Abstract
Background
Rotator cuff disease is a common disorder leading to shoulder pain and loss of function. Its etiology in atraumatic cases is uncertain and likely extends beyond repetitive micro-trauma or overuse. Our objective was to determine whether there is a genetic or familial predisposition to rotator cuff disease.
Methods
A literature search of PubMed and EMBASE databases identified 251 citations. After reviewing the titles, abstracts, and full articles, seven met our inclusion/exclusion criteria.
Results
Four studies assessed familial predisposition to rotator cuff disease. One of these demonstrated that siblings of an individual with a rotator cuff tear were more likely to develop a full-thickness tear and more likely to be symptomatic. A five-year follow-up showed that the relative risks were increased for the siblings to have a full-thickness tear, for a tear to progress in size, and for being symptomatic. Another study demonstrated that a significantly higher number of individuals with tears had family members with a history of tears or surgery than those without tears. The other three studies investigated whether a genetic predisposition to rotator cuff disease exists and found significant association of haplotypes in DEFB1, FGFR1, FGFR3, ESRRB, and FGF10, and two single nucleotide polymorphisms within SAP30BP and SASH1.
Conclusion
Prior studies provide preliminary evidence for genetic and familial predisposition to rotator cuff disease. However, there is a lack of large genome-wide studies that can provide more definitive information and guide early detection of individuals at risk, prophylactic rehabilitation, and potential gene therapies and regenerative medicine interventions.
Level of Evidence
Systematic Review
Keywords: Rotator cuff disease, rotator cuff tears, tendinopathy, genetic predisposition, familial predisposition, epidemiology
Tendon disorders account for over 30% of all musculoskeletal office visits1. Rotator cuff disease is a common disorder and affects 30–50% of the population older than 50 years of age17. It includes a spectrum of pathology ranging from tendinopathy to partial or complete tears19. Rotator cuff disease is associated with shoulder pain and loss of function28. There were an estimated 272,148 ambulatory surgeries performed for rotator cuff tears in the United States in 20069.
The cause of atraumatic rotator cuff tears has only been studied by a limited number of investigators and remains unknown. The pathophysiology of rotator cuff tearing is described as intrinsic defects of tendons, including increased tendon cell death, a higher proportion of fat composition, aberrant microstructure of structural fibers, and abnormal nutrient vessels4, 14. This suggests that atraumatic rotator cuff tears are not purely due to repetitive micro-trauma or overuse. It is possible that the biological changes are regulated by genes. Identifying genes associated with rotator cuff disease and rotator cuff tears can help early recognition of individuals at higher risk of developing this pathology. This could warrant application of primary or secondary prevention strategies for this specific population.
The purpose of this study was to perform a systematic review on the genetic and familial predisposition to rotator cuff disease.
Materials and Methods
The term rotator cuff disease is used loosely in the literature. This term can encompass disorders ranging from impingement to tendinopathy to rotator cuff tearing. The transition from rotator cuff tendinopathy to rotator cuff tear was described as a continuum by Neer19. Hence, in our study we used the umbrella term rotator cuff disease and included studies on impingement syndrome and rotator cuff tendinopathy/tear.
A systematic literature search on familial or genetic predisposition to rotator cuff disease of PubMed and EMBASE databases was performed from their years of inception through March 2016. The database search was performed with the help of a trained librarian, and the keywords used included “rotator cuff disease,” “genetics,” “polymorphism,” and “family.” The full search criteria can be found in the appendix (Appendix A). Initially, 251 citations were identified, and two of the authors (C.G. and N.B.J.) independently reviewed the titles and abstracts for relevance. The full texts of 17 of the citations were then reviewed, and 10 studies were found not to be relevant to our topic. Bibliographies of full text articles that met our inclusion criteria were also reviewed for additional articles. No additional articles were gained from the bibliography search.
The studies included in this review were assessed with the Methodological Index for Non-Randomized Studies (MINORS) and were scored accordingly25. The maximum possible score was 24. When required, authors of included articles were contacted for additional information. We used the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) methodology for reporting our manuscript13.
Results
The initial literature search produced 251 articles which were assessed for relevance by their title and abstract. Of these, 234 were excluded due to lack of relevance to our topic. After the remaining 17 full texts were reviewed, seven studies were found to meet the inclusion criteria and were thus included in our final analysis (Figure 1).
Figure 1.
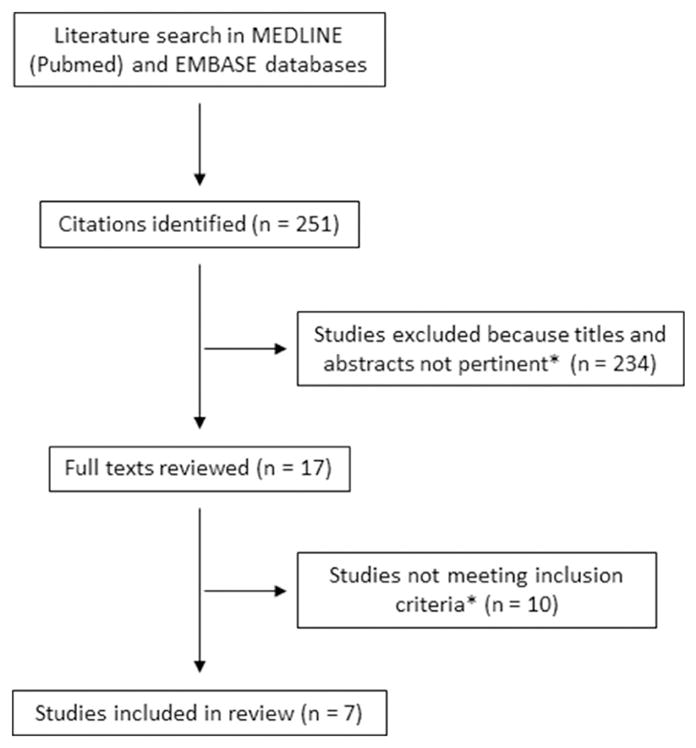
PRISMA diagram of the literature search and study selection
*Inclusion criteria: studies on familial predisposition or genetic epidemiology of rotator cuff disease
Four studies investigated whether there is a familial predisposition to rotator cuff disease. One of these studies (n = 129) demonstrated that siblings of an individual with a rotator cuff tear were twice as likely to develop a full-thickness tear and nearly five times more likely to suffer symptoms, when compared to spouses of these individuals (Table I)7. A five-year follow-up (n = 62) showed that the relative risk for the siblings to have a full-thickness tear was 2.85 (95% CI 1.75–4.64) compared to the control population, the relative risk for a tear to progress in size was 2.08 (95% CI 1.58–2.7), and the relative risk of having a symptomatic tear was 1.44 (95% CI 2.04–8.28) (Table I)6. Tashjian et al’s 2009 study (n = 3,091) used the Genealogical Index of Familiality to demonstrate a significant excess relatedness when all generations were used but not when only looking at more distant relationships (Table I)29. When only individuals diagnosed before age 40 (n = 652) were studied, significant excess relatedness was found when both all generations and only more distant relationships were used (Table I). Close relationships were defined as those between first- and second-degree relatives, while distant relationships were those with a genetic path length of at least three. Excess relatedness was used interchangeably with excess familial clustering or heritable predisposition. Tashjian et al’s 2014 study (n = 92) demonstrated that a significantly higher number of individuals with rotator cuff tears (32.3%) also had family members with a history of rotator cuff tears or surgery when compared to individuals without rotator cuff tears (18.3%) (Table I)31.
Table I.
Studies on familial predisposition to rotator cuff disease
| Study | Methods | Participants | Group Definitions | Results | Source of Funding | Notes |
|---|---|---|---|---|---|---|
| Harvie et al.6 (2004) | Case-control | Cases: n = 129; mean age 63.1 (41 to 85) Controls: n = 150; mean age 62.4 (43 to 85) |
Case: Included siblings of individuals diagnosed with a rotator cuff tear by ultrasound. Excluded those who were not available for review, were not full first-degree relatives of individuals with rotator cuff tears, or had concomitant systemic disease affecting the function of the shoulder. Control: Included spouses of individuals diagnosed with a rotator cuff tear by ultrasound. |
Relative to Controls, Cases participants had more than twice the risk of developing full-thickness tears (p < 0.0001), and nearly five times the risk of suffering symptoms (p < 0.0001). No significant differences were found in other measures between Cases and Controls. |
Girdlestone Memorial Scholarship in Orthopaedic Surgery from The Lord Nuffield Orthopaedic Centre Trust | |
| Gwilym et al.5 (2009) | Case-control | Cases: n = 62; mean age 66.6 (46 to 88) Controls: n = 68; mean age 66.1 (52 to 82) |
Case: Included siblings of individuals diagnosed with a rotator cuff tear by ultrasound. Excluded individuals who had shoulder surgery or had a systemic disease affecting shoulder function. Control: Included spouses of individuals diagnosed with a rotator cuff tear by ultrasound. Excluded individuals who had shoulder surgery or had a systemic disease affecting shoulder function. |
69.2% of Cases had full-thickness tears compared to 22.1% of Controls (p = 0.0001). The relative risk estimate for Cases to have a full-thickness tear was 2.85 (95% CI 1.75–4.64, p = 0.0001). The relative risk progression in Cases compared to Controls was 2.08 (95% CI 1.58–2.7) (p = 0.007). The relative risk of pain associated with a full-thickness tear in Cases compared to Controls was 1.44 (95% CI 2.04–8.28) |
None stated. | 5-year follow-up to Harvie et al.6 (2004) with a loss of follow up of about half the original cohort |
| Tashjian et al.24 (2009) | Population-based case-control | Cases: n = 3,091 Case Subgroup: n = 652 Controls: n = 15,455 |
Case: Included individuals who went to the University of Utah Hospital and Clinics, had an ICD-9 diagnosis code or CPT-4 procedure code, and had at least three generations of genealogical data. Case Subgroup: Included individuals in Case group who were diagnosed before age 40. Control: Included individuals who were in the Utah Population Database and had genealogical data. |
The overall Genealogical Index of Familiality shows a significant excess relatedness for Cases (p < 0.001), but the distance tests shows that excess relatedness observed is not significant when close relationships are ignored (p = 0.848). First degree relatives of Cases had p < 0.0001, relative risk of 2.44, and 95% CI 2.06–2.89. Second degree relatives had p = 0.0177, relative risk of 1.24, and 95% CI 1.04–1.48. Third degree relatives had p = 0.2866, relative risk of 1.08, and 95% CI 0.94–1.24. For Case Subgroup participants, both the overall (p = 0.001) and distance Genealogical Index of Familiality (p = 0.004) tests show significant excess relatedness. First degree relatives of Case Subgroup participants had p = 0.2614, relative risk of 1.73, and 95% CI 0.69–4.37. Second degree relatives had p = 0.0076, relative risk of 3.66, and 95% CI 1.47–9.11. Third degree relatives had p = 0.0479, relative risk of 1.81, and 95% CI 1.05–3.11. |
National Institutes of Health-National Library of Medicine University of Utah Huntsman Cancer Institute |
|
| Tashjian et al.26 (2014) | Case-control | Cases: n = 92; mean age 58.24 +/− 7.4 Controls: n = 92; mean age 58.42 +/− 8.5 |
Case: Included individuals who had magnetic resonance imaging (MRI)-confirmed, symptomatic, full-thickness rotator cuff tears. Control: Included individuals who were over age 18 and had no shoulder pain or prior shoulder injury or surgery. |
32.3% of Cases reported having family members with a history of rotator cuff tears or surgery, compared with 18.3% of Controls (p = 0.035). 22.8% of Cases reported having family members with tendon problems or surgery, compared with 17.5% of Controls (p = 0.407). 38.7% of Cases reported having a history of other tendon problems, compared with 19.3% of Controls (p = 0.005). 18.3% of Cases reported having other prior tendon surgeries, compared with 13.6% of Controls (p = 0.605). |
None stated. | Control matching between group participants was based on age to within five years |
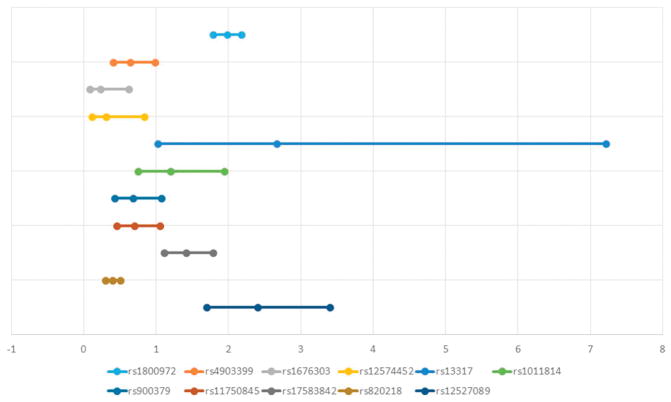
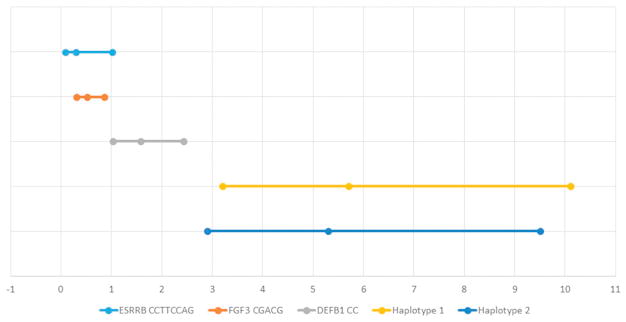
Three studies investigated the genes associated with rotator cuff disease by using association analyses to map genes to rotator cuff disease. One of these studies (n = 203) found a significant association of certain haplotypes in DEFB1, FGFR1, FGFR3, and ESRRB with rotator cuff disease (Table II)17. After adjusting for ethnic group and sex, another association was found for FGF10 (Table II). A second study (n = 175) discovered two haplotypes in ESRRB that significantly increased the risk of tears (Table II)32. The third study (n = 311) found two single nucleotide polymorphisms (SNPs) within genes SAP30BP and SASH1 associated with rotator cuff tears (Table II)30. The specific SNPs and haplotypes associated with rotator cuff disease from these three prior studies are presented in Table III. The accompanying forest plots are in Figures 2 and 3.
Table II.
Studies on genetic predisposition to rotator cuff disease
| Study | Methods | Participants | Group Definitions | Results | Source of Funding | Notes |
|---|---|---|---|---|---|---|
| Motta et al.13 (2014) | Case-control | Cases: n = 203; mean age 51.8 +/− 5.1 Controls: n = 207; mean age 53.5 +/− 5 |
Case: Included individuals diagnosed with rotator cuff disease by clinical examination, radiography, and MRI. Excluded individuals who were older than 60 or younger than 45; or had a history of trauma, bursitis, rheumatoid arthritis, autoimmune diseases, pregnancy, chronic systemic corticoid use, or hyperlaxity. Control: Included individuals who had no history of shoulder pain, a negative specific test result for impingement syndrome, and absence of tendinopathy in other joints. |
Whites (p = 0.002) and women (p = 0.001) had a higher prevalence of rotator cuff disease. Based on odds ratio calculation, the risk in women (OR 2.07, 95% CI 1.30–3.30) and whites (OR 1.88, 95% CI 1.21–2.90) was two times higher than in group 2. Cases had a higher incidence of high blood pressure (p < 0.001). Controls had a higher prevalence of systemic diseases (p < 0.0001), medication use (p = 0.01), and calcium supplementation (p = 0.01). A significant association of certain haplotypes in DEFB1, FGFR1, FGFR3, and ESRRB was observed with RCD. Adjusted by ethnic group and sex revealed another association in FGF10 |
None stated. | |
| Teerlink et al.27 (2015) | Case-control | Cases: n = 175 Controls: n = 2,595 |
Case: Included individuals who had a full-thickness supraspinatus or infraspinatus rotator cuff tear documented on MRI after age 30 and before age 80. Excluded individuals who had a partial-thickness rotator cuff tear, tendinopathy only, or significant glenohumeral arthritis; or had prior surgery on the involved shoulder. | There was a significant association between ESRRB genetic variants and rotator cuff disease. Two haplotypes constructed from 22 SNPs spanning ESRRB both significantly increased the risk of rotator cuff tearing | Veterans Administration Merit Review Grant (Number 1157449) U.S. Department of Veterans Affairs | |
| Tashjian et al.25 (2016) | GWAS | Cases: n = 311 Controls: n = 2,641 |
Case: Included individuals who had a full-thickness supraspinatus or infraspinatus rotator cuff tear documented on MRI, and who were older than 30 and younger than 80. Excluded individuals who had a partial-thickness rotator cuff tear, tendinopathy only, or significant glenohumeral arthritis; or had prior surgery on the involved shoulder. | Two SNPs within genes SAP30BP (rs820218) and SASH1 (rs12527089) were significantly associated with rotator cuff tears | Veterans Administration Merit Review Grant (No. 1157449), U.S. Department of Veterans Affairs | Teerlink et al.27 (2015) found an associated SNP in ESRRB, which was further confirmed in this study |
Table III.
Associations between rotator cuff disease and specific SNPs
| Study | Gene | SNP | Chromosome | Base Pair Position | P value | Odds Ratio (95% CI) | Haplotypes | P value | Odds Ratio (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| Motta et al.13 (2014) | DEFB1 | rs1800972 | 8 | 6735423 | 3.4x10−40 | 1.98 (1.79–2.18) | DEFB1 CC | 3.7x10−2 | 1.58 (1.03–2.43) |
|
|
|||||||||
| ESRRB | rs4903399 | 14 | 76775202 | 4.9x10−2 | 0.64 (0.41–0.98) | ESRRB CCTTCCAG | 5.4x10−2 | 0.30 (0.09–1.02) | |
|
|
|||||||||
| rs1676303 | 14 | 76992164 | 6.4x10−3 | 0.23 (0.08–0.62) | |||||
|
|
|||||||||
| FGF3 | rs12574452 | 11 | 69631731 | 2.7x10−2 | 0.31 (0.11–0.84) | FGF3 CGACG | 1.3x10−2 | 0.52 (0.31–0.86) | |
|
|
|||||||||
| FGFR1 | rs13317 | 8 | 38269514 | 5.3x10−2 | 2.67 (1.02–7.21) | ||||
|
|
|||||||||
| FGF10 | rs1011814 | 5 | 44335820 | 4.6x10−1 | 1.20 (0.75–1.94) | ||||
|
|
|||||||||
| rs900379 | 5 | 44369656 | 9.9x10−2 | 0.68 (0.43–1.07) | |||||
|
|
|||||||||
| rs11750845 | 5 | 44373060 | 9.6x10−2 | 0.70 (0.46–1.05) | |||||
|
|
|
|
|||||||
| Teerlink et al.27 (2015) | ESRRB | rs17583842 | 14 | 76050858 | 4.9x10−3 | 1.41 (1.11–1.79) | Haplotype 1 | 3.4x10−9 | 5.7 (3.2–10.1) |
|
|
|||||||||
| rs7157192 | 14 | 75936713 | NA | NA | Haplotype 2 | 5.9x10−8 | 5.3 (2.9–9.5) | ||
|
|
|
|
|||||||
| Tashjian et al.25 (2016) | SAP30BP | rs820218 | 17 | 73687545 | 4.3x10−10 | 0.4 (0.3–0.5) | |||
|
|
|||||||||
| SASH1 | rs12527089 | 6 | 148787159 | 8.4x10−7 | 2.4 (1.7–3.4) | ||||
|
|
|||||||||
| ESRRB | NA | NA | NA | 1.9x10−2 | NA | ||||
SNP = Single Nucleotide Polymorphism
NA = not available
Figure 2.
Forest plot depicting the odds ratios and confidence intervals of specific SNPs
Figure 3.
Forest plot depicting the odds ratios and confidence intervals of specific haplotypes
The results of bias assessment according to the MINORS criteria are in Table IV25. All of the studies had a clearly stated aim, prospective collection of data, end points appropriate to the aim of the study, loss to follow-up <5%, and contemporary groups. Six studies included consecutive patients6, 7, 17, 29, 30, 32, two stated they had unbiased assessments of study end points6, 7, one had a prospective calculation of the study size17, three had adequate control groups6, 7, 29, five had baseline equivalence of groups6, 7, 17, 29, 31, and five had adequate statistical analyses7, 17, 29, 30, 32. The lowest score was 1631, and the highest was 227.
Table IV.
MINORS scores
| Criteria | Score | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Harvie et al.6 (2004) | Gwilym et al.5 (2009) | Tashjian et al.24 (2009) | Tashjian et al.26 (2014) | Motta et al.13 (2014) | Teerlink et al.27 (2015) | Tashjian et al.25 (2016) | |
| A clearly stated aim | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Inclusion of consecutive patients | 2 | 2 | 2 | 0 | 2 | 2 | 2 |
| Prospective collection of data | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| End points appropriate to the aim of study | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Unbiased assessment of the study end point | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| Follow-up period appropriate to the aim of study | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Loss to follow-up <5% | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Prospective calculation of the study size | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| Adequate control group | 2 | 2 | 2 | 1 | 2 | 1 | 1 |
| Contemporary group | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Baseline equivalence of groups | 2 | 2 | 2 | 2 | 2 | 1 | 1 |
| Adequate statistical analysis | 2 | 1 | 2 | 1 | 2 | 2 | 2 |
|
| |||||||
| Total | 22 | 21 | 20 | 16 | 22 | 18 | 18 |
MINORS = Methodological Index for Non-Randomized Studies
Discussion
Rotator cuff disease is a common tendon disorder that is associated with shoulder pain and functional disability. The pathogenesis of rotator cuff disease is not completely understood. Identifying a possible genetic association could help our understanding of the disease process that leads to rotator cuff pathology. This systematic review summarized studies on familial and genetic predisposition to rotator cuff disease. Although there were only a limited number of studies on this topic, they do in general constitute a consensus that rotator cuff disease is a heritable trait.
Rotator cuff disease is a generic term that can be used to describe impingement syndrome, subacromial/subdeltoid bursal pathology, rotator cuff tendinopathy, and rotator cuff tear. An issue with prior literature is the absence of a uniform definition and diagnostic criteria for rotator cuff disease. Even in studies limited to rotator cuff tears, the case definition is variable. One study used clinical diagnosis29 by a physician as their criterion, whereas other studies used ultrasound6, 7 and MRI17, 30–32 for diagnosis. One of the studies used a criterion of whether the patient underwent a surgical rotator cuff repair29. Rotator cuff pathology is a clinical syndrome since structural defects found on imaging have been demonstrated in asymptomatic individuals.16, 24 Hence the case definition of rotator cuff pathology needs to account for both clinical presentation and structural deficit.
Harvie et al compared the rates of symptomatic and asymptomatic tears in siblings and spouses of individuals with rotator cuff tears, and determined that both were higher in siblings7. A follow-up study demonstrated that rotator cuff tears in siblings also had a higher risk of progressing6. Another study reported differing results when analyzing all of their subjects versus analyzing only those diagnosed before the age of 4029. The entire cohort did not demonstrate excess relatedness when only distant relationships were studied, implying that perhaps environmental factors were playing a confounding role. In contrast, the subgroup of younger patients showed excess relatedness in both close and distant relationships.
The genetic association studies observed associations between rotator cuff disease and SNPs in seven candidate genes (Table III, Figure 2, Figure 3)17, 30, 32. DEFB1 (Defensin, Beta 1) encodes the protein antimicrobial peptide defensin β-1, which aids in preventing epithelial surfaces from being colonized by microbes. The rs1800972 C>G variant was significantly more frequent in individuals with rotator cuff disease17. This base change is thought to lead to a decreased production of defensin β-1 production and higher expression levels.20, 21 The G allele is also more common in individuals with severe acute pancreatitis and less predominant in individuals with diabetes and S. aureus nasal colonization8, 20, 21.
ESRRB (estrogen-related receptor beta) encodes a protein similar to the estrogen receptor and is believed to have an inhibitory effect on estrogen signaling27. Mutations in this gene have also been associated with hearing impairment and dental decay35. In addition, upregulation of ESRRB has been linked with the progression of endometriosis3. In vitro studies have demonstrated a correlation between estrogen deficiency and poor tendon healing34, implying a possible role ESRRB may have in rotator cuff disease.
FGF3 (fibroblast growth factor 3) and FGF10 (fibroblast growth factor 10) encode fibroblast growth factor proteins and are involved in a number of processes such as cell growth and tissue repair, including tendons, and could thus be associated with the pathogenesis of rotator cuff disease. Mutations in FGF3 have been linked with improper embryonic development of the inner ear33. Mutations in FGF10 can lead to aplasia of lacrimal and salivary glands5. FGFR1 encodes one of the receptors also associated with fibroblast growth factor; however, this gene is more specific to limb development. Mutations have been associated with cleft lip and cleft palate, Pfeiffer syndrome, and osteoglophonic dysplasia18, 22, 36.
SAP30BP is implicated in cell death. SASH1 is a tumor suppressor gene implicated in a number of cancers23, 37. Thus, many of the SNPs associated with rotator cuff disease have a potential biologic mechanism for their association with rotator cuff disease, but further research is needed in this area.
A few studies on familial and genetic predisposition to rotator cuff disease have used controls from the general population, and have used cases and controls genotyped on different platforms in different experiments30–32. This is problematic since these studies assume that the prevalence of rotator cuff disease in the general population is low. This can also cause bias if the cases and the controls vary in other characteristics related to risk of rotator cuff disease, or if there are systematic differences in genotyping error between platforms. However, asymptomatic rotator cuff tears are demonstrated in 40% of persons over the age of 50 years, 54% in those over 60 years, and 65% in persons over 7016, 24. Many of these patients will progress to a symptomatic rotator cuff tear in the future15. Hence, true controls are individuals from the general population that are asymptomatic and do not have structural evidence for a rotator cuff tear.
The findings of previous studies provide evidence that there may be an important relationship between genes and rotator cuff disease. However, data on this issue is still limited. To our knowledge, only one study performed a genome wide association analysis (GWAS), which found two SNPs within genes SAP30BP and SASH1 associated with rotator cuff tears30. Both gene products, SAM and SH3 domain-containing protein 1 (SASH1) and SAP30-binding protein (SAP30BP), were reported in the process of cell apoptosis30. The effort to identify susceptibility genes of common multifactorial traits could lead to some insight into the pathogenesis mechanism that would, in turn, facilitate the development of better therapeutic and prophylactic approaches. However, many significant GWAS signals associated with multifactorial traits, e.g. type 2 diabetes, were mapped outside the coding gene sequence, which imposes a barrier to understanding their identities and function as well as limits their usefulness in experimental studies2, 10. It is equally important to assess the reproducibility of reported genotype-phenotype association, as more of them failed to replicate. This was attributed to several issues, including inappropriate reliance on standard significant thresholds, small samples, and genotype and phenotype heterogeneity11, 12. At the advent of GWAS, it appeared a promising proposal to estimate disease-risk by capturing the profile of common genetic variants. Nevertheless, the majority of common variants identified by GWAS only possess a very moderate effect size, and even the sum of these genetic effects only accounts for a minor portion of estimated trait heritability26. To address this issue, the effort has been made to identify the variants with lower frequency but higher penetrance by using imputed variants as well as to explore gene-gene and gene-environment interactions26. Further such studies would be beneficial, with the awareness that GWAS studies are designed to find common SNPs associated with complex traits and thus may not reveal conclusive information. Even if multiple SNPs are identified, they may still not fully explain the relationship between genes and rotator cuff disease. GWAS studies that identify SNPs would also need to be replicable and use a consistent definition of the clinical phenotype of rotator cuff tears.
Conclusions
There is data suggesting a genetic predisposition to rotator cuff disease. A large GWAS study with adequate controls could discover SNPs associated with symptomatic rotator cuff tears. The results from such a study could assist with early detection of individuals at risk of developing non-traumatic tears or suggest mechanisms of idiopathic rotator cuff disease. This may lead to medical treatments or prophylactic rehabilitation therapies to avoid development of symptomatic rotator cuff tears.
Acknowledgments
Source of Funding: This study was funded by the Rehabilitation Research Experience for Medical Students (RREMS) Program from the Association of Academic Physiatrists (AAP).
Dr. Jain is supported by funding from National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) 1K23AR059199 and the Vanderbilt’s Institute for Clinical and Translational Research (VICTR).
The project was supported by CTSA award No. UL1TR00045 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Andarawis-Puri N, Flatow EL, Soslowsky LJ. Tendon basic science: Development, repair, regeneration, and healing. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 2015;33:780–784. doi: 10.1002/jor.22869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Human molecular genetics. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 3.Cavallini A, Resta L, Caringella AM, Dinaro E, Lippolis C, Loverro G. Involvement of estrogen receptor-related receptors in human ovarian endometriosis. Fertility and sterility. 2011;96:102–106. doi: 10.1016/j.fertnstert.2011.04.032. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhury S, Carr AJ. Lessons we can learn from gene expression patterns in rotator cuff tears and tendinopathies. Journal of shoulder and elbow surgery. 2012;21:191–199. doi: 10.1016/j.jse.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 5.Entesarian M, Matsson H, Klar J, Bergendal B, Olson L, Arakaki R, et al. Mutations in the gene encoding fibroblast growth factor 10 are associated with aplasia of lacrimal and salivary glands. Nature genetics. 2005;37:125–127. doi: 10.1038/ng1507. [DOI] [PubMed] [Google Scholar]
- 6.Gwilym SE, Watkins B, Cooper CD, Harvie P, Auplish S, Pollard TC, et al. Genetic influences in the progression of tears of the rotator cuff. The Journal of bone and joint surgery British volume. 2009;91:915–917. doi: 10.1302/0301-620X.91B7.22353. [DOI] [PubMed] [Google Scholar]
- 7.Harvie P, Ostlere SJ, Teh J, McNally EG, Clipsham K, Burston BJ, et al. Genetic influences in the aetiology of tears of the rotator cuff. Sibling risk of a full-thickness tear. The Journal of bone and joint surgery British volume. 2004;86:696–700. doi: 10.1302/0301-620X.86B5.14747. [DOI] [PubMed] [Google Scholar]
- 8.Huang YP, Wang TY, Wang W, Sun HZ. Association between Genetic Polymorphisms in DEFB1 and Susceptibility to Digestive Diseases. Medical science monitor : international medical journal of experimental and clinical research. 2015;21:2240–2250. doi: 10.12659/MSM.893453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain NB, Higgins LD, Losina E, Collins J, Blazar PE, Katz JN. Epidemiology of musculoskeletal upper extremity ambulatory surgery in the United States. BMC musculoskeletal disorders. 2014;15(4) doi: 10.1186/1471-2474-15-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, et al. Defining functional DNA elements in the human genome. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraft P, Zeggini E, Ioannidis JP. Replication in genome-wide association studies. Statistical science : a review journal of the Institute of Mathematical Statistics. 2009;24:561–573. doi: 10.1214/09-STS290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li A, Meyre D. Challenges in reproducibility of genetic association studies: lessons learned from the obesity field. International journal of obesity. 2013;37:559–567. doi: 10.1038/ijo.2012.82. [DOI] [PubMed] [Google Scholar]
- 13.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. Journal of clinical epidemiology. 2009;62:e1–34. doi: 10.1016/j.jclinepi.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Longo UG, Berton A, Papapietro N, Maffulli N, Denaro V. Epidemiology, genetics and biological factors of rotator cuff tears. Medicine and sport science. 2012;57:1–9. doi: 10.1159/000328868. [DOI] [PubMed] [Google Scholar]
- 15.Mall NA, Kim HM, Keener JD, Steger-May K, Teefey SA, Middleton WD, et al. Symptomatic progression of asymptomatic rotator cuff tears: a prospective study of clinical and sonographic variables. The Journal of bone and joint surgery American volume. 2010;92:2623–2633. doi: 10.2106/JBJS.I.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milgrom C, Schaffler M, Gilbert S, van Holsbeeck M. Rotator-cuff changes in asymptomatic adults. The effect of age, hand dominance and gender. The Journal of bone and joint surgery British volume. 1995;77:296–298. [PubMed] [Google Scholar]
- 17.da Motta GR, Amaral MV, Rezende E, Pitta R, Vieira TC, Duarte ME, et al. Evidence of genetic variations associated with rotator cuff disease. Journal of shoulder and elbow surgery. 2014;23:227–235. doi: 10.1016/j.jse.2013.07.053. [DOI] [PubMed] [Google Scholar]
- 18.Muenke M, Schell U, Hehr A, Robin NH, Losken HW, Schinzel A, et al. A common mutation in the fibroblast growth factor receptor 1 gene in Pfeiffer syndrome. Nature genetics. 1994;8:269–274. doi: 10.1038/ng1194-269. [DOI] [PubMed] [Google Scholar]
- 19.Neer CS., 2nd Impingement lesions. Clinical orthopaedics and related research. 1983:70–77. [PubMed] [Google Scholar]
- 20.Nemeth BC, Varkonyi T, Somogyvari F, Lengyel C, Fehertemplomi K, Nyiraty S, et al. Relevance of alpha-defensins (HNP1-3) and defensin beta-1 in diabetes. World journal of gastroenterology. 2014;20:9128–9137. doi: 10.3748/wjg.v20.i27.9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nurjadi D, Herrmann E, Hinderberger I, Zanger P. Impaired beta-defensin expression in human skin links DEFB1 promoter polymorphisms with persistent Staphylococcus aureus nasal carriage. The Journal of infectious diseases. 2013;207:666–674. doi: 10.1093/infdis/jis735. [DOI] [PubMed] [Google Scholar]
- 22.Riley BM, Mansilla MA, Ma J, Daack-Hirsch S, Maher BS, Raffensperger LM, et al. Impaired FGF signaling contributes to cleft lip and palate. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rimkus C, Martini M, Friederichs J, Rosenberg R, Doll D, Siewert JR, et al. Prognostic significance of downregulated expression of the candidate tumour suppressor gene SASH1 in colon cancer. British journal of cancer. 2006;95:1419–1423. doi: 10.1038/sj.bjc.6603452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sher JS, Uribe JW, Posada A, Murphy BJ, Zlatkin MB. Abnormal findings on magnetic resonance images of asymptomatic shoulders. The Journal of bone and joint surgery American volume. 1995;77:10–15. doi: 10.2106/00004623-199501000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Slim K, Nini E, Forestier D, Kwiatkowski F, Panis Y, Chipponi J. Methodological index for non-randomized studies (minors): development and validation of a new instrument. ANZ journal of surgery. 2003;73:712–716. doi: 10.1046/j.1445-2197.2003.02748.x. [DOI] [PubMed] [Google Scholar]
- 26.Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–383. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanida T, Matsuda KI, Yamada S, Hashimoto T, Kawata M. Estrogen-related Receptor beta Reduces the Subnuclear Mobility of Estrogen Receptor alpha and Suppresses Estrogen-dependent Cellular Function. The Journal of biological chemistry. 2015;290:12332–12345. doi: 10.1074/jbc.M114.619098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tashjian RZ. Epidemiology, natural history, and indications for treatment of rotator cuff tears. Clinics in sports medicine. 2012;31:589–604. doi: 10.1016/j.csm.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Tashjian RZ, Farnham JM, Albright FS, Teerlink CC, Cannon-Albright LA. Evidence for an inherited predisposition contributing to the risk for rotator cuff disease. The Journal of bone and joint surgery American volume. 2009;91:1136–1142. doi: 10.2106/JBJS.H.00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tashjian RZ, Granger EK, Farnham JM, Cannon-Albright LA, Teerlink CC. Genome-wide association study for rotator cuff tears identifies two significant single-nucleotide polymorphisms. Journal of shoulder and elbow surgery. 2016;25:174–179. doi: 10.1016/j.jse.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Tashjian RZ, Saltzman EG, Granger EK, Hung M. Incidence of familial tendon dysfunction in patients with full-thickness rotator cuff tears. Open access journal of sports medicine. 2014;5:137–141. doi: 10.2147/OAJSM.S63656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teerlink CC, Cannon-Albright LA, Tashjian RZ. Significant association of full-thickness rotator cuff tears and estrogen-related receptor-beta (ESRRB) Journal of shoulder and elbow surgery. 2015;24:e31–35. doi: 10.1016/j.jse.2014.06.052. [DOI] [PubMed] [Google Scholar]
- 33.Tekin M, Hismi BO, Fitoz S, Ozdag H, Cengiz FB, Sirmaci A, et al. Homozygous mutations in fibroblast growth factor 3 are associated with a new form of syndromic deafness characterized by inner ear agenesis, microtia, and microdontia. American journal of human genetics. 2007;80:338–344. doi: 10.1086/510920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torricelli P, Veronesi F, Pagani S, Maffulli N, Masiero S, Frizziero A, et al. In vitro tenocyte metabolism in aging and oestrogen deficiency. Age. 2013;35:2125–2136. doi: 10.1007/s11357-012-9500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber ML, Hsin HY, Kalay E, BroZkova DS, Shimizu T, Bayram M, et al. Role of estrogen related receptor beta (ESRRB) in DFN35B hearing impairment and dental decay. BMC medical genetics. 2014;15:81. doi: 10.1186/1471-2350-15-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White KE, Cabral JM, Davis SI, Fishburn T, Evans WE, Ichikawa S, et al. Mutations that cause osteoglophonic dysplasia define novel roles for FGFR1 in bone elongation. American journal of human genetics. 2005;76:361–367. doi: 10.1086/427956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeller C, Hinzmann B, Seitz S, Prokoph H, Burkhard-Goettges E, Fischer J, et al. SASH1: a candidate tumor suppressor gene on chromosome 6q24. 3 is downregulated in breast cancer. Oncogene. 2003;22:2972–2983. doi: 10.1038/sj.onc.1206474. [DOI] [PubMed] [Google Scholar]