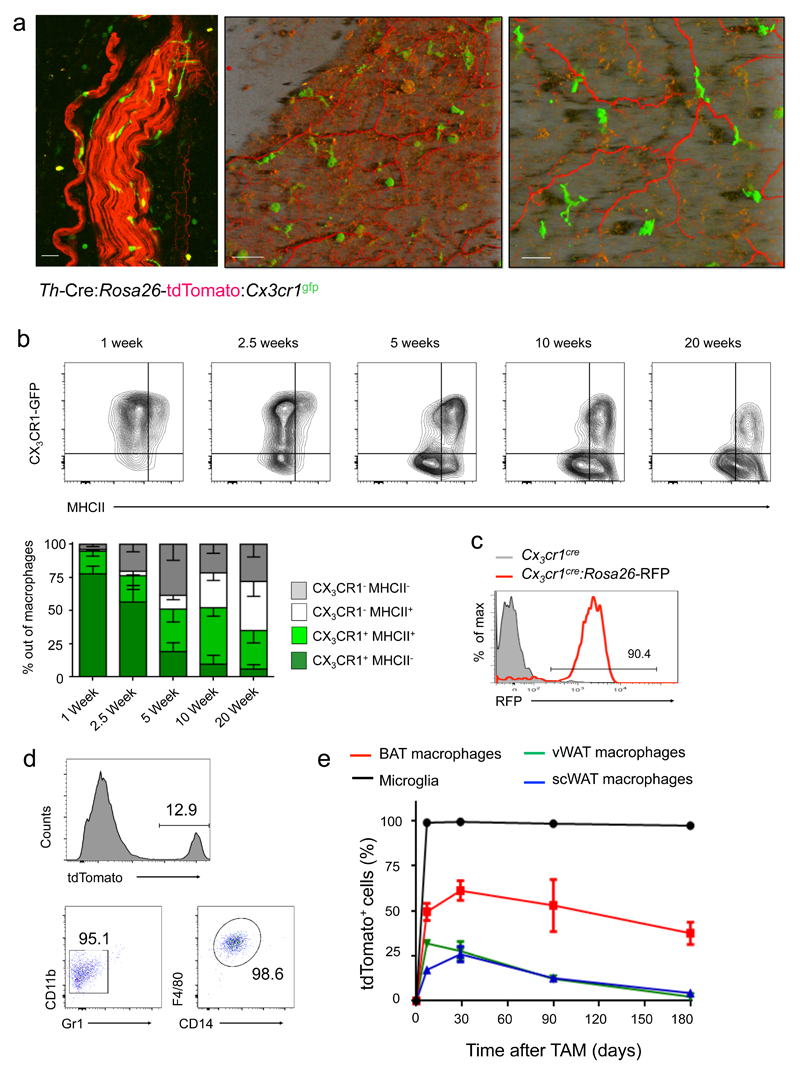
Two-photon microscopy analysis of iBAT of Th-Cre:Rosa26-tdTomato:Cx3cr1gfp mice revealed dense sympathetic innervation of this tissue and Cx3cr1+ macrophages, some of which were intimately associated with the axons (Fig. 6a). Flow cytometric analysis showed that iBAT harbors macrophages displaying the canonic surface marker signature 31, including CD11b, F4/80, CD14, CD64, MerTK and MHCII (Supplementary Fig. 4a). Analysis of Cx3cr1gfp reporter animals and staining for surface Cx3cr1 using a Cx3cl1-Fc fusion protein 32 revealed that iBAT macrophages comprise two subpopulations, as also confirmed by histological analysis (Supplementary Fig. 4b, c). Moreover, Cx3cr1+ macrophages represented the majority of Cx3cr1+ cells in iBAT (Supplementary Fig. 4d, e). Analysis of iBAT of mice at different ages showed early predominance of Cx3cr1+MHCII– cells, but BAT macrophages progressively diversified into four distinct subsets according to Cx3cr1 and MHCII expression; percentages of Cx3cr1+ cells dropped from >90% at the perinatal stage to 50% by 5 weeks of age concomitant with MHCII induction (Fig. 6b). Fate mapping with Cx3cr1cre:Rosa26-Rfp reporter animals established that Cx3cr1- iBAT macrophages derive from Cx3cr1+ precursors (Fig. 6c) 5. In TAM-treated Cx3cr1creER:Rosa26-tdTomato mice, the Cx3cr1+ population showed reporter gene induction (Fig. 6d), comprising half of the iBAT macrophages at time of TAM treatment (6 weeks) (Fig. 6e, Supplementary Fig. 5a). Persistence of this labeled macrophage subpopulation established its extended half-life (> 8 months) and independence from ongoing hematopoiesis and monocytes 5(Fig. 6e, Supplementary Fig. 5b). Cx3cr1+ iBAT macrophages mirror in this respect microglia 33; in contrast WAT harbored a less prominent Cx3cr1+ macrophage population that displayed higher turnover (Fig. 6e). Collectively, this establishes that both the constitutive Cx3cr1cre and the TAM-inducible Cx3cr1creER mutagenesis system, which requires Cx3cr1 expression and longevity 33, target a Cx3cr1+ iBAT macrophage subset.
Fig. 6. Characterization of brown adipose tissue macrophages.
(a) Two-photon microscopy of iBAT obtained from Th-Cre:Rosa26-tdTomato:Cx3cr1gfp mice. Original image size was: 424.27 µm, y: 424.27 µm, z: 23.69 µm; Z-Stack of 24 images was acquired; Shadow Projection mode (left and middle figures) and Maximum Intensity Projection (MIP) mode (right Fig.) of Zen Imaging Software were used in order to render a represented images of both tdTomato and GFP channels. Data are representative of 6 mice. Scale bars from left to right: 50 μm, 50 μm and 25 μm.
(b) FACS analysis of BAT macrophages obtained from Cx3cr1gfp mice at different ages for Cx3cr1/GFP and MHC II expression. n=4 for each age. Bar diagram summarizing data.
(c) FACS analysis of iBAT macrophages of Cx3cr1Cre:Rosa26-RFP mouse; cells are gated as in (b). Data are representative of three mice.
(d) FACS analysis of iBAT macrophages of TAM-treated Cx3cr1creER:Rosa26-tdTomato mouse, 6 months post TAM treatment. Data are representative of three mice.
(e) Fate mapping of indicated macrophage populations in TAM-treated Cx3cr1creER:Rosa26-tdTomato mice. n=3.
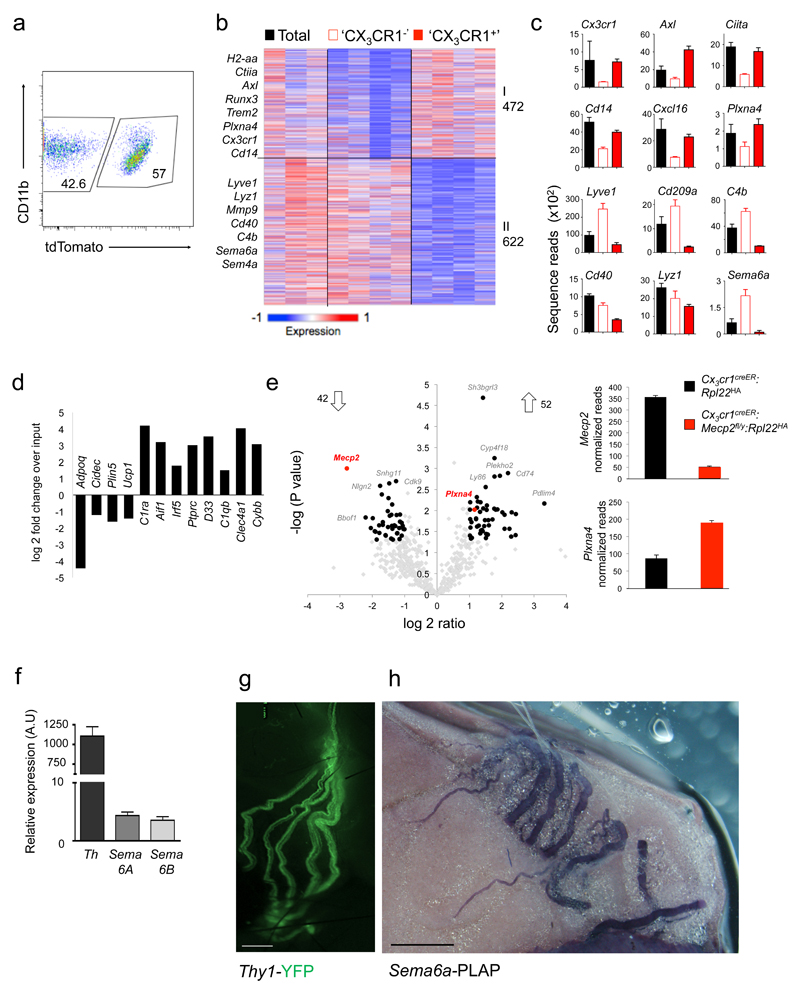
When integrated with reported tissue macrophage expression profiles 34, iBAT macrophages segregated within the transcriptomic macrophage landscape (Supplementary Fig 6a,b, Supplementary Table 1). To retrieve transcriptomes specific for the two BAT macrophage subsets we profiled tdTomato+ and tdTomato– cells obtained from TAM-treated Cx3cr1creER:Rosa26-tdTomato mice. These two macrophage populations - defined by presence or absence of Cx3cr1 expression at time of TAM treatment and termed 'CX3CR1+' and 'CX3CR1–' hereafter - displayed distinct transcriptomes (Fig. 7a, b). Specifically, 472 genes were found enriched in 'CX3CR1+' cells, including Cx3cr1, csf2rb2, Axl, H2-Aa and Plxna4 (Fig. 7b, c, Supplementary Table 2). 622 genes were enriched in 'CX3CR1–' macrophages, including genes encoding complement protein 4b, Lyve1, the C-type lectin CD209a and Semaphorin family members (Fig. 7b, c, Supplementary Table 3). Gene ontology analysis by the DAVID online software 35 highlighted enrichment of the 'MHCII-mediated antigen presentation pathway', 'TGF-β signaling' and 'positive regulation of cell-cell adhesion' in 'CX3CR1+' iBAT macrophages. Genes associated with 'complement activation', 'metal ion homeostasis' and 'chemokine mediated signaling pathways' were overrepresented in 'CX3CR1–' iBAT macrophages (all p<0.001). Collectively, iBAT hosts two macrophage populations, which display distinct expression profiles suggesting discrete functions.
Fig. 7. Mecp2-mutant macrophages overexpress PlexinA4 and might interact with Sema6A-expressing sympathetic axons in BAT.
(a) FACS plot indicating sorting gates for tdTomato+ and tdTomato– population, isolated from TAM-treated Cx3cr1creER:Rosa26-tdTomato mice. Data are representative of four mice.
(b) K-means clustering (K = 8) of 1094 genes differentially expressed by 'CX3CR1–' and 'CX3CR1+' BAT macrophages.
(c) Bar diagrams illustrating RNA expression of selected genes across macrophage populations presented in (B). ntotal=3, n’cx3cr1-‘=4, n’cx3cr1+’=4.
(d) Bar graph showing enrichment of macrophage genes and de-enrichment of adipocyte genes as determined by RNA-seq of ribosome-attached-mRNAs retrieved by IP of BAT from Cx3cr1CreER:Rpl22HA mice, representing fold change over input (mRNA of total tissue) (n=2)
(e) Volcano plot of statistical significance (-log10(p value)) against log 2 ratio between translatomes of 'CX3CR1' macrophages of Cx3cr1creER:Rpl22HA (n=2) and Cx3cr1creER:Mecp2fl/y:Rpl22HA mice (n=4), based on the RNA-seq data. Significantly changed genes (at least 2 fold change (1 log2 ratio), P value <0.05)) are indicated by red box (left). Bar diagrams illustrating raw normalized reads of Mecp2 and Plexna4 mRNA expression in Cx3cr1creER:Rpl22HA animals (black bars) and Cx3cr1creER:Mecp2fl/y:Rpl22HA animals (red bars) as determined by RNA-seq (right).
(f) RT-PCR analysis of stellate ganglia for Tyrosine hydroxylase and Semaphorin expression. n=3.
(g) Micrograph of main sympathetic neuronal fibers innervating the BAT, as visualized in Thy1-YFP mouse. Scale bar, 450 μm.
(h) Micrograph of BAT of Sema6a-PLAP (human placental alkaline phosphatase) mouse, illustrating Sema6a expression by sympathetic nerves according to PLAP reporter expression. Scale bar, 100 μm.
Mecp2 deficient macrophages dysregulate PlexinA4 expression
Cx3cr1CreER-mediated mutagenesis introduces mutations in the 'CX3CR1+' macrophage population and the mecp2 deficiency of these cells is likely causative for the observed BAT phenotype and metabolic alterations. Total BAT macrophage numbers of TAM-treated Cx3cr1creER:Mecp2fl/y mice were unaltered, as was the abundance of Cx3cr1+ iBAT macrophages in the mutants (Supplementary Fig. 7a, b). Moreover, iBAT of pre-obese mice did not show signs of alternative macrophage polarization or inflammation that have been reported for experimental BAT challenges 23 (Supplementary Fig. 7c). Likewise, neither mRNAs encoding iNOS or TNF, nor IL-1β protein levels, as measured by ELISA, were found increased in mutant iBAT, as compared to controls (Supplementary Fig. 7d, e). To directly investigate the impact of the mecp2 mutation on the expression profile of the 'CX3CR1+' iBAT macrophage population we resorted to a 'ribotag' approach 36. Specifically, we generated Cx3cr1creER:Mecp2fl/y:rpl22HA mice and Cx3cr1creER:Mecp2+/y:rpl22HA littermate controls that allow to retrieve after TAM treatment the translatomes of mutant and WT 'CX3CR1+' macrophages by immuno-precipitation of epitope-tagged ribosomes from crude iBAT extracts. RNAseq analysis of the ribosome-attached mRNA precipitated from Cx3cr1creER:rpl22HA iBAT extracts confirmed de-enrichment of adipocyte genes and enrichment of macrophage genes (Fig. 7d). In line with reported subtle effects of Mecp2 on gene expression 37, comparison of the translatomes retrieved from TAM-treated Cx3cr1creER:Mecp2fl/y:rpl22HA mice and controls revealed significant 2 fold up- and down-regulation of 52 and 42 genes, respectively (Fig. 7e, Supplementary Table 4). Mecp2 mRNA was reduced in mutant cells over WT controls, confirming the validity of the approach. Moreover, interestingly, Mecp2-deficient macrophages showed upregulation of plxna4 expression. PlexinA4 belongs to family of receptors that alone or in complex with neuropilin members, signal in response to guidance cues from semaphorins on multiple axonal tracks 38,39. In addition, PlexinA4 can act as ligand and trigger 'reverse signalling' through selected transmembrane semaphorins, an activity first reported in Drosophila, but now extended to mammalian systems 40. Specifically, Sema6a reverse signaling was shown to reduce axonal outgrowth of primary neurons in vitro in response to the Plexin ecto-domain 41. Given the reduced sympathetic innervation of mutant iBAT, we propose that PlexinA4 overexpression of Mecp2-deficient 'CX3CR1+' macrophages might inhibit axon outgrowth of Sema6a-positive nerves and BAT innervation. Indeed, stellate ganglion (SG) cells, which represent the major input to the iBAT pads 42 expressed both Sema6a and Sema6b, which was also shown to bind PlexinA4 43, as determined by RT-PCR (Fig. 7f). Each murine iBAT lobe displays postganglionic sympathetic innervation 26, as can be visualized in Thy1-YFP animals (Fig. 7g). Confirming Sema6a expression by these sympathetic nerves, they were found labeled in reporter animals that harbor a PLAP gene under control of the Sema6a promoter 44 (Fig. 7h). Collectively, these data establish that the Mecp2 mutagenesis results in increase of PlexinA4 expression that could act to repel Sema6a-expressing sympathetic axons in the iBAT and thereby reduce BAT innervation.
Discussion
Here we provide evidence for a unique role of macrophages in maintaining the sympathetic innervation of brown adipose fat tissue that is critical for balanced homeostatic energy expenditure in the adult. Specifically, we introduced a restricted genetic deficiency either in all tissue macrophages (due to their derivation from Cx3cr1+ precursors) (Cx3cr1Cre:Mecp2fl/y mice), or a macrophage subpopulation characterized by Cx3cr1 expression and longevity (TAM-treated Cx3cr1creER :Mecp2fl/y mice) 5,33. Mutant mice kept on normal chow developed spontaneous alterations of body composition and obesity. The phenotype could be linked to iBAT dysfunction, without significant alteration of hypothalamus performance. Rather, mutant iBAT macrophages that overexpress PlexinA4 seem to impede sympathetic innervation of the tissue and thereby impair the tonic local NE signal required for steady state Ucp1 expression and BAT thermogenesis.
Reports on functional contributions of adipose tissue macrophages have so far been restricted to conditions of disrupted homeostasis, such as HFD-induced obesity or cold challenge, that are associated with local inflammation and leukocyte recruitment 24,45,46. Moreover, most of these studies, that were also extended to other immune cells, have focused on 'beiging' WAT, rather than BAT. In contrast, the alterations we observe in Cx3cr1cre and TAM-treated Cx3cr1creER:Mecp2fl/y mice develop in animals kept on normal chow and in absence of signs of inflammation.
The metabolic changes that are associated with the genetic perturbation of the iBAT macrophage subset reveal a novel physiological role of these cells in controlling tissue innervation. Specifically, our data suggest that PlexinA4 overexpression by macrophages contributes to the repulsion of Sema6A-expressing sympathetic nerves. Of note, PlexinA4 is also expressed by WT Cx3cr1+ BAT macrophages, but not their Cx3cr1-negative counterpart. The latter cells do however express transmembrane semaphorins, suggesting a potential involvement of the Plexin-Semaphorin axis in macrophage crosstalk that requires further exploration. Moreover, steady state PlexinA4 expression by the BAT macrophage subset suggests that WT CX3CR1+ macrophages might locally impact on autonomous innervation, which is critical for BAT function. Indeed, also brown adipocytes have been reported to interfere with innervation by nerve growth factor production in relation to their thermogenic state 47. Brain macrophages, i.e. microglia, are known to communicate with neurons in the central nervous system, both in steady state and during pathology. Likewise, crosstalk of muscularis macrophages and enteric neurons was reported to regulate gastrointestinal motility 48. However, reports on interactions of macrophages and peripheral autonomic nervous system during homeostasis are scarce.
The 'methylation reader' Mecp2 has been suggested to promote chromatin compaction, gene activation and repression49. How the mecp2 mutation results in dysregulated plxna4 expression remains to be elucidated. Interestingly, Plexina4 expression was also reported to be de-repressed in Mecp2-deficient neurons 50, and, spanning 400 kb, plxna4 belongs to the group of long genes proposed to be preferentially targeted by Mecp2 50.
In iBAT of TAM-treated Cx3cr1creER:Mecp2fl/y mice, only the Cx3cr1+ macrophage subpopulation in the adult is mutated. Notably though, these animals develop obesity comparable to Cx3cr1cre:Mecp2fl/y mice suggesting that Cx3cr1– BAT macrophages cannot compensate for the deficiency. This fact highlights further functional specialization within the macrophage compartment of a single tissue, an issue that should be explored in the future.
Collectively, we establish a novel homeostatic function of a tissue macrophage population through controlling sympathetic organ innervation. Impaired BAT innervation disrupts the tonic NE signal required for normal BAT thermogenesis, thereby altering body composition and ultimately causing obesity. Our results point at a possible role of the Plexin-Semaphorin axis in the macrophage-neuron crosstalk. Future studies should investigate, whether macrophages also impact autonomous innervation and steady state functions of other organs. Finally, our data support the emerging concept that tissue macrophages comprise discrete subpopulations with distinct functional activities that are critical for organismal homeostasis.
Methods
Mice
Animals were maintained in a specific pathogen-free (SPF), temperature-controlled (22°C ± 1°C) mouse facility on a reverse 12-hour light, 12-hour dark cycle at the Weizmann Institute of Science. Food and water were given ad libitum. Mice were fed regular chow diet (Harlan Biotech Israel Ltd, Rehovot, Israel. The following mouse strains were used in this study: Cx3cr1gfp mice (JAX stock 005582 B6.129P-Cx3cr1tm1Litt/J 32; Cx3cr1cre (JAX stock 025524 B6J.B6N(Cg)-Cx3cr1tm1.1(cre)Jung/J) and Cx3crcCreER mice (JAX stock 020940 B6.129P2(C)-Cx3cr1tm2.1(cre/ERT2)Jung/J) 5; Rosa26-RFP (JAX stock 006067 129 Gt(ROSA)26Sor<tm2(CAG-Dsred2/EGFP)Luo>/J) 51; tdTomato reporter mice (JAX stock 007908 B6.129S6-Gt(ROSA)26Sor<tm14(CAG-tdTomato)Hze>/J) 52; Mecp2fl mice (JAX stock 007177 B6.129P2-Mecp2<tm1Bird>/J) 6; Th-Cre mice 53; Thy1-YFP 56; RiboTag mice (B6.129-Rpl22tm1.1Psam/J) 36; Sema6a-PLAP mice 44. For high-fat diet experiments, mice were fed with food pellets containing 60% Kcal fat (Research diet, USA). All mice studied were males on C57BL/6JOlaHsd background and handled under protocols approved by the Weizmann Institute Animal Care Committee (IACUC) in accordance with international guidelines.
Tamoxifen (TAM) administration
TAM (Sigma-Aldrich) was dissolved in 100% ethanol to a 1 g/ml solution and was 10-fold diluted in corn oil (Sigma-Aldrich) to a 100 mg/ml final solution for oral or s.c. administration. To induce Cre-mediated gene recombination in 5- to 7-wk-old cx3cr1CreER mice, 5 mg TAM was administered orally for 5 consecutive days (25 mg total) by gavage or subcutaneous injections.
Pyrogen challenge
To induce a fever response, mice were administered 2mg/kg of LPS (Sigma) i.p and monitored for two hours.55.
Glucose and insulin tolerance tests
Single-caged mice were food deprived for 4 hours prior to test. For GTT, mice were weighed and injected with 2g /kg D-Glucose (Sigma). For ITT, mice were weighed and injected with 0.75 units/kg of human recombinant insulin (Beit Ha’emek, Israel). For both tests, blood glucose levels were measured using the Accu-check performa glucometer (Roche Diagnostics) by tail bleeding 0, 15, 30, 60, 90 and 120 minutes after glucose or insulin injection.
Leptin administration
Overnight starved mice were housed individually prior to leptin stimulation, injected IP with leptin (4μg/g body weight) or saline for 45 min and then sacrificed by cervical dislocation. Hypothalami were dissected and snap frozen in liquid nitrogen before analysis by Western blot.
Metabolic studies
Indirect calorimetry and food and water intake, as well as locomotor activity were measured using the Labmaster system (TSE-Systems, Bad Homburg, Germany) 56. Data were collected after 48 hours of adaptation in acclimated singly housed mice. Body composition was assessed using the Bruker minispec mq7.5 Live Mice analyzer.
Cold challenge
Seven days before challenge, mice were anesthetized with isoflurane, and an implantable programmable temperature transponder 300 (iPTT 300; Bio medic data system, USA) was transplanted subcutaneously. At the day of the cold challenge, the mice were single-caged and transferred to a 4°C room. Mice were then monitored every 30 minutes for their body temperature changes using the SP-6005 data acquisition probe (Bio medic data system, USA) over a period of 8 hours. After the cold challenge the mice were sacrificed and tissues were collected.
Tissue denervation
Denervation was performed as previously described 57. Briefly, mice were anesthetized by injection of 10% ketamine/ 10% xylazine in PBS, 10μl per g body weight. After anesthesia, the skin above the interscapular region was removed and the two lobes of the iBAT were exposed under a binocular. For unilateral denervation, axon bundles innervating the left lobe were severed, while the right lobe was left intact.
Flow cytometry
For adipose tissues (BAT, vWAT and scWAT) the tissues were collected, shredded with scissors and then incubated for 30 minutes with DMEM medium (Beit Ha’emek, Israel) containing 1 mg/ml collagenase-2 (Sigma-Aldrich), 2% BSA (Sigma-Aldrich) and 12.5Mm HEPES buffer (Beit-Ha’emek, Israel). The resulting cell suspension was filtered through a 70 μm mesh and erythrocytes were removed by ACK lysis. Following cell suspension, cells were incubated in FACS buffer (PBS with 1%BSA, 2mM EDTA and 0.05% sodium azide) in the presence of staining antibody. For intracellular staining, the CytoFix/Cytoperm Kit (BD) was used according to the manufacturer's instruction. Cells were acquired on FACSCanto, LSRII, and LSRFortessa systems (BD) and analyzed with FlowJo software (Tree Star). For cell sorting, the FACSAria (BD) was used. For the Cx3cl1-Fc stain, cell suspensions were blocked with goat IgG for 10 mins, incubated with the Cx3cl1 -Fc fusion protein 32 for 20 mins followed by staining with a Cy5-goat anti human IgG was added, for additional 20 minutes. Antibodies used throughout the study are listed in Supplementary Table 5.
Histology
For hematoxylin and eosin (H & E), tissues were embedded in paraffin and serially sectioned. For immunofluorescence, tissues fixed in 4% paraformaldehyde overnight at 4°C, incubated with 30% sucrose for 48h and flash-frozen with isopentane before sectioning by cryostat. Samples were blocked with PBS containing 0.05% Tween (PBS-T), 0.3% triton and 20% normal horse serum (NHS) before incubation with PBS-T containing 0.3% triton, 2% NHS and primary antibodies in 4°C, followed by incubation with secondary antibody in PBS at RT for 1 hour and final stain with Hoechst (1:25,000). For immunohistochemistry in paraffin embedded sections: paraffin sections of brown adipose tissue were de-paraffinized and rehydrated by washes in 100% xylene, 100% ethanol, 95% ethanol, 70% ethanol, and then washed in phosphate-buffered saline (PBS). Heat-induced antigen retrieval of sections was carried out using citrate buffer pH 6.0 and then washed in PBS. Sections were then blocked for 1 hour in blocking buffer (20% Normal Horse Serum (NHS), 0.2% Triton, in PBS), then incubated with primary antibody diluted in antibody dilution buffer (2% NHS, 0.2% Triton in PBS) overnight at 23°C, followed by 24 more hours at 4°C. Sections were then incubated with Biotin anti rabbit antibody for 90 minutes at RT, washed, and incubated with streptavidin conjugated to Cy3 or Alexa Fluor 488 diluted in PBS for 1hour at RT. Finally sections were incubated with Hoechst for 2 minutes. Slides were then washed thoroughly and coverslips were mounted with aqueous glue. Sections were observed the following day by fluorescent microscopy. Anti-Th antibody used was identical to the one used in the western blot section.
PLAP staining
BAT was dissected into PBS solution enriched with 2mM MgCl2 followed by fixation in 4% PFA in PBS for 2h at 4°C. Tissue was then thoroughly rinsed with PBS and heated to 65°C for 2h. Tissue was rinsed x3 in alkaline buffer (100mM Tris-HCl pH 9.5, 100mM NaCl, 50mM MgCl2) and AP activity was detected by the formation of precipitants after an overnight incubation at room temp with nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate substrate (NBT/BCIP Roche #1168145100, 1:50 in alkaline buffer). Finally, tissue was rinsed in 20mM EDTA in PBS for 2-4 hours.
RiboTag approach
Samples were extracted from mice, flash frozen in liquid nitrogen and stored at -80C until use. Samples were homogenated on ice in ice-cold homogenization buffer (Tris 50mM pH 7.4, KCl 100mM, MgCl2 12mM, NP-40 1%, DTT 1mM, Protease inhibitor 1:100 (Sigma), RNasin 200 units/ml (Promega), cycloheximide 0.1mg/ml (Sigma) in RNase free DDW) 10% w/v with dounce homogenizer (Sigma) until suspension was homogeneous. To remove cell debris, 1ml of the homogenates was transferred to eppendorf tubes and centrifuged at 10,000g, 4°C for 10 minutes. Supernatant was transferred to a new eppendorf tube on ice, 50μl were taken for “Input” analysis, and 5μl of anti-HA (sigma) or 5μl of mouse IgG (sigma) were added to the sup, followed by a 4h incubation on slow rotation in cold room at 4°C. Meanwhile, Dynabeads® Protein G (Thermo Fisher Scientific), 100μl per sample, were equilibrated to homogenization buffer by washing 3 times. At the end of 4h incubation with antibody, beads were added to each sample and incubation was let overnight in cold room at 4°C. After not more than 12h, samples were washed with high salt buffer (Tris 50mM, KCl 300mM, MgCl2 12mM, NP-40 1%, DTT 1mM, Protease inhibitor 1:200, RNasin 100 units/ml, cycloheximide 0.1 mg/ml in RNase free DDW) 3 times, 5 min each wash, incubated in cold room on rotator. At the end of washes, beads were magnetized, excess buffer was removed, 350μl of RLT buffer was added to the beads and RNA was extracted with RNeasy Mini Kit (Qiagen). RNA was eluted in 12μl, concentration was read in Qubit 3.0 Fluorometer (Thermo Fisher Scientific), and sequenced.
RNA Isolation, Library Construction, and Analysis
RNA-Seq was performed as described previously 34. In brief, 103–105 cells from each population were sorted into 50μl of lysis/binding buffer (Life Technologies) and stored at -80°C. mRNA was captured with Dynabeads oligo(dT) (Life Technologies) according to manufacturer’s guidelines. We used a derivation of MARS-seq 58 to produce expression libraries with a minimum of two replicates per population. 4 million reads per library were sequenced and aligned to the mouse reference genome (NCBI 37, mm9) using HISAT v0.15 with default parameters. Expression levels were calculated and normalized for each sample to the total number of reads using HOMER software (http://homer.salk.edu) 59. RNA-seq analysis focused on genes in 25th percentile of expression with a 2-fold differential between at least two populations. The value of k for the K-means clustering (matlab function kmeans) was chosen by assessing the average silhouette (matlab function evalclusters; higher score means more cohesive clusters) for a range of possible values with correlation as the distance metric
Two-photon microscopy
iBAT was isolated from indicated animals and subjected to two-photon microscopy using an upright LSM 880 NLO combined confocal-multi-photon system, (Zeiss, Germany) with Chameleon Ultra tunable Ti:Sapphire laser, Coherent (USA) with Plan-Apochromat 20x/0.8 M27 lens. For excitation of tdTomato and GFP the laser was tuned to 900 nm and the emission filter set was 578 – 638 nm and 519 – 561 respectively. Image acquisition and further visualization was performed using ZEN Imaging software (Zeiss, Germany).
Norepinephrine measurement
iBAT lobes were homogenized by TissueLyser LT (Qiagen) in lysis buffer (0.16M acetic acid, 7mM phosphoric acid) containing 6ng/μl D6-norepinephrine (C.D.N Isotopes) as internal standard. Extracts were centrifuged for 15min 20,000g and the supernatants were collected. Derivatization was performed using Aqc reagent in 150mM borate buffer and derivatized samples were analyzed by Liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described in 60.
Genomic DNA extraction
Sorted cells were lysed and digested in TES buffer (10mM Tris buffer, pH=8, 5mM EDTA, 0.1 M NaCL, 0.5% SDS and 100 μg PK) overnight in 56°C. DNA was precipitated in 70% ethanol for 30 mins in RT, centrifuged twice at top speed and the tubes were left to dry. The pellet was reconstituted with TE buffer for subsequent PCR. Genomic PCR for mecp2 was performed using the following primers: TGGTAAAGA CCCATGTGACCCAAG, GGCTTGCCACATGACAAGAC, TCCACCTAG CCTGCCTGTACTTTG.
ELISA
IL-1β was measured using the DueSet kit (R&D). Leptin was measured using the Quantikine kit (Cystal Chem). Insulin was measured using the Ultra Mouse Insulin ELISA Kit (Crystal Chem). ELISA was performed according to the manufacturer’s instructions.
Quantitative real time PCR
Total RNA was extracted with RNeasy Mini Kit (QIAGEN). RNA was reverse transcribed with a mixture of random primers and oligo-dT with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). PCRs were performed with SYBR Green PCR Master Mix kit (Applied Biosystems). Real-time PCRs were carried out on a 7500 real-time PCR system using fluorescent SYBR Green technology (Applied Biosystems). Quantification of the PCR signals of each sample was performed by comparing the cycle threshold values (Ct), in duplicate, of the gene of interest with the Ct values of the GAPDH housekeeping gene. DNA primers used in the research are detailed in Supplementary Table 6.
Western blot
Whole BAT was extracted in 300ul Buffer A (25 mmol/l Tris-HCl [pH 7.4], 10 mmol/l sodium orthovanadate, 10 mmol/l sodium pyrophosphate, 100 mmol/l sodium fluoride, 10 mmol/l EDTA, 10 mmol/l EGTA, and 1 mmol/l phenylmethylsulfonyl fluoride) containing one metal bead using pre-cooled TissueLyser for 3min’s max strength. Then 1% Triton X was added to solution and further agitated for 2h at 4°C. Extracts were then centrifuged (20,000 g; 15min) and intermediate solution was transferred to a new tube. Protein concentration was measured by Bradford and extracts were supplemented with 1x Laemmli sample buffer and boiled for 5 min. Samples were then resolved by SDS-PAGE and Western blotted with the indicated antibodies.
Statistical analysis
Results are expressed as means ± SEM. Statistical analysis was performed using Student’s T test, Mann-Whitney’s U test, ANOVA or repeated-measurements 2-way ANOVA with post hoc Student's t tests or paired Student's t tests, as appropriate using the GraphPad Prism (San Diego, CA).
Data availability
The accession numbers for the RNA-seq datasets reported in this paper can be found at GEO: GSE95859. Additional data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgement
We would like to thank the members of the Jung lab for helpful discussion, R. Goldner for technical assistance, and A. Rudich (Ben Gurion University, Beer Sheva) and Martin Jastroch (Helmholtz Zentrum, Munich) for advice and comments on the manuscript. We thank G. Shakhar and R. Eilam for help with imaging and immunofluorescence, Z. Kam for writing an algorithm for the axon counting, and the Amit laboratory for advice concerning the profiling. The Jung laboratory was supported by the Israeli Science Foundation (ISF), the European Research Council (340345) and the University of Michigan – Israel Partnership for Research. N.C. was supported by a scholarship of the Lombroso fund. Y.K is supported by the Perlman Family Foundation, and is the incumbent of the Sarah and Rolando Uziel Research Associate Chair.
Footnotes
Author Contributions
Y.W., S. B.-H. and S.J. designed the study and wrote the manuscript; Y.W., S. B.-H. performed most of the experimental work; N. C. performed neuron quantification; Y. K, V, K., A.B. provided expert help and advice with metabolic, imaging and mass spectometry analysis; E. D. provided bioinformatic analysis; Z. H. established the RiboTag approach; Y. S.H. and L. C.-M. performed gene expression profiling; H. S. S. and A. Y. provided reagents and critical advice for the neuronal analysis.
Competing Financial Interests
The authors declare no competing financial interests.
Accession codes: The accession numbers for the RNA-seq datasets reported in this paper can be found at GEO: GSE95859.
References
- 1.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nature Publishing Group. 2014;14:392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- 3.Okabe Y, Medzhitov R. Tissue biology perspective on macrophages. Nat Immunol. 2016;17:9–17. doi: 10.1038/ni.3320. [DOI] [PubMed] [Google Scholar]
- 4.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 7.Schafer DP, et al. Microglia contribute to circuit defects in Mecp2 null mice independent of microglia-specific loss of Mecp2 expression. Elife. 2016;5:185. doi: 10.7554/eLife.15224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 9.Hesselbarth N, et al. Tamoxifen affects glucose and lipid metabolism parameters, causes browning of subcutaneous adipose tissue and transient body composition changes in C57BL/6NTac mice. Biochemical and Biophysical Research Communications. 2015;464:724–729. doi: 10.1016/j.bbrc.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 10.Horvath TL. The hardship of obesity: a soft-wired hypothalamus. Nat Neurosci. 2005;8:561–565. doi: 10.1038/nn1453. [DOI] [PubMed] [Google Scholar]
- 11.Rousso-Noori L, et al. Protein tyrosine phosphatase epsilon affects body weight by downregulating leptin signaling in a phosphorylation-dependent manner. Cell Metabolism. 2011;13:562–572. doi: 10.1016/j.cmet.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 12.Valdearcos M, et al. Microglia dictate the impact of saturated fat consumption on hypothalamic inflammation and neuronal function. CellReports. 2014;9:2124–2138. doi: 10.1016/j.celrep.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richard D, Picard F. Brown fat biology and thermogenesis. Front Biosci (Landmark Ed) 2011;16:1233–1260. doi: 10.2741/3786. [DOI] [PubMed] [Google Scholar]
- 14.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological Reviews. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 15.Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metabolism. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 16.de Jong JMA, Larsson O, Cannon B, Nedergaard J. A stringent validation of mouse adipose tissue identity markers. Am J Physiol Endocrinol Metab. 2015;308:E1085–105. doi: 10.1152/ajpendo.00023.2015. [DOI] [PubMed] [Google Scholar]
- 17.Seale P, et al. Transcriptional Control of Brown Fat Determination by PRDM16. Cell Metabolism. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chechi K, Carpentier AC, Richard D. Understanding the brown adipocyte as a contributor to energy homeostasis. Trends Endocrinol Metab. 2013;24:408–420. doi: 10.1016/j.tem.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 19.López M, Alvarez CV, Nogueiras R, Diéguez C. Energy balance regulation by thyroid hormones at central level. Trends Mol Med. 2013;19:418–427. doi: 10.1016/j.molmed.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 21.Liang H, Ward WF. PGC-1alpha: a key regulator of energy metabolism. Adv Physiol Educ. 2006;30:145–151. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 22.Enerbäck S, et al. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. 1997;387:90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen KD, et al. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature. 2011;480:104–108. doi: 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu Y, et al. Eosinophils and Type 2 Cytokine Signaling in Macrophages Orchestrate Development of Functional Beige Fat. Cell. 2014;157:1292–1308. doi: 10.1016/j.cell.2014.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryu V, Garretson JT, Liu Y, Vaughan CH, Bartness TJ. Brown Adipose Tissue Has Sympathetic-Sensory Feedback Circuits. Journal of Neuroscience. 2015;35:2181–2190. doi: 10.1523/JNEUROSCI.3306-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartness TJ, Vaughan CH, Song CK. Sympathetic and sensory innervation of brown adipose tissue. Int J Obes Relat Metab Disord. 2010;34:S36–S42. doi: 10.1038/ijo.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bachman ES, et al. betaAR signaling required for diet-induced thermogenesis and obesity resistance. Science. 2002;297:843–845. doi: 10.1126/science.1073160. [DOI] [PubMed] [Google Scholar]
- 28.Klingenspor M, Meywirth A, Stöhr S, Heldmaier G. Effect of unilateral surgical denervation of brown adipose tissue on uncoupling protein mRNA level and cytochrom-c-oxidase activity in the Djungarian hamster. J Comp Physiol B Biochem Syst Environ Physiol. 1994;163:664–670. doi: 10.1007/BF00369517. [DOI] [PubMed] [Google Scholar]
- 29.Bartness TJ, Wade GN. Effects of interscapular brown adipose tissue denervation on body weight and energy metabolism in ovariectomized and estradiol-treated rats. Behavioral Neuroscience. 1984;98:674–685. doi: 10.1037//0735-7044.98.4.674. [DOI] [PubMed] [Google Scholar]
- 30.Dulloo AG, Miller DS. Energy balance following sympathetic denervation of brown adipose tissue. Can J Physiol Pharmacol. 1984;62:235–240. doi: 10.1139/y84-035. [DOI] [PubMed] [Google Scholar]
- 31.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung S, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Molecular and Cellular Biology. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldmann T, et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nature Publishing Group. 2013;16:1618–1626. doi: 10.1038/nn.3531. [DOI] [PubMed] [Google Scholar]
- 34.Lavin Y, et al. Tissue-Resident Macrophage Enhancer Landscapes Are Shaped by the Local Microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yaron A, Huang P-H, Cheng H-J, Tessier-Lavigne M. Differential Requirement for Plexin-A3 and -A4 in Mediating Responses of Sensory and Sympathetic Neurons to Distinct Class 3 Semaphorins. Neuron. 2005;45:513–523. doi: 10.1016/j.neuron.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Suto F, et al. Interactions between plexin-A2, plexin-A4, and semaphorin 6A control lamina-restricted projection of hippocampal mossy fibers. Neuron. 2007;53:535–547. doi: 10.1016/j.neuron.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 40.Kruger RP, Aurandt J, Guan K-L. Semaphorins command cells to move. Nat Rev Mol Cell Biol. 2005;6:789–800. doi: 10.1038/nrm1740. [DOI] [PubMed] [Google Scholar]
- 41.Perez-Branguli F, et al. Reverse Signaling by Semaphorin-6A Regulates Cellular Aggregation and Neuronal Morphology. PLoS ONE. 2016;11:e0158686–24. doi: 10.1371/journal.pone.0158686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison SF, Madden CJ, Tupone D. Central neural regulation of brown adipose tissue thermogenesis and energy expenditure. Cell Metabolism. 2014;19:741–756. doi: 10.1016/j.cmet.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suto F, et al. Plexin-a4 mediates axon-repulsive activities of both secreted and transmembrane semaphorins and plays roles in nerve fiber guidance. J Neurosci. 2005;25:3628–3637. doi: 10.1523/JNEUROSCI.4480-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leighton PA, et al. Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature. 2001;410:174–179. doi: 10.1038/35065539. [DOI] [PubMed] [Google Scholar]
- 45.Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J Clin Invest. 2011;121:2111–2117. doi: 10.1172/JCI57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagareddy PR, et al. Adipose Tissue Macrophages Promote Myelopoiesis and Monocytosis in Obesity. Cell Metabolism. 2014;19:821–835. doi: 10.1016/j.cmet.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nisoli E, Tonello C, Benarese M, Liberini P, Carruba MO. Expression of nerve growth factor in brown adipose tissue: implications for thermogenesis and obesity. Endocrinology. 1996;137:495–503. doi: 10.1210/endo.137.2.8593794. [DOI] [PubMed] [Google Scholar]
- 48.Muller PA, et al. Crosstalk between Muscularis Macrophages and Enteric Neurons Regulates Gastrointestinal Motility. Cell. 2014;158:300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyst MJ, Bird A. Rett syndrome: a complex disorder with simple roots. Nat Rev Genet. 2015;16:261–275. doi: 10.1038/nrg3897. [DOI] [PubMed] [Google Scholar]
- 50.Gabel HW, et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522:89–93. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 52.Madisen L, et al. nn.2467. Nat Neurosci. 2009;13:133–140. [Google Scholar]
- 53.Lindeberg J, et al. Transgenic expression of Cre recombinase from the tyrosine hydroxylase locus. genesis. 2004;40:67–73. doi: 10.1002/gene.20065. [DOI] [PubMed] [Google Scholar]
- 54.Feng G, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 55.Shaked I, et al. MicroRNA-132 Potentiates Cholinergic Anti-Inflammatory Signalingby Targeting Acetylcholinesterase. Immunity. 2009;31:965–973. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 56.Greenman Y, et al. Postnatal ablation of POMC neurons induces an obese phenotype characterized by decreased food intake and enhanced anxiety-like behavior. Mol Endocrinol. 2013;27:1091–1102. doi: 10.1210/me.2012-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee Y-H, Petkova AP, Konkar AA, Granneman JG. Cellular origins of cold-induced brown adipocytes in adult mice. The FASEB Journal. 2015;29:286–299. doi: 10.1096/fj.14-263038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heinz S, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boughton BA, et al. Comprehensive profiling and quantitation of amine group containing metabolites. Anal Chem. 2011;83:7523–7530. doi: 10.1021/ac201610x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the RNA-seq datasets reported in this paper can be found at GEO: GSE95859. Additional data that support the findings of this study are available from the corresponding author upon request.