Abstract
Ankylosing spondylitis (AS) is a complex and chronic inflammatory disease with a high heritage. Previous study has shown that IL-1A and IL-1B involved in inflammatory reaction. But little is known about single nucleotide polymorphisms (SNPs) of IL-1A and IL-1B associated with AS. We conducted a case-control study among 267 AS cases and 297 healthy controls from China. In the genetic model analysis, we found the “T” genotype of rs3783550 was associated with decreased AS risk in the dominant model (p = 0.044) and log-additive model (p = 0.023); the “C” genotype of rs3783546 was significantly associated with decreased AS risk based in the dominant model (p = 0.044) and log-additive model (p = 0.023). Additionally, the minor allele “A” of rs2853550 may also reduce the risk of AS in dominant (p = 0.025) and log-additive model (p = 0.024). Our results suggested that the polymorphisms of IL-1A and IL-1B are associated with the AS susceptibility in the Chinese Han population. Further studies are needed to characterize the functional sequences that cause AS.
Keywords: ankylosing spondylitis (AS), single nucleotide polymorphism (SNP), IL-1A, IL-1B, case-control study
INTRODUCTION
Ankylosing spondylitis (AS) is a complex and chronic inflammatory disease with a high heritage. The overall prevalence of AS in Chinese is 0.2–0.54% [1]. The condition affects predominantly the axial skeleton, including the spine and sacroiliac joints, and causes pain, stiffness, and, eventually, bony ankylosis. Joints and tendon insertions (entheses) elsewhere are also commonly involved, and approximately one-third of patients develop acute anterior uveitis.
Although AS is also a complex multifactorial disease resulting from gene and environment interaction, the exact mechanism of AS is still unclear. As we all know, Genetic factors have an importance role in AS susceptibility and expression [2]. HLA–B27 is the major gene associated with the susceptibility of AS, which is present in > 95% of Caucasians of northern European ancestry, yet only 1%–5% of HLA-B27 carriers develop AS, so that HLA-B27 carriage alone does not explain the pattern of disease recurrence in families [3]. Therefore, we provide a hypothesis that HLA-B27 is almost essential but not adequate to cause AS and that other genes interact with HLA-B27 to cause the condition. Recent genome-wide screening identified non-MHC regions with linkage with AS [4, 5]. The interleukin-1 (IL-1) cluster on chromosome 2q has emerged as a robust non-MHC susceptibility locus for AS in several groups study [5–7]. Those studies, however, always have conflicting results in different populations, making it difficult to put forward a conclusion about the relationship between IL-1 gene cluster and AS. Due to the IL-1 gene cluster also play a prominent role in inflammation and host defense against infection. The prototypic members of the IL-1 family gene cluster are the genes IL-1A, IL-1B, and IL-1RN [5], which were related to AS, but IL-1A and IL-1B were less reported [4, 5, 8, 9]. IL-1A and -B encode proinflammatory cytokines involved in host defense against infection [5]. The genes polymorphisms have been related to a number of chronic inflammatory diseases such as Crohn's disease and inflammatory bowel disease (IBD) [10, 11]. Rs16944 in IL-1B was identified associated with AS in European ancestry [5], but was not replicated in another study in a Han Chinese population, and few information is found about other SNPs on IL-1A and L-1B associated with AS risk. Besides, recent report has showed that association particularly with haplotypes of single nucleotide polymorphisms (SNPs) in the IL1RN gene [4], suggesting that the true associated polymorphism lay on the associated haplotype but was not one of the individual markers genotype.
Thus, in consideration of these circumstances, we sought to test whether IL-1 genes were associated with AS in Chinese Han population, which is previously associated with AS in Europe population.
RESULTS
A total of 276 cases and 296 controls were enrolled in our study. The demographic characteristics of the study population are showed in Table 1, which showed the significant difference in age and gender distribution between the case and control groups (p < 0.001).
Table 1. Number of individuals for ankylosing spondylitis case and controls.
| Variables Case (n = 267) | Control (n = 297) | Total | P* |
|---|---|---|---|
| Sex < 0.001a | |||
| Female | 67 (25.1%) | 198 (66.7%) | 265 |
| Male | 200 (74.9%) | 99 (33.3%) | 299 |
| Age, yr (mean ± SD) | 31.58 ± 12.28 | 56.35 ± 9.72 | <0.001b |
aPearson’s test;
bWelch’s test.
Table 2 summarized the minor allelic frequency (MAF) of tested SNPs among the individuals in the case and control groups. After Hardy–Weinberg equilibrium (HWE) p value screening, we excluded one SNP (rs2853550). The allelic frequency of SNPs in the controls group was similar to those of the HapMap Asian population. Through the χ2 test, we found that no SNP was significantly associated with AS risk.
Table 2. Allele frequencies in cases and controls and odds ratio estimates for ankylosing spondylitis.
| SNP ID | Genes | Role | AllelsAa/B | HWE P-value | MAF | ORs | 95 % CI | Pb | |
|---|---|---|---|---|---|---|---|---|---|
| Case | Control | ||||||||
| rs3783550 | IL1A | Intron | T/G | 0.790 | 0.337 | 0.320 | 1.08 | 0.84–1.39 | 0.539 |
| rs3783546 | IL1A | Intron | C/G | 0.894 | 0.337 | 0.319 | 1.08 | 0.85–1.39 | 0.523 |
| rs2856838 | IL1A | Intron | A/G | 0.748 | 0.253 | 0.235 | 1.10 | 0.84–1.45 | 0.482 |
| rs1609682 | IL1A | Intron | T/G | 0.789 | 0.327 | 0.321 | 1.08 | 0.84–1.38 | 0.564 |
| rs3783521 | IL1A | Promoter | G/A | 0.790 | 0.306 | 0.320 | 1.08 | 0.84–1.39 | 0.539 |
| rs2853550 | IL1B | Downstream | A/G | 0.032# | 0.082 | 0.082 | 1.01 | 0.66–1.55 | 0.963 |
| rs1143643 | IL1B | Intron | C/T | 0.350 | 0.444 | 0.475 | 0.88 | 0.70–1.12 | 0.302 |
| rrs3136558 | IL1B | Intron | G/A | 1.000 | 0.380 | 0.380 | 1.00 | 0.79–1.27 | 0.998 |
| rs1143630 | IL1B | Intron | T/G | 0.654 | 0.180 | 0.153 | 1.21 | 0.88–1.66 | 0.231 |
| rs1143627 | IL1B | Promoter | G/A | 0.412 | 0.466 | 0.486 | 0.92 | 0.73–1.17 | 0.500 |
| rs16944 | IL1B | Promoter | A/G | 0.416 | 0.470 | 0.478 | 0.97 | 0.77–1.22 | 0.787 |
| rs1143623 | IL1B | Promoter | G/C | 0.398 | 0.399 | 0.408 | 0.96 | 0.76–1.22 | 0.762 |
HWE: Hardy-Weinberg equilibrium; MAF: Minor allelic frequency
OR: odds ratio; 95%CI: 95%confidence interval;
SNP: Single-nucleotide polymorphism;
aMinor allele;
bp ≤ 0.05 indicates statistical significance;
# HWE p ≤ 0.05 was excluded.
The minor allele of each SNP was assumed a risk allele compared to the wild-type allele. MAF in cases and controls are listed in Table 3. Five models (co-dominant model, over-dominant model, log-additive model additive model, dominant model and recessive model) were applied for analyzing the association between polymorphisms and AS, which was modified by the age of the subjects. We found that the risk allele “T” of rs3783550 was associated with a decreased risk of AS based on dominant model (OR = 0.56, 95% CI = 0.32–0.99, p = 0.044), and log-additive model (OR = 0.61, 95% CI = 0.39–0.94, p = 0.023). The minor allele “C” of rs3783546 was associated with a decreased AS risk under dominant model (OR = 0.56, 95% CI = 0.32–0.99, p = 0.044), and log-additive model (OR = 0.61, 95% CI = 0.39–0.94, p = 0.023). Additionally, the minor allele “A” of rs2853550 may also reduce the risk of AS, based on the dominant (OR = 0.36, 95% CI = 0.15–0.91, p = 0.025) and log-additive model (OR = 0.41, 95% CI = 0.18–0.91, p = 0.024). However, no statistically significant evidence suggested that the other polymorphisms tested were associated with AS risk.
Table 3. Logistic regression analysis of the association between the SNPs and ankylosing spondylitis disease (adjusted sex and age).
| SNP | Model | Genotype | Case | Control | OR (95% CI) | p* | AIC | BIC |
|---|---|---|---|---|---|---|---|---|
| rs3783550 | Codominant | G/G | 136 (45.8%) | 117 (43.8%) | 1.00 | 0.073 | 323.2 | 344.8 |
| G/T | 132 (44.4%) | 120 (44.9%) | 0.63 (0.34–1.14) | |||||
| T/T | 29 (9.8%) | 30 (11.2%) | 0.35 (0.13–0.99) | |||||
| Dominant | G/G | 136 (45.8%) | 117 (43.8%) | 1.00 | 0.044* | 322.3 | 339.7 | |
| G/T-T/T | 161 (54.2%) | 150 (56.2%) | 0.56 (0.32–0.99) | |||||
| Recessive | G/G-G/T | 268 (90.2%) | 237 (88.8%) | 1.00 | 0.090 | 323.5 | 340.8 | |
| T/T | 29 (9.8%) | 30 (11.2%) | 0.43 (0.16–1.17) | |||||
| Overdominant | G/G-T/T | 165 (55.6%) | 147 (55.1%) | 1.00 | 0.300 | 325.3 | 342.7 | |
| G/T | 132 (44.4%) | 120 (44.9%) | 0.74 (0.41–1.32) | |||||
| Log-additive — | — | — | 0.61 (0.39–0.94) | 0.023* | 321.2 | 338.5 | ||
| rs3783546 | Codominant | G/G | 136 (46%) | 117 (43.8%) | 1.00 | 0.074 | 323.1 | 344.8 |
| G/C | 131 (44.3%) | 120 (44.9%) | 0.63 (0.34–1.14) | |||||
| C/C | 29 (9.8%) | 30 (11.2%) | 0.35 (0.13–0.99) | |||||
| Dominant | G/G | 136 (46%) | 117 (43.8%) | 1.00 | 0.044* | 322.3 | 339.6 | |
| G/C-C/C | 160 (54%) | 150 (56.2%) | 0.56 (0.32–0.99) | |||||
| Recessive | G/G-G/C | 267 (90.2%) | 237 (88.8%) | 1.00 | 0.090 | 323.5 | 340.8 | |
| C/C | 29 (9.8%) | 30 (11.2%) | 0.43 (0.16–1.17) | |||||
| Overdominant | G/G-C/C | 165 (55.7%) | 147 (55.1%) | 1.00 | 0.310 | 325.3 | 342.6 | |
| G/C | 131 (44.3%) | 120 (44.9%) | 0.74 (0.41–1.32) | |||||
| Log-additive — | — | — | 0.61 (0.39–0.94) | 0.023* | 321.2 | 338.5 | ||
| rs2853550 | Codominant | G/G | 251 (85.4%) | 227 (85%) | 1.00 | 0.076 | 322 | 343.6 |
| A/G | 38 (12.9%) | 36 (13.5%) | 0.38 (0.15–1.00) | |||||
| A/A | 5 (1.7%) | 4 (1.5%) | 0.22 (0.02–3.31) | |||||
| Dominant | G/G | 251 (85.4%) | 227 (85%) | 1.00 | 0.025* | 320.1 | 337.4 | |
| A/G-G/G | 43 (14.6%) | 40 (15%) | 0.36 (0.15–0.91) | |||||
| Recessive | G/G-A/G | 289 (98.3%) | 263 (98.5%) | 1.00 | 0.290 | 324 | 341.3 | |
| A/A | 5 (1.7%) | 4 (1.5%) | 0.26 (0.02–3.61) | |||||
| Overdominant | G/G-A/A | 256 (87.1%) | 231 (86.5%) | 1.00 | 0.050 | 321.3 | 338.6 | |
| A/G | 38 (12.9%) | 36 (13.5%) | 0.40 (0.15–1.02) | |||||
| Log-additive — | — | — | 0.41 (0.18–0.91) | 0.024* | 320 | 337.4 | ||
AIC: Akaike's Information criterion;
BIC: Bayesian Information criterion;
*p value ≤ 0.05 indicates statistical signifcance.
Two blocks were detected in studied IL-1A and IL-1B SNPs by haplotype analyses (Figure 1). The results of the association between the IL-1A and IL-1B haplotype andthe risk of AS were listed in Table 4. Haplotype “TCG” of IL-1A in Block were found to be associated with decrease risk of AS (OR = 0.36, 95% CI = 0.16–0.81 p = 0.014).
Figure 1. LD analysis of the association between all the SNPs of the IL-1A and IL-1B gene and ankylosing spondylitis.
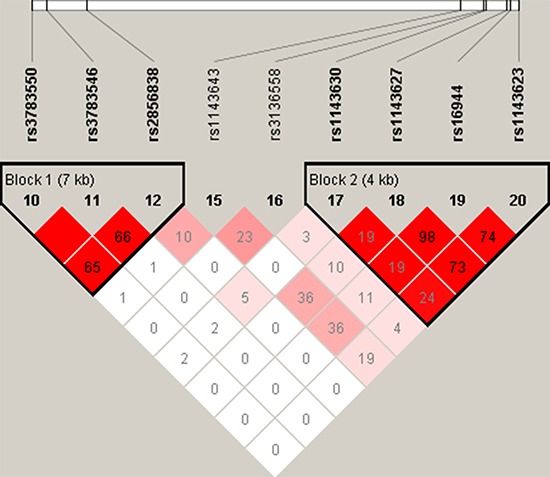
Table 4. IL-1A and IL-1B haplotype frequencies and the association with the risk of ankylosing spondylitis in case and control patients (adjusted sex and age).
| Gene | SNPs | Haplotype | Freq (case) | Freq(control) | OR(95% CI) | P-value* |
|---|---|---|---|---|---|---|
| IL-1A | rs3783550/rs3783546/rs2856838 | GGG | 0.662 | 0.680 | 1(referent) | — |
| TCA | 0.252 | 0.237 | 0.72(0.44–1.18) | 0.19 | ||
| TCG | 0.086 | 0.083 | 0.36(0.16–0.81) | 0.014* | ||
| IL-1B | rs1143630/rs1143627/rs16944/rs1143623 | CAGC | 0.530 | 0.517 | 1(referent) | — |
| GGAG | 0.223 | 0.255 | 1.08(0.68–1.73) | 0.74 | ||
| TGAG | 0.178 | 0.151 | 0.89(0.52–1.52) | 0.67 | ||
| GGAC | 0.066 | 0.070 | 0.68(0.30–1.56) | 0.37 |
*p value ≤ 0.05 indicates statistical signifcance.
DISCUSSION
In this Chinese Han population-based case–control study, we investigated the associations between the 12 SNPs and risk of AS. We found the associations between IL-1A and IL-1B SNPs and the risk of AS, rs3783550, rs3783546 and rs2853550 showed a protective effect on AS risk.
The haplotype “TCG” of IL-1A were significance associated with a decreased risk of AS, which can act as a protect factor for AS. Although the individual polymorphism investigated showed no association with AS adjusted by age and sex, it is contribute to our hypothesis that substantially stronger haplotype associations with SNPs of IL-1A than with individual SNPs.
IL-1A and IL-1B are one of the family of IL-1 and are proinflammatory cytokine that are mainly produced by activated macrophages, whose actions include augmentation of activation of T and B lymphocytes and monocyte/macrophages, and induce fibroblast proliferation leading to synovial pannus formation [12], which also produce IL-1α and IL-1β, respectively [13, 14]. The genes for the proinflammatory cytokines IL-1α and IL-1β are located on the long arm of human chromosome 2, in close linkage with another gene of the IL-1 gene family, the gene encoding their endogenous antagonist IL-lRa [15]. Randomized clinical trials in rheumatoid arthritis with recombinant human IL-1ra demonstrated modest anti-inflammatory effects and significant slowing in the rate of joint damage [16]. This may suggest that in certain patients the lack of production of IL-1ra, the natural antagonist of IL-1β, contributes to the chronicity of the inflammation. Furthermore, IL1A and IL-1B may induce inflammation in AS through intracellular effects [17], it is also possible that IL1A and IL-1B are involved in the initiation of disease, but has a less significant role in established disease.
A previous study found that rs3783550 was significantly associated with reduced AS risk in Canadian populations [8]. Rs3783546 and rs2853550 were associated with erythematosus [18] and rheumatoid arthritis [19], respectively. So that we speculated that rs3783550, rs3783546 and rs2853550 play an important role in chronic inflammatory disease, but three SNPs were not significantly associated with AS risk, only were associated with AS in different model in our study. That result might be due to differences among different regions of the world, which needs us to do further research to verify the results.
Despite this study showed three SNPs (rs3783550, rs3783546 andrs2853550) associated with AS susceptibility in the different model, some limitations should be considered. First, as the incidence of ankylosing spondylitis is low, so that the sample size of our study was relatively small. There may be some false positive results, but it has been able to explain the trend of IL-1A and IL-1B in patients with ankylosing spondylitis. In the future we will recruit more samples included in our future studies to verify our results. Second, the heterogeneity in living conditions was not evaluated in this study, which has contributed to the progress in elucidating the pathogenesis of AS.
In conclusion, our study has described the association between rs3783550 (IL-1A), rs3783546 (IL-1A) and rs2853550 (IL-1B) and AS risk and between a new haplotype, “TCG”, of rs3783550, rs3783546 and rs2853550 and AS in Chinese Han population. Therefore, the results provide additional evidence for the relationships between genetic variants and AS susceptibility in the Hainan population. Moreover, a large number of genetic risk factors for AS should be investigated and validated to fully understand the pathogenesis of AS. Further studies will focus on the functional experiments based on the relevant genes on animal models, to investigate detailed mechanism involved.
MATERIALS AND METHODS
Study participants
We recruited a total of 267 patients diagnosed with AS and 297 controls from 2014 to 2016 among Han Chinese. All the subjects were treated by the Second Affiliated Hospital of Inner Mongolia Medical University. All participants signed informed written consent. All cases were verified, and patients were recruited without age, or disease stage restriction and who were previously healthy. AS was defined by the modified New York diagnostic criteria [20]. In all cases the diagnosis of AS was confirmed by a qualified rheumatologist and was followed by either examination or telephone interview. In cases with an atypical history or no previous radiographic evidence, pelvic and lumbosacral spine radiographs were obtained and attending general practitioners were contacted to confirm diagnosis. All cases were HLA-B27 positive. To reduce the potential environmental and therapeutic factors impacting the variation of complex human diseases, we performed detailed recruitment and set exclusion criteria to exclude subjects with chronic disease and conditions involving vitalorgans (brain, liver, heart, and lung). Controls were selected unrelated and randomly. Among control individuals, none had any chronic diseases. The use of samples was approved by the Human Research Committee of the Second Affiliated Hospital of Inner Mongolia Medical University for Approval of Research Involving Human Subjects. The personal characteristics of the participants are listed in Table 1.
SNP selection and genotyping
Among the 12 SNPs we selected, rs16944 in IL-1B [5] was chosen from previously published polymorphisms associated with AS, others were randomly chosen from the published genes (IL-1A and IL-1B) associated with AS. Minor allele frequencies of all SNPs were > 5%, in the HapMap of the Chinese Han CHB population. We adopted Gold Mag-Mini Whole Blood Genomic DNA Purification Kit (GoldMagCo. Ltd. Xi’an City, China) to extract DNA that was from whole blood. Quantification of the extracted DNA was performed using NanoDrop2000. Genotyping was done with the Sequenom MassARRAY RS1000 system using the standard protocol recommended by the manufacturer. The multiplexed SNP Mass EXTENDED assay was designed using Sequenom Mass-ARRAY Assay Design 3.0 Software [21]. Data management and analysis was done using SequenomTyper 4.0 Software. We use the Sequenom MassARRAY RS1000 system to analysis genotype, which was conform to the standard protocol recommended by the manufacturer.
Statistical analysis
We used Microsoft Excel and SPSS 18.0 (SPSS, Chicago, IL, USA) to perform statistical analyses. In this study, all p values were two-sided, and p ≤ 0.05 was regarded as getting statistical significance. Control genotype frequencies for each SNP were tested for departure from Hardy-Weinberg equilibrium (HWE) using Fisher's exact test. The χ2 test was used to compare the distribution of marker alleles and genotypes in cases and controls [22]. Unconditional logistic regression analysis adjustment with age was used to test odds ratios (ORs) and 95% confidence intervals (CIs) [23]. Associations between SNPs and risks of ASwere tested in genetic models by analysis with SNP Stats software, obtained from http://bioinfo.iconcologia.net. Values of OR and 95% CI were calculated as above.Akaike's InformationCriterion (AIC) and Bayesian InformationCriterion (BIC) were applied to choose the best-fit model for each SNP. Finally, we used the SHEsis software (http://analysis.bio-x.cn/myAnalysis.php) to estimate the pairwise linkage disequilibrium (LD) and haplotype construction [24].
Footnotes
CONFLICTS OF INTEREST
All Authors declare that they have no conflicts of interest.
REFERENCES
- 1.Myrand S, Sekiguchi K, Man M, Lin X, Tzeng R, Teng C, Hee B, Garrett M, Kikkawa H, Lin C. Pharmacokinetics/genotype associations for major cytochrome P450 enzymes in native and first-and third-generation Japanese populations: comparison with Korean, Chinese, and Caucasian populations. Clin Pharmacol Ther. 2008;84:347–361. doi: 10.1038/sj.clpt.6100482. [DOI] [PubMed] [Google Scholar]
- 2.Tsui FW, Tsui HW, Akram A, Haroon N, Inman RD. The genetic basis of ankylosing spondylitis: new insights into disease pathogenesis. The application of clinical genetics. 2014;7:105–115. doi: 10.2147/TACG.S37325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown MA, Edwards S, Hoyle E, Campbell S, Laval S, Daly AK, Pile KD, Calin A, Ebringer A, Weeks DE, Wordsworth BP. Polymorphisms of the CYP2D6 gene increase susceptibility to ankylosing spondylitis. Hum Mol genet. 2000;9:1563–1566. doi: 10.1093/hmg/9.11.1563. [DOI] [PubMed] [Google Scholar]
- 4.Maksymowych WP, Reeve JP, Reveille JD, Akey JM, Buenviaje H, O’Brien L, Peloso PM, Thomson GT, Jin L, Russell AS. High-throughput single-nucleotide polymorphism analysis of the IL1RN locus in patients with ankylosing spondylitis by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. Arthritis Rheumatol. 2003;48:2011–2018. doi: 10.1002/art.11037. [DOI] [PubMed] [Google Scholar]
- 5.Timms AE, Crane AM, Sims AM, Cordell HJ, Bradbury LA, Abbott A, Coyne MR, Beynon O, Herzberg I, Duff GW, Calin A, Cardon LR, Wordsworth BP, et al. The interleukin 1 gene cluster contains a major susceptibility locus for ankylosing spondylitis. Am J Hum Genet. 2004;75:587–595. doi: 10.1086/424695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGarry F, Neilly J, Anderson N, Sturrock R, Field M. A polymorphism within the interleukin 1 receptor antagonist (IL-1Ra) gene is associated with ankylosing spondylitis. Rheumatology. 2001;40:1359–1364. doi: 10.1093/rheumatology/40.12.1359. [DOI] [PubMed] [Google Scholar]
- 7.van der Paardt M, Crusius JB, Garcia-Gonzalez MA, Baudoin P, Kostense PJ, Alizadeh BZ, Dijkmans BA, Pena AS, van der Horst-Bruinsma IE. Interleukin-1beta and interleukin-1 receptor antagonist gene polymorphisms in ankylosing spondylitis. Rheumatology. 2002;41:1419–1423. doi: 10.1093/rheumatology/41.12.1419. [DOI] [PubMed] [Google Scholar]
- 8.Maksymowych WP, Rahman P, Reeve JP, Gladman DD, Peddle L, Inman RD. Association of the IL1 gene cluster with susceptibility to ankylosing spondylitis: an analysis of three Canadian populations. Arthritis Rheumatol. 2006;54:974–985. doi: 10.1002/art.21642. [DOI] [PubMed] [Google Scholar]
- 9.Sims AM, Timms AE, Bruges-Armas J, Burgos-Vargas R, Chou CT, Doan T, Dowling A, Fialho RN, Gergely P, Gladman DD, Inman R, Kauppi M, Kaarela K, et al. Prospective meta-analysis of interleukin 1 gene complex polymorphisms confirms associations with ankylosing spondylitis. Ann rheum dis. 2008;67:1305–1309. doi: 10.1136/ard.2007.081364. [DOI] [PubMed] [Google Scholar]
- 10.van der Pouw Kraan CT, Baggen JM, van Bodegraven AA, Mulder CJ, Zwiers A, Geerts D, Kraal G, Horrevoets AJ, Bouma G. Defective IL-1A expression in patients with Crohn’s disease is related to attenuated MAP3K4 signaling. Hum immunol. 2012;73:912–919. doi: 10.1016/j.humimm.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Nemetz A, Toth M, Garcia-Gonzalez MA, Zagoni T, Feher J, Pena AS, Tulassay Z. Allelic variation at the interleukin 1beta gene is associated with decreased bone mass in patients with inflammatory bowel diseases. Gut. 2001;49:644–649. doi: 10.1136/gut.49.5.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson K, Hashimoto S, Lotz M, Pritzker K, Terkeltaub R. Interleukin-1 induces pro-mineralizing activity of cartilage tissue transglutaminase and factor XIIIa. Am J Pathol. 2001;159:149–163. doi: 10.1016/S0002-9440(10)61682-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bioque G, Crusius JB, Koutroubakis I, Bouma G, Kostense PJ, Meuwissen SG, Pena AS. Allelic polymorphism in IL-1 beta and IL-1 receptor antagonist (IL-1Ra) genes in inflammatory bowel disease. Clin Exp Immunol. 1995;102:379–383. doi: 10.1111/j.1365-2249.1995.tb03793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N, Lanyon G, Martin M, Fraumeni JF, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 15.Nicklin MJ, Weith A, Duff GW. A physical map of the region encompassing the human interleukin-1 alpha, interleukin-1 beta, and interleukin-1 receptor antagonist genes. Genomics. 1994;19:382–384. doi: 10.1006/geno.1994.1076. [DOI] [PubMed] [Google Scholar]
- 16.Bresnihan B. Treatment of rheumatoid arthritis with interleukin 1 receptor antagonist. Ann Rheum Dis. 1999;58:I96–98. doi: 10.1136/ard.58.2008.i96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werman A, Werman-Venkert R, White R, Lee JK, Werman B, Krelin Y, Voronov E, Dinarello CA, Apte RN. The precursor form of IL-1alpha is an intracrine proinflammatory activator of transcription. P Natl Acad Sci USA. 2004;101:2434–2439. doi: 10.1073/pnas.0308705101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solus JF, Chung CP, Oeser A, Li C, Rho YH, Bradley KM, Kawai VK, Smith JR, Stein CM. Genetics of serum concentration of IL-6 and TNFalpha in systemic lupus erythematosus and rheumatoid arthritis: a candidate gene analysis. Clin Rheumatol. 2015;34:1375–1382. doi: 10.1007/s10067-015-2881-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaidya B, Pearce SH, Charlton S, Marshall N, Rowan AD, Griffiths ID, Kendall-Taylor P, Cawston TE, Young-Min S. An association between the CTLA4 exon 1 polymorphism and early rheumatoid arthritis with autoimmune endocrinopathies. Rheumatology. 2002;41:180–183. doi: 10.1093/rheumatology/41.2.180. [DOI] [PubMed] [Google Scholar]
- 20.van der Linden S, Valkenburg HA, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheumatol. 1984;27:361–368. doi: 10.1002/art.1780270401. [DOI] [PubMed] [Google Scholar]
- 21.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Current protocols in human genetics/editorial board, Jonathan L Haines et al. 2009. p. 12. Chapter 2: Unit 2. [DOI] [PubMed]
- 22.Adamec C. [Example of the Use of the Nonparametric Test. Test X2 for Comparison of 2 Independent Examples] Ceskoslovenske zdravotnictvi. 1964;12:613–619. [PubMed] [Google Scholar]
- 23.Bland JM, Altman DG. Statistics notes. The odds ratio. Bmj. 2000;320:1468. doi: 10.1136/bmj.320.7247.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]


