Abstract
SP6 is a salmonella phage closely related to coliphage K1-5. K1-5 is notable in that it encodes two polysaccharide-degrading tailspike proteins, an endosialidase that allows it to infect E. coli K1, and a lyase that enables it to infect K5 strains. SP6 is similar to K1-5 except that it encodes a P22-like endorhamnosidase tailspike, gp46, allowing it to infect group B Salmonella. We show here that SP6 can also infect Salmonella serogroups C2 and C3 and that a mutation in a putative second tailspike, gp47, eliminates this specificity. Gene 47 was fused to the coding region of the N-terminal portion of the Pseudomonas aeruginosa R2 pyocin tail fiber and expressed in trans such that the fusion protein becomes incorporated into pyocin particles. These pyocins, termed AvR2-SP47, killed serogroups C2 and C3 Salmonella. We conclude that SP6 encodes two tail proteins providing it a broad host range among Salmonella enterica.
1Introduction
Phage SP6 is a lytic T7-like phage of the Podoviridae first isolated using Salmonella enterica serovar typhimurium as a host (Zinder, 1961). Genome sequence analysis indicated that SP6 is closely related to phage K1-5 (Scholl et al, 2004; Dobbins et al., 2004). Both SP6 and K1-5 are biologically similar to T7, but have diverged considerably at the sequence level and have therefore been classified the “SP6 group” as a subset of the larger “T7 supergroup” (Scholl et al., 2004). One distinction of the SP6 group is that the phages do not encode a single tail fiber analogous to that of T7 but instead encode one or two distinct polysaccharide-degrading tailspikes that are part of the phage virion structure (Scholl et al., 2001; Scholl et al., 2004). The genes encoding these tailspikes are located in a genetic cassette at one end of the phage genome, and the protein products are connected to the virion by a non-covalent interaction with a separate peptide that is homologous to the N-terminal portion of T7-like tail fibers, gp37 (Scholl et al., 2004; Leiman et al., 2007). In the case of K1-5, these two tailspikes are a K5 capsule-specific lyase, gp46, and a K1 capsule-specific endosialidase, gp471. Both of these proteins are structural; each forms homotrimers of which there are six arranged on the tail structure (Scholl et al., 2001, Leiman et al., 2007). SP6 encodes two different proteins in the analogous positions in the genome, with little sequence similarity to those of K1-5. One, gp46, is a P22-like endorhamnosidase. The P22 endorhamnosidase tailspike has been very well studied and allows P22 to infect group B and D serovars, all of which have a similar O-antigen backbone structure (Steinbacher et al., 1997). A second putative SP6 tailspike, encoded by gene 47, was found also to be structural and was speculated to be a second tailspike protein; however its exact specificity was unknown (Scholl et al., 2004).
R-type pyocins are phage tail-like bacteriocins produced by some Pseudomonas aeruginosa strains as a defense against other strains of the same species (Michel-Briand and Baysse, 2002). R-type pyocins are evolutionarily related to the tail structures of P2-like phages, which are members of the Myoviridae, and are composed of a contractile sheath, core, baseplate, and tail fibers but no capsid or genetic material (Kagayama et al., 1964; Nakayama et al., 2000, Ge et al., 2015). Pyocin genes are encoded in the bacterial chromosome and are induced by bacterial stress, such as DNA damage. This results in production of 100–200 pyocin particles/cell, followed by lysis of the producer cells and release of the particles. These structures kill target cells by first binding to a surface structure via the six tail fibers followed by a sheath contraction and insertion of the core, which results in a depolarization of the cell membrane and death (Uratani and Hoshino, 1984). Binding of a single particle to a cell can be sufficient to cause death. We have shown that it is possible to retarget the spectrum of R-type pyocins by making chimeric tail fiber fusions between the N-terminal portion of the pyocin tail fiber and tail fibers or tailspikes of phages (Williams et al, 2008; Scholl et al 2009; Scholl et al, 2012, Gebhart et al., 2015). These engineered pyocins bind and kill cells that express the specific receptors that are the targets for the donor tailspike, and can thus be used as tools to identify unknown tailspike specificities.
In this work we find that, in addition to serogroups B and D, SP6 can infect C2 and C3 Salmonella serogroups, which include serovars kentucky and newport, and we show that gp47 confers this host specificity. We also show that the binding specificity of the tailspike can be used to retarget R-type pyocins of P. aeruginosa to these specific strains of Salmonella enterica.
Results
Host range of SP6
The host range of SP6, propagated on strain ATCC 27198 (typhimurium LT2), was studied by plaque assaying on a panel of different Salmonella enterica serovars. The results are shown in Table 1. Besides typhimurium, SP6 infects other serogroup B strains including saintpaul, and heidelberg, as well as serogroup D strains, enteritidis and javiana. This is consistent with the fact that it encodes a tailspike (gp46) with high sequence similarity to that of P22, which also infects these strains. All of these strains have a common O-antigen backbone (Luk and Lindberg, 1991; Figure 1).
Table 1.
Sensitivities of various Salmonella enterica strains to bacteriophage SP6 and the engineered R-type pyocins. Plaquing efficiency is based on reference strain 27198.
| Strain | Serovar | Serogroup | SP6 Plaquing efficiencya | SP6 Plaquing efficiencyb | AvR2-SP47 killing | AvR2-SP46 killing |
|---|---|---|---|---|---|---|
| ATCC 27198 | typhimurium | B | 1.0 | 1.0 | No | Yes |
| ATCC 700720 | typhimurium | B | 0.52 | No | Yes | |
| ATCC 9712 | saintpaul | B | 0.15 | No | Yes | |
| ATCC 8326 | heidelberg | B | 0.25 | No | Yes | |
| ATCC 13076 | enteritidis | D1 | 0.40 | No | Yes | |
| ATCC BAA-1593 | javiana | D1 | 0.67 | No | Yes | |
| ATCC 27869 | newport | C2 | 0.06 | 0.14 | Yes | No |
| ATCC 9263 | kentucky | C3 | 0.04 | Yes | No | |
| ATCC 8391 | thompson | C1 | No plaques | No | No |
SP6 stock was propagated on ATCC 27198 has the host.
SP6 stock was propagated on ATCC 27869.
Figure 1.

O-antigen structures of Salmonella serogroups examined in this study. (From Luk and Lindberg, 1991).
SP6 can also infect serovar newport (serogroup C2). This was somewhat surprising since the O-antigen structure of C2 is quite distinct from that of serogoups B and D (Figure 1). There are no common sugar residues between these two O-antigen backbones that suggest gp46 is targeting and degrading these structures. SP6 also infects serovar kentucky which belongs to serogroup C3. The C2 and C3 O-antigen structures differ only in the position of a side residue; the backbone structures are identical (Figure 1). SP6 did not infect the one serogroup C1 strain tested (serovar thompson); however, the O-antigen structure of this strain is quite different from C2 and C3. Plaquing efficiency was less on C2 and C3 serorvars vs B serovars; this changed only slightly when the phage stock was propagated on a newport strain vs typhimurium.
SP6 host range mutants
It was previously shown that the two tailspikes of phage K1-5 could be independently mutated to distinguish the two receptor binding activities (Scholl et al., 2001). We followed the same strategy for SP6 using selective binding/enrichment experiments. SP6 particles were bound to log phase Salmonella heidelberg (ATCC 8326) cells and the bound cells were filtered before a phage burst could occur. The filtrate was expected to be enriched with phage particles that cannot bind to strain 8326. This population of phages was then then amplified on Salmonella newport (ATCC 27869) by infecting and allowing for phage replication and burst. The lysate from this enrichment was then re-bound to strain 8326 and the selective filtration/enrichment repeated a total of 3 times. This regiment was designed to select for phage mutants that can no longer bind to strain 8326 but can still bind and replicate on strain 27869. A plaque was picked from the lawn of strain 27869 and tested for growth by spotting on both strains. At a spot of ~105 pfu; killing was observed on strain 27869 but not strain 8326 (data not shown). This phage has labelled SP6H−. The converse experiment was also performed in which the phage were bound to strain 27869 and enriched on strain 8326 to select mutants that could replicate on heidelberg but not newport. This mutant was termed SP6N−. Both SP6H− and SP6N− tailspike regions genes 46 and 47 were cloned and sequenced. SP6H− had a single point mutation in gp46, W101H. SP6N− had a single point mutation in gp47, D246Y.
Pyocin tail fiber/tailspike fusions
The above mutation studies provide some evidence that gp47 determines the host specificity to serogroups C2 and C3. To test this hypothesis we made a fusion between the gene encoding amino acids 10–495 of SP6 gene 47 to amino acids 1–164 of R2 tail fiber (see materials and methods and Williams et al., 2008; Scholl et al., 2009). This tail fiber fusion was expressed in P. aeruginosa PAO1 Δprf15 (strain MΔ) simultaneously with induction of the R2 pyocin genes by mitomycin C such that the tail fiber fusion protein was incorporated into the R2 pyocin particle. The resulting pyocin particles were purified and tested for bactericidal activity against the panel of Salmonella strains (Table 1) using the spot plate assay method (Figure 2). This engineered pyocin, termed AvR2-SP47, killed serogroup C2 and C3 strains but not serogroups B or D. A similar tail fiber construct was made by fusing the coding region of gp46 (amino acids 2–550) to amino acids 1–164 of the R2 tail fiber. Pyocin particles, termed AvR2-SP46 that incorporated this fusion had bactericidal activity against heidleberg and typhimurium and not newport or kentucky (Figure 2) which is expected since this tailspike is related to that of P22.
Figure 2.
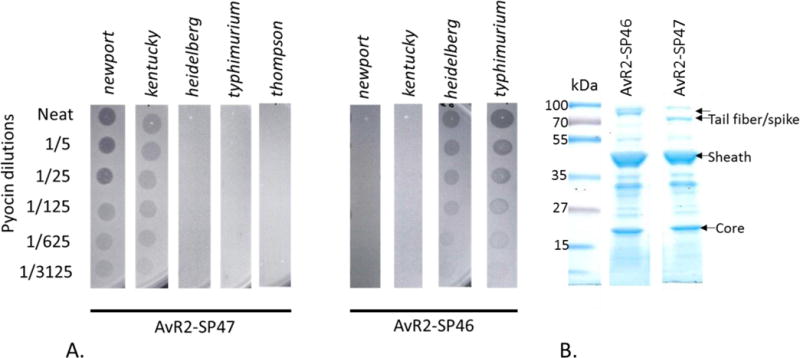
A. Spot activity assays of AvR2-SP47 and AvR2-SP46 on lawns of Salmonella enterica strains. AvR2-SP47 only has bactericidal activity against newport (C2) and kentucky (C3) where AvR2-SP46 has activity only on heidelberg and typhimurium (serogroup B). B SDS PAGE gels of purified pyocin particles. Note the different tail fiber/spike sizes. (Lanes were cut and pasted from different positions from the same gel).
Discussion
In addition to infecting Salmonella enterica serogroups B and D as predicted by its P22-like tailspike (gp46), SP6 also infects serogroups C2 and C3. These latter 2 serogroups share an O-antigen backbone structure that is quite distinct from the B and D strains, predicted to be recognized by gp46. Accordingly, one would predict that a different protein would be needed to target these serogroup C2 and C3 cells. We provide evidence here that gene 47 encodes this protein. Because Podoviridae phage tailspikes often bind to O-antigens, we speculate that the common repeating sugar units of the O-antigen backbone of C2 and C3 strains is the receptor.
We previously predicted that gp47 is likely a second tail spike (Scholl et al., 2004) but at the time it shared little sequence similarity with any other known protein and relatively little host range data was known for SP6. More recently, complete genome sequences of Salmonella enterica newport have been reported and deposited in the genome databases (Cao et al., 2012). Several contain prophage or prophage remnants that encode bifunctional tailspikes that have a gp47-like C-terminal domain. These prophage tailspikes fall into two types, those that have a head-binding domain with high sequence similarity to P22-like tailspikes and those that have a head binding domain related to phiV10/epsilon 15-like tailspikes (Scholl et al., 2009; Kropinsky et al., 2007; Figure 3). This is another example where an evolutionarily related C-terminal carbohydrate domain can be found as part of the tailspike structure(s) of much more distantly related phage types (Scholl and Merril, 2005; Stummeyer et al., 2006). Interestingly one strain, CMV 19470, contains both P22 and phiV10-like prophages within its genome, each with a putative gp47-like domain. The fact that some newport genomes contain prophage tailspikes related to SP6 gp47 further supports the findings in this work since prophages that are present in the genome may have also utilized this O-antigen structure as a receptor to infect.
Figure 3.

Comparison of SP6 gp47-like bacteriophage tailspikes. Salmonella enterica newport strain 19470 harbors two Podoviridae prophage elements. One is related to P22 and encodes a P22-like tailspike protein (gene 20999). The other is much more closely related to phage phiV10 and encodes a phiV10-like tailspike (gene 12751). Both of these putative tailspikes are modular in structure with head binding domains (HBD) corresponding to their respective phage type but with receptor binding portions that are related to SP6 gp47.
The plaquing efficiency of SP6 was lower on C2 and C3 strains than on B and D serogroups. This was true even when the phage stock was produced from a C2 host strain, indicating that it is unlikely to be caused by a host modification to the DNA. This could be explained by differences in adsorption to the different O-antigens and/or different levels of O-antigen expression on a given bacterial isolate.
The fact that R-type pyocins can be retargeted to Salmonella enterica using an SP6 tailspikes illustrates the flexibility of this bactericidal platform. It also supports the idea that the primary spectrum determinant of these Myoviridae-like tail structures is the receptor binding protein (tailspikes and tail fibers) and that the general mechanism of killing by R-type pyocin is less specific, showing bactericidal activity against a wide range of Enterobacteriaceae, provided the appropriate receptor binding function is present. This platform can be used as a tool to directly examine phage tailspike and other receptor binding functions separate from other aspects of phage biology that influence host range. Together the two pyocin constructs described here kill serogroups B, D, and C2/3. Jones et al., 2008 reported that the top five human disease isolates consist of typhimurium, enteritidis, newport, heidelberg, and javiana, all of which are killed by one of the two engineered R-type pyocins. This combination could potentially be used as an anti-Salmonella agent in animal health and food safety applications.
Materials and Methods
Salmonella strains were propagated in Luria Broth (LB) at 37°C with 250 rpm shaking. SP6 phage stocks were prepared by growing the host strain in LB (1/5 flask volume) to an OD600 of ~0.2 and adding phage at an MOI of 0.1, the cultures were allowed to lyse overnight. Cell debris was removed by centrifugation (25,000×g) and the lysates were passed through 0.2 μM cellulose acetate filters. Plaque assays were performed as previously described (Scholl et al., 2001) Briefly phage stocks were diluted serially in LB medium, 100 μl of each dilution was added to 200 μl of logarithmically growing cells (OD ~0.5–1.0). This was added to molten, tempered 0.5% overlay agar (LB) and poured onto a 1.5% LB agar plate. They were incubated at 37°C overnight and plaques were enumerated the following day.
To generate AvR2-SP47, gene 47 encoding amino acids 10–495 was amplified from an agar plug of SP6 using primers 5′-cctttcaagcttacacagtggttgcaacctgc-3′ and 5′-cctcccgaattctcacttgaatatgtagctgccag-3′, which contain HindIII and EcoR1 sites respectively. The product was cloned directly into plasmid DG100 (Williams et al., 2008) digested with HindIII/EcoRI to create plasmid DG268. DG268 was transformed into P. aeuruginosa strain MΔ, which is derived from PAO1 but deleted of the wild type R2 pyocin tail fiber gene, prf15 (see Williams et al., 2008). To generate AvR2-SP46, gene 46 was amplified using primers 5′-ttcgtctcgcgcgtttctatgacaaacctctcaaagg-3′ and 5′-atgcttccggctcgtagtcttctccgccattctgat-3′. The product was digested with BsmBI (primer was designed to leave a HindIII compatible end) and EcoRI and cloned into DG100 to generate DG263. This was transformed into the production strain as above. The engineered bacteriocins were expressed and purified as described in Williams et al 2008. SDS gels (4–20% Tris-glycine) of 5 μg of the purified particles are shown in Figure 2.
Bactericidal activity was assessed by the spot method. Diluted bacteriocin (in LB medium) was directly spotted onto a set, seeded 0.5% LB agar overlay lawn, and the spots were allowed to absorb into the surface. The plates were then incubated overnight at 37°C. Zones of clearing at the location of the spot application indicate bactericidal activity.
Highlights.
SP6 is a “dual specificity” bacteriophage that encodes two different receptor binding proteins giving it a broad host range.
These receptor binding proteins can be used to re-target the spectrum of R-type bacteriocins to Salmonella enterica.
Both SP6 and the engineered R-type bacteriocins can kill the Salmonella serovars most associated with human disease making them attractive for development as antimicrobial agents.
Acknowledgments
This work was funded in part by NIAID grant 5R21AI085318. We thank Richard Calendar and Jeff Miller for helpful discussions and comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
In this report, SP6 and K1-5 genes 37, 46 and 47 and their protein products are as defined by Scholl et al. (2004) and Genbank AY370673.1 and AY370674.1 (NC_008152.1). The same nomenclature is used for SP6 in Dobbins et al. (2004); however, in Genbank AY288927.2 (NC_004831.2), the corresponding SP6 genes are numbered 38, 49, and 50, respectively.
References
- Cao G, Zhao S, Strain E, Luo Y, Timme R, Wang C, Brown E, Meng J, Allard M. Draft genome sequences of eight Salmonella enterica serotype newport strains from diverse hosts and locations. J Bacteriol. 2012;194:5146. doi: 10.1128/JB.01171-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbins AT, George M, Jr, Basham DA, Ford ME, Houtz JM, Pedulla ML, Lawrence JG, Hatfull GF, Hendrix RW. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. J Bacteriol 2004. 2004;186:1933–44. doi: 10.1128/JB.186.7.1933-1944.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P, Scholl D, Leiman PG, Yu X, Miller JF, Zhou ZH. Atomic structures of a bactericidal contractile nanotube in its pre- and post-contraction states. Nat Struct Mol Biol. 2015;22:377–382. doi: 10.1038/nsmb.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhart D, Lok S, Clare S, Tomas M, Stares M, Scholl D, Donskey C, Lawley T, Govoni G. A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. mBio. 2015:e02368–14. doi: 10.1128/mBio.02368-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TF, Ingram LA, Cieslak PR, Vugia DJ, Tobin-D’Angelo M, Hurd S, Medus C, Cronquist A, Angulo FJ. Salmonellosis outcomes differ substantially by serotype. J Infect Dis. 2008;198:109–14. doi: 10.1086/588823. [DOI] [PubMed] [Google Scholar]
- Kageyama M, Ikeda K, Egami F. Studies of a pyocin. III. Biological properties of the pyocin. J Biochem. 1964;55:59–64. doi: 10.1093/oxfordjournals.jbchem.a127841. [DOI] [PubMed] [Google Scholar]
- Kropinski AM, Kovalyova IV, Billington SJ, Patrick AN, Butts BD, Guichard JA, Pitcher TJ, Guthrie CC, Sydlaske AD, Barnhill LM, Havens KA, Day KR, Falk DR, McConnell MR. The genome of epsilon15, a serotype-converting, Group E1 Salmonella enterica-specific bacteriophage. Virology. 2007;369:234–44. doi: 10.1016/j.virol.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Battisti AJ, Bowman VD, Stummeyer K, Muhlenhoff M, Gerardy-Schahn R, Scholl D, Molineux IJ. The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. J Mol Biol. 2007;371:836–849. doi: 10.1016/j.jmb.2007.05.083. [DOI] [PubMed] [Google Scholar]
- Luk JMC, Lindberg AA. Anti-Salmonella lipopolysaccharide monoclonal antibodies: Characterization of Salmonella BO-, CO-, DO-, and EO-specific clones and their diagnostic usefulness. J Clin Microbiol. 1991;29:2424–2433. doi: 10.1128/jcm.29.11.2424-2433.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel-Briand Y, Baysse C. The pyocins of Pseudomonas aeruginosa. Biochimie. 2002;84:499–510. doi: 10.1016/s0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol. 2000;38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- Scholl D, Rogers S, Adhya S, Merril CR. Bacteriophage K1-5 encodes two different tail fiber proteins allowing it to infect and replicate on both K1 and K5 strains of E. coli. J Virol. 2001;75:2509–2515. doi: 10.1128/JVI.75.6.2509-2515.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl D, Adhya S, Merril CR. Bacteriophage SP6 is closely related to phages K1-5 K5 and K1E but encodes a tail protein very similar to that of the distantly related P22. J Bacteriol. 2002;184:2833–2836. doi: 10.1128/JB.184.10.2833-2836.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl D, Kieleczawa J, Kemp P, Rush J, Richardson CC, Merril CR, Adhya S, Molineux I. Genome sequences of bacteriophages SP6 and K1-5 estranged members of the T7 group. J Mol Biol. 2004;335:1151–1171. doi: 10.1016/j.jmb.2003.11.035. [DOI] [PubMed] [Google Scholar]
- Scholl D, Cooley M, Williams S, Gebhart D, Martin D, Bates A, Mandrell R. An engineered R-type pyocin is a highly specific and sensitive bactericidal agent for the foodborne pathogen, Escherichia coli O157:H7. Antimicrob Agents Chem. 2009;53:3074–3080. doi: 10.1128/AAC.01660-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl D, Gebhart D, Williams SR, Bates A, Mandrell R. Genome sequence of E. coli O104:H4 leads to rapid development of a targeted antimicrobial agent against this emerging pathogen. PloS One. 2012;7:e33637. doi: 10.1371/journal.pone.0033637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, Huber R. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J Mol Biol. 1997;267:865–880. doi: 10.1006/jmbi.1997.0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stummeyer K, Schwarzer D, Claus H, Vogel U, Gerardy-Schahn R, Mühlenhoff M. Evolution of bacteriophages infecting encapsulated bacteria: lessons from Escherichia coli K1-specific phages. Mol Microbiol. 2006;60:1123–35. doi: 10.1111/j.1365-2958.2006.05173.x. [DOI] [PubMed] [Google Scholar]
- Uratani Y, Hoshino T. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J Bacteriol. 1984;57:632–636. doi: 10.1128/jb.157.2.632-636.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S, Gebhart D, Martin DW, Scholl D. Re-targeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microbiol. 2008;74:3868–3876. doi: 10.1128/AEM.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder ND. A bacteriophage specific for F- Salmonella strains. Science. 1961;133:2069. doi: 10.1126/science.133.3470.2069. [DOI] [PubMed] [Google Scholar]


