Abstract
Background
Around 70–90% of peptic ulcer disease (PUD) is due to Helicobacter pylori and requires treatment with antimicrobials to which these bacteria are susceptible. Common H. pylori diagnostic tests do not provide drug susceptibility data. Using the GenoType HelicoDR PCR test designed for gastric biopsies for simultaneous detection of H. pylori and its resistance to clarithromycin (CLA)/fluoroquinolones (FLQ), we present evidence for stool as an optional test specimen and also provide data on prevalence of H. pylori resistance to CLA and FLQ in Uganda.
Methods
Stool from 142 symptomatic PUD patients at three hospitals in Kampala was screened for H. pylori using a rapid antigen test. The GenoType HelicoDR test was run on all H. pylori antigen positives to determine PCR positivity and resistance to CLA/FLQ.
Results
Thirty-one samples (22%) were H. pylori antigen positive, and 21 (68%) of these were H. pylori PCR positive. Six of the 21 (29%) were resistant to CLA and eight to FLQ (42%), while two gave invalid FLQ resistance results.
Conclusion
Stool is a possible specimen for the GenoType HelicoDR test for rapid detection of H. pylori and drug resistance. In Uganda, Helicobacter pylori is highly resistant to CLA and FLQ.
1. Introduction
Helicobacter pylori is a motile Gram-negative, oxidase-, catalase-, and urease-positive, and microaerophilic bacillus [1]. Being microaerophilic, it needs only about 4% oxygen, 5% carbon dioxide, and 5% hydrogen for its growth and survival. This bacterium has been reported to be the cause of 90% of duodenal and 70% of gastric ulcer cases and may lead to stomach and/or duodenal perforation and fatal bleeding, and it is also associated with gastric cancer [2–4]. It is estimated that half of the world's population is infected with H. pylori, making it the most widespread infection in the world [5–7]. The actual burden of infection varies from nation to another. In Western Europe, North America, Australia, and parts of Asia, the proportion of infected people is estimated to be about 25% [7]. Developing countries have much higher infection rates, and in Africa, it is estimated that 61–100% of the population is infected and at risk of PUD and the other associated diseases [2, 8–11].
Effective management of PUD caused by H. pylori requires a combination of antimicrobials and proton pump inhibitors (PPIs, which inhibit secretion of acid by the stomach). The antimicrobial agents commonly used in first-line treatment include clarithromycin (CLA), metronidazole, and amoxicillin, while others such as fluoroquinolones (FLQ) and tetracycline are used in second-line regimens. A number of studies have shown that H. pylori has developed resistance to the most commonly used antimicrobial agents used in PUD treatment. Work reported by Lee et al. from 2003 to 2012 found that resistance of H. pylori to CLA increased from 17 to 24% while for the FLQ (levofloxacin and moxifloxacin) from 5% to 28% and amoxicillin from 6% to 15% [12]. Another study by Kumala and Rani in 2006 found that H. pylori resistance to CLA was 28%, to amoxicillin at 19%, and to FLQ (ciprofloxacin, norfloxacin, and ofloxacin at 7%, sparfloxacin and gatifloxacin at 3%, and levofloxacin and moxifloxacin at 1%) [13]. Similarly, the prevalence of resistance in 128 H. pylori strains isolated in 2004-2005 to FLQ (ciprofloxacin MIC > 1 mg/L) was 17% (22 strains) [14], while for CLA the pretreatment and posttreatment prevalence of resistance were 19% (23/123) and 69% (9/13), respectively [15]. This CLA resistance was relatively high compared to that reported as the national prevalence of resistance in France, which stood at 13% [16]. Results of a study of 2204 patients in Europe showed that H. pylori resistance rates were 16% to CLA, 14% to levofloxacin, and 35% to metronidazole. Resistance was significantly higher for CLA and levofloxacin in Western, Central, and Southern Europe (>20%) than in Northern European countries (<10%) [17]. Most of these studies recommended that in addition to accurate detection of H. Pylori, susceptibility tests (resistance diagnosis) should be done prior to treatment.
The diagnosis of H. pylori infection is currently carried out using four major methods [18, 19]: (i) breath test, in which the patient drinks 13C- or 14C-labelled urea, which the bacterium metabolizes producing labelled carbon dioxide that can be detected in the breath (the test is quick in the initial detection of the bacteria but requires complex equipment and cannot be used to detect drug resistance); (ii) stool antigen test, which detects H. pylori in the faeces for the purpose of initial detection of the bacteria and recurrences after antibiotic therapy (just like breath test, the H. pylori stool antigen test cannot detect drug resistance); (iii) blood antibody test, which detects antibodies to H. pylori in blood samples for purpose of detecting prior exposure but cannot also detect drug resistance; and (iv) gastric biopsy examination test (for culture, rapid urease test, or histological examination), which confirms H. pylori infection in PUD cases. These test approaches have before been regarded as reference tests but they are invasive, expensive, and not easily adopted in routine diagnostics in resource-poor settings. Culture and phenotypic antimicrobial susceptibility testing of H. pylori requires up to 10–14 days, on expensive special media and special microaerophilic environments, and it is hardly performed in routine clinical laboratories in the resource-poor settings [20].
The GenoType HelicoDR test (GTHDT, Hain Lifescience Nehren, Germany) was developed recently. This is a multiplex PCR-based assay for simultaneous detection of H. pylori and its resistance to two-key antibiotics, CLA and FLQ [21]. The test was originally designed for use on gastric biopsy or culture materials to detect mutations in gyrA gene codons 87 and 91 for resistance to FLQ and in the 23S rRNA genes for resistance to CLA. The test involves DNA extraction, amplification, and solid-phase reverse hybridization onto probes fixed on a nitrocellulose membrane. Several studies evaluating the GTHDT for diagnosis of CLA/FLQ-resistant H. pylori have been conducted on biopsy specimens or cultured isolates, but as a diagnostic test for H. pylori performed directly on stool specimens the data is limited. In 2009, Cambau et al. evaluated the GTHDT using gastric biopsy and clinical strains, and the sensitivity and specificity for detecting resistance were high, that is, 94% and 99% for CLA and 87% and 98.5% for FLQ, respectively [22].
However, the requirement of stomach or duodenal biopsy specimen is a major limitation to use of this test in poor countries where endoscopic facilities as well as the other tests remain extremely scarce and expensive. Obtaining these biopsy specimens involves an invasive and expensive endoscopic procedure not usually acceptable by patients. Consequently, this potentially useful test has attracted little use in routine patient care, particularly in the developing countries where H. pylori infection is highest, yet resources for endoscopy are very limited. Hence, very few data on H. pylori resistance have come from these resource-poor countries. An alternative noninvasive, less costly, easy to collect specimen such as stool, which is also more acceptable to patients, would be needed for use with the GTHDT in routine PUD patient care particularly in the detection of H. pylori and diagnosis of drug resistance. This study was undertaken to determine the performance of the GTHDT on stool as a starting material for molecular diagnosis of H. pylori and its resistance to CLA and FLQ simultaneously and determine the prevalence of H. pylori resistance to CLA/FLQ in Uganda.
2. Materials and Methods
2.1. Study Design and Approval
This was a cross-sectional study carried out from November 2012 to July 2013. This study was approved by the School of Biomedical Sciences Research and Ethics Committee (SBS-REC) of Makerere University College of Health Science (SBS 076) and by the Mulago Hospital Research and Ethics Committee (MREC: 374).
2.2. Study Site and Setting
The study was conducted at three sites (Mulago, Mengo, and Case Medical Centre Hospitals) in Kampala, Uganda. The Mulago National Referral Hospital has a gastrointestinal (GI) unit with both inpatient and outpatient services. The outpatient service runs weekly with about 40 patients seen every week, about one-half due to either established or suspected PUD cases, some of which get admitted into the wards. Mengo Hospital and Case Medical Centre are tertiary referral Nongovernmental Organisation and private hospitals within Kampala City, respectively. They provide care for many patients with various ailments including gastroenterological cases. Recruitment of the study participants and sample collections were done at these three hospitals. All laboratory assays were performed at MBN Clinical Laboratories, Kampala, Uganda, which offer advanced medical laboratory diagnostic services for patient care and research.
2.3. Study Population and Inclusion Criteria
The study included patients with suspected PUD who were at least 13 years old attending the GIT clinics at the study hospitals. Inclusion was based on having symptoms and signs of PUD as judged by the attending physician and consent to participate in the study. These symptoms included burning pain in the epigastrium with or without any of the following: bloating, nausea, dark or black stool, vomiting blood, or weight loss. The patients were individually approached to obtain informed consent (or assent with legal guardian consent) and consecutively recruited into the study. A predesigned case report form was used to collect both demographic and clinical data.
2.4. Sample Collection
Upon consenting, each participant was provided with a sterile dry stool container and instructed to collect three scoopful of the stool into the stool container. The stool specimen was then placed in a cool box and transported to the laboratory within 2 hours of sample collection.
2.5. Laboratory Methods
2.5.1. H. pylori Stool Antigen Test
All stool samples were tested for H. pylori antigen using an immunochromatopgraphy assay, the Taytec H. pylori stool antigen rapid diagnostic kit (Taytec Enterprises Canada) whose manufacturer's claimed sensitivity and specificity were above 95% [23, 24]. In the assay, the H. pylori antigen in stool reacts with conjugated-red latex particles sensitised with anti-H. pylori monoclonal antibodies. The H. pylori-conjugate complex migrates along the membrane by capillarity and binds to the specific antibody molecules fixed at the reaction zone. The excess of the complex keeps migrating through the membrane until reaching the control (C) zone where it binds to another specific antibody coated to the membrane forming a green band. Presence of the green band confirms the functionality of the test. In the procedure, about 50 mg of each stool sample was picked using the applicator stick and emulsified in the buffer and four drops of the emulsion were dispensed into the well of the cassette. The interpretations of the results (bands) for the H. pylori stool antigen test were as follows: a positive result was indicated by two bands (control = green and test = red), negative by only the control band (green), and invalid by either only red or no band [24]. The detailed procedure is found in the Taytec, Aldercrest Dr. Mississauga, Canada, H. pylori stool antigen test kit insert [24].
2.5.2. GTHDT
Only H. pylori antigen positive stool samples were subjected to this test.
DNA Extraction from Stool. This was performed using the QIAamp® DNA Stool Mini Kit (Qiagen, Hilden Germany, Catalogue Number: 51504). The procedures are detailed in the QIAamp DNA Stool Handbook [25]. Briefly, 180 to 220 mg of each H. pylori antigen positive stool specimen was transferred to 1.5 mL microcentrifuge tube and the ASL buffer was added to lyse the bacterial cells. The suspension was then heated for 5 min at 70°C and then vortexed and centrifuged to pellet debris. The supernatant was then transferred into a new 1.5 mL microcentrifuge followed by addition of the InhibitEX tablet and incubated at 65°C for 5 minutes to allow inhibitors to be adsorbed to the InhibitEX matrix. After centrifugation, 200 μL of the resultant supernatant was transferred to 15 μL proteinase K in another 1.5 mL microcentrifuge tube. Then 200 μL of absolute ethanol was added to the lysate to purify the DNA. This was then washed with 500 μL of buffer AW1 followed by the same amount of buffer AW2. It was then eluted using 50 μL of elution buffer AE after incubation at 65°C for 10 minutes. Eight microlitres of the pure DNA was added to 45 μL PCR reagent mix ready for amplification.
Multiplex PCR Amplification of Extracted DNA. As recommended by the manufacturer, the composition of each reaction mix was 35 μL of Primer Nucleotide Mix (PNM, containing biotin labelled primer sequences), 5 μL of 10x PCR buffer, 5 μL of 25 mM MgCl2, 0.2 μL of HotStarTaq polymerase (1 U), and 8 μL of the extracted DNA solutions. The PCR reaction tubes were loaded and amplified on the GTQ Cycler 96 (Hain LifeScience Nehren, Germany). The amplification program included phase 1, one cycle at 95°C for 15 min; phase 2, 10 cycles at 95°C for 30 s and at 58°C for 2 min; phase 3, 25 cycles at 95°C for 30 s, 53°C for 40 s, and 70°C for 40 s; and phase 4, one cycle at 70°C for 8 min. Details of the PCR reagent recipe, primer sequences, what they detect, and detailed procedure are found in the GTHDT kit insert, Lot number JF0045 [21].
Detection of Amplification Products. This was performed using a solid-phase reverse hybridization technique on the TwinCubator according to the manufacturer's instructions outlined in the GTHDT kit insert [21]. The hybridization process entailed chemical denaturation of double stranded DNA at room temperature for 5 min, hybridization of single stranded DNA with complementary DNA probes fixed on a nitrocellulose membrane strip in the hybridization buffer (HYB, green) at 45°C for 30 min, stringent washing, conjugation at room temperature for 30 min, and enzymatic detection of the reaction products [21].
Quality Control and Interpretation of the Results. During the entire experimental processes, measures were taken to avoid contamination and ensure validity of the results. Also, positive and negative controls were included at all steps from sample processing to hybridization. Data were cleaned before analysis. A result was considered valid if the Helicobacter pylori (HP), amplification control (AC), and conjugate control (CC) bands were all present. In addition, the locus controls (gyrA, 23S and RNA) must have stained positive when the HP zone indicated the presence of H. pylori. If neither the locus control probe nor the respective wild type or mutation probes of one of the two genes examined had developed, the result was considered invalid for interpretation of resistance to the respective drug. The bands formed were interpreted accordingly as shown in the prototype in Figure 1, and details are outlined in the GTHDT kit insert [21].
Figure 1.
Prototype of the GTHDT result (downloaded from http://www.hain-lifescience.de/en/products/microbiology/helicobacter/genotype-helicodr.html).
2.5.3. Data Management
The collected data was entered into a Microsoft excel spread sheet and exported to SPSS software v.11 for the analysis. The proportions of positivity of each test and the proportion of H. pylori resistance to CLA and FLQ were computed using SPSS v.16 software.
3. Results
3.1. Baseline Characteristics of the Studied Participants
A total of 142 participants, 105 (74%) from Mulago, 22 (15%) from Mengo, and 15 (1%) from Case Medical Centre, were recruited. Of these, 79/142 (55.6%) were females. The participants were aged between 13 and 85 (mean age = 40, SD = 18). A total of 135 participants had had the symptoms of PUD for more than two weeks. In addition, at least 42% (59/142) admitted to have taken one or more antibiotic(s) during the course of illness. Furthermore, 114/142 (80%) had taken a drug within one week before the screening. Detailed data are shown in Table 1.
Table 1.
Baseline characteristics of studied patients (N = 142).
| ITEM | Number (n = 142) | % |
|---|---|---|
| Hospitals of recruitment | ||
| Mulago | 105 | 74 |
| Mengo | 22 | 15 |
| Case Medical Centre | 15 | 1 |
|
| ||
| Gender | ||
| Female | 79 | 56 |
| Male | 62 | 44 |
| Not given | 1 | 7 |
|
| ||
| Duration of illness | ||
| Up to 2 weeks | 7 | 5 |
| 3-4 weeks | 8 | 6 |
| 2 months to 1 year | 40 | 28 |
| 2–5 years | 61 | 43 |
| More than 5 years | 26 | 18 |
|
| ||
| Taken drug(s) | ||
| Yes | 140 | 99 |
| No | 2 | 1 |
|
| ||
| The time the drug was last taken | ||
| Up to one week | 114 | 83 |
| Within a month | 16 | 11 |
| More than a month | 10 | 7 |
| NA | 2 | 1 |
|
| ||
| Drug(s) taken | ||
| One antibiotic plus proton pump inhibitors (PPI) | 41 | 29 |
| Proton pump inhibitors (PPI) only | 38 | 27 |
| Triple therapy | 28 | 20 |
| Single antibiotic | 19 | 13 |
| Two antibiotics | 12 | 9 |
| Never taken peptic ulcer drugs | 3 | 2 |
| Herbal | 1 | 0 |
|
| ||
| Specific antibiotics taken in last 2 weeks | ||
| FLQ taken | ||
| No | 135 | 95 |
| Yes | 7 | 5 |
| CLA taken | ||
| No | 138 | 97 |
| Yes | 4 | 3 |
| ∗Other antibiotics taken | ||
| Yes | 94 | 66 |
| No | 48 | 34 |
CLA = clarithromycin, FLQ = fluoroquinolones. ∗Other antibiotics included amoxicillin, metronidazole, and ceftrioxone.
3.2. Taytec H. pylori Antigen Test Results
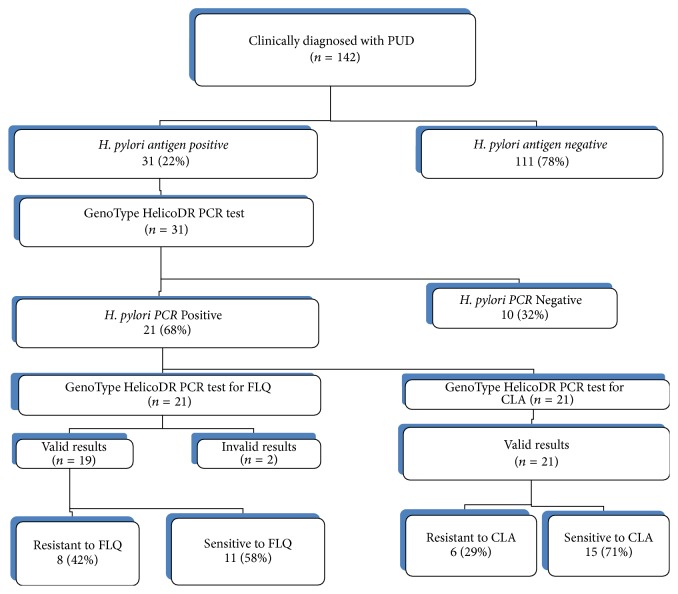
Out of the 142 stool samples, 31 were positive for the H. pylori antigen giving a prevalence of 22% in patients clinically diagnosed with PUD as shown in Figure 2.
Figure 2.
Study flowchart and summary of the results. Invalid results: lack of H. pylori control bands in the gyrA gene locus, CLA: clarithromycin, and FLQ: fluoroquinolones. Of the two FLQ invalid results, one was CLA susceptible and the other was resistant.
3.3. GTHDT Results
In Figure 3 in the supplementary materials (available online at https://doi.org/10.1155/2017/5430723), we have shown representative results of hybridization on the nitrocellulose strips from our laboratory.
Of the 31 H. pylori stool antigen positive samples, 21 (68%) tested positive with the GTHDT, as shown in Table 2 and Figure 3 in the supplementary materials. Among the 10 GTHDT negative samples, 7 were from patients who had taken antibiotics such as amoxicillin, metronidazole, and ceftrioxone, and one of these had also taken ciprofloxacin.
Table 2.
Results of the Taytec H. pylori stool antigen and GTHDT (N = 31).
| Serial number | Sex | Age (years) | Antibiotics taken within last two weeks | Taytec antigen test | GTHDT results | ||||
|---|---|---|---|---|---|---|---|---|---|
| FLQ | CLA | ∗Other antibiotics | H. pylori | CLA | FLQ | ||||
| (1) | M | 30 | No | No | Yes | P | P | R | S |
| (2) | M | 29 | No | No | No | P | P | S | S |
| (3) | M | 40 | No | No | No | P | P | S | R |
| (4) | M | 76 | No | No | No | P | N | NA | NA |
| (5) | M | 35 | No | No | Yes | P | N | NA | NA |
| (6) | M | 45 | No | No | Yes | P | P | R | Invalid |
| (7) | M | 25 | No | No | No | P | P | S | S |
| (8) | F | 58 | No | No | Yes | P | N | NA | NA |
| (9) | F | 23 | No | No | No | P | P | S | S |
| (10) | F | 85 | No | No | Yes | P | P | S | S |
| (11) | M | 20 | Yes | No | Yes | P | N | NA | NA |
| (12) | F | 80 | No | No | Yes | P | P | S | S |
| (13) | F | 16 | No | No | Yes | P | N | NA | NA |
| (14) | F | 44 | No | No | Yes | P | P | R | R |
| (15) | F | 32 | No | No | No | P | P | S | S |
| (16) | F | 75 | No | No | No | P | N | NA | NA |
| (17) | M | 57 | No | No | Yes | P | P | S | R |
| (18) | F | 55 | No | No | Yes | P | P | S | S |
| (19) | M | 24 | No | No | No | P | P | R | R |
| (20) | F | 35 | No | No | Yes | P | N | NA | NA |
| (21) | F | 42 | No | No | Yes | P | P | S | R |
| (22) | F | 43 | No | No | No | P | P | R | R |
| (23) | M | 17 | No | No | No | P | P | S | S |
| (24) | M | 33 | No | No | No | P | P | S | Invalid |
| (25) | M | 40 | No | No | Yes | P | N | NA | NA |
| (26) | UNK | UNK | No | No | No | P | P | R | R |
| (27) | F | 28 | No | No | No | P | N | NA | NA |
| (28) | F | 42 | No | No | Yes | P | N | NA | NA |
| (29) | M | 38 | No | No | No | P | P | S | S |
| (30) | F | 17 | No | No | No | P | P | S | S |
| (31) | M | 20 | No | No | Yes | P | P | S | R |
|
| |||||||||
| Total | M = 15 | No = 30 | No = 31 | Yes = 16 | P = 31 | P = 21 | R = 6 | R = 8 | |
| S = 15 | S = 11 | ||||||||
| F = 15 | Yes = 1 | Yes = 0 | No = 15 | N = 10 | NA = 10 | Invalid = 2 | |||
| NA = 10 | |||||||||
CLA = clarithromycin, F = female, FLQ = fluoroquinolone, M = male, N = negative, NA = not tested, P = positive, S = sensitive, and UNK = unknown. ∗Other antibiotics included amoxicillin, metronidazole, and ceftrioxone.
3.4. H. pylori Resistance to CLA and FLQ
3.4.1. CLA
All the 21 samples which tested positive for H. pylori with the GTHDT gave valid CLA test results, and 6 (29%) were resistant to CLA as shown in Figure 3 in the supplementary materials.
3.4.2. FLQ
Nineteen of the 21 antigen positive samples gave interpretable results for FLQ resistance testing, of which 8 (42%) were resistant as shown in Table 2 and Figure 2. Invalid results were due to lack of the gyrA gene locus control bands for H. pylori.
4. Discussion
In this study, we explored new simple to collect stool specimens as starting materials for use with the GTHDT for simultaneous detection for H. pylori and its resistance to CLA and FLQ. We also determined the proportions of H. pylori resistance to FLQ and CLA. We anticipate that our findings have the potential to inform clinicians on the level of H. pylori resistance to the selected antibiotics, which will help to improve on overall management of patients with suspected peptic or gastroduodenal ulcers. Additionally, with stool as specimen for this test, we anticipate high patient acceptability and reduce the costs to the patient/health systems of testing for H. pylori.
In this preliminary study only H. pylori antigen positive stool samples were subjected to the study test. Out of the H. pylori stool antigen positive specimens, the GTHDT showed a high positivity score of 68% (21/31). By study design, we did not aim at determining sensitivity or specificity of the GTHDT at this stage of new test evaluation, and another study to do this has been planned. However, the high positivity score obtained in this study provides strong evidence for stool as a possible specimen for same day molecular diagnosis of H. pylori and its resistance to CLA and FLQ. Our results are similar to those obtained in other studies on H. pylori diagnostics. In a study in 2016, Brennan et al. used the GenoType HelicoDR test to detect 80.3% (53/66) of H. pylori infection in stool samples [26]. In another study by Lottspeich et al., 63% of the confirmed stool samples turned positive for H. pylori when a real-time PCR assay was used [23]. Data from various studies indicate that antigen tests for H. pylori on stool samples have a high sensitivity and specificity [23]. The low performance of the PCR-based test in our study compared with the antigen test is disappointing but might be improved if antigen positive PCR-negative samples were retested, and/or the amount of stool/DNA used for the PCR-based test was increased as reported by Schabereiter-Gurtner et al. 2004 [27]. By repeating the stool antigen tests before PCR, Schabereiter-Gurtner and colleagues reported that the real-time PCR test was highly accurate in the detection of H. pylori infection in stool and allowed for culture-independent clarithromycin susceptibility testing [27]. Furthermore, in a systematic review and meta-analysis study in patients with bleeding peptic ulcer by Gisbert et al. in 2006 [28], the sensitivities of the different test methods were stool antigen test 87% (6 studies and 377 patients), rapid urease test 67% (16 studies and 1,417 patients), urea breath test 93% (8 studies and 520 patients), serology 88% (9 studies and 803 patients), histology 70% (10 studies and 827 patients), and culture 45% (3 studies and 314 patients). While our study did not perform formal computation of sensitivity of the GTHDT, the high positivity rate of 68% among the H. pylori antigen positive stool samples indicates this test to be technically promising and warranties well-designed prospective studies to determine the diagnostic accuracy of the GTHDT on stool specimens against an established gold standard. In our study, 32% of the hitherto H. pylori Ag positive stool samples remained negative at PCR. It is likely that nonspecific PCR inhibitors were responsible for this negativity. Optimization of DNA extraction methods or repeat testing as proposed by Makristathis and Hirschl may help to solve this false negativity problem but it should be noted that repeat testing is an expensive approach in routine patient care [29]. This could also be associated with mixed populations of the bacteria as reported by Schabereiter-Gurtner where it led to failure of PCR to detect the resistant genotype in the biopsy DNA, stool DNA, or both in one case [27].
4.1. Drug Resistant H. pylori
The 4th edition of the Maastricht consensus recommended a threshold of 15–20% to separate regions of high and low CLA resistance [30]. In our study, H. pylori resistance to CLA was high at 29%. Uganda can thus be considered an area of high CLA resistance. Our findings are in conformity with several global reports. The study by Lee et al. reported H. pylori resistance to CLA at 24.7% [12]. Another study by Kumala and Rani in 2006 found that H. pylori resistance to CLA was 28% [13]. In addition, CLA resistance in children is reported to be at 34% in Austria, 37% in France, 39% in Portugal, and 49.2% in Spain [31]. Our findings also agree with the 37% resistance to CLA reported in a Pakistan study [32], 20% in Southern European studies [33], 52% in Brazil [34], and 28% in Japan [35]. However, in USA, CLA resistance was found in only 8.6% [36, 37]. Some researchers have reported that a higher prevalence of resistance to both CLA and FLQ was detected in samples from stool compared to biopsy, and they suggested that this could be related to the existence of mixed infections with both resistant and susceptible strains [38]. Whereas this could partly explain our findings, it also could imply that when patients harbouring mixed infections receive antibiotics, the resistant strains naturally become selected for making the drug resistant problem even worse.
In Uganda clarithromycin is often used in first-line anti-PUD regimens, the so-called triple therapy without any susceptibility data or sometimes even a prescription. It is most likely that patients who tend to get recurrent disease might be harbouring super selected drug resistant H. pylori. Future studies need to focus on this area. In low clarithromycin resistance regions, treatment regimens containing clarithromycin are recommended as a first-line empirical treatment. In areas of high clarithromycin resistance such as in Uganda as from this study, first-line empirical treatment should utilize quadruple treatment regimens, that is, bismuth subsalicylate, proton pump inhibitors (PPI), tetracycline, and metronidazole [39]. After failure of the latter regimen, a fluoroquinolone-containing triple therapy should ideally be recommended [39].
The FLQ are usually considered safe oral antibiotics used as second-line drug for H. pylori infection treatment. The 42% resistance to FLQ in Uganda is not only very high but also worrying. In Uganda and many other developing countries with insufficient controls on sale of antibiotics, FLQ are used as first-line antibiotics for other diseases such as enteric upsets, cough, and urinary tract infections (UTIs). This could have led to super selection of the resistant strains and thus the relatively high resistance of H. pylori (42%) reported in the current study. Our findings are in agreement with previous studies where resistance to FLQ was found in 48%, 56%, and 62%, in Japan, China, and Pakistan, respectively [32, 40–42]. Furthermore, it is in agreement with the Lee et al. work carried out from 2003 to 2012 which reported an increased resistance rate of H. pylori to FLQ (levofloxacin and moxifloxacin) from 5% to 28% [12]. However, Hung et al. in their 2009 study in Taiwan found resistance in only 11% [43]. Additionally, Kumala and Rani study of 2006 also reported lower resistance of H. pylori to FLQ (ciprofloxacin, norfloxacin, and ofloxacin at 6.9%, sparfloxacin and gatifloxacin at 2.8%, and levofloxacin and moxifloxacin at 1.4%) [13]. In the developed countries, FLQ are reserved as second-line antibiotics for H. pylori treatment. In Africa, FLQ are first-line antibiotics for many other ailments increasing antibiotic resistance selection pressure, and it remains worrying what options will be there for the 42% patients with FLQ-resistant H. pylori.
5. Conclusions
This preliminary study has shown that stool is a possible specimen for use with the GTHDT in direct detection of H. pylori and its resistance to CLA/FLQ. We have further shown that H. pylori resistance to CLA and FLQ in Uganda is high. We recommend a prospectively designed study to determine the sensitivity and specificity of the GTHDT for diagnosis of H. pylori in stool samples against an established standard reference test and also determine the prevalence of H. pylori resistance to CLA and FLQ on larger sample sizes to guide patient care.
6. Limitations
The main limitation of our study is that we did not have resources for endoscopic biopsy and confirmatory diagnosis of H. pylori as we could not carry out the urease breath test, and we studied a biased population of stool antigen positive samples. However, we strongly believe that the evidence presented in this paper will ignite more studies to be conducted on this topic.
Supplementary Material
Figure 3 (supplementary materials) shows representative hybridization results of the GTHDT on the nitrocellulose strips as obtained in our laboratory. In strip (a) the H. pylori strain was sensitive to both FQL and CLA because of presence of probe bands in the gyrA and 23S rRNA wild type loci but no mutation bands. In strip (b) the H. pylori strain was resistant to both FQL and CLA because of presence of mutation bands in both the gyrA 91 (MUT1) and 23S (MUT3) probe loci. In strip (c) the H. pylori strain was resistant to FQL because of presence of mutation band in the gyrA 91 (MUT1) but sensitive to CLA because of presence of the 23S wildtype probe bands but no mutation bands in the 23S (MUT1-MUT3) probe loci. In strip (d) (negative control) only the conjugate and amplification control bands are seen and no other bands because only PCR water was tested.
Kit insert for the Taytec® Helicobacter pylori antigen test in stool.
Acknowledgments
The authors acknowledge the management and clinicians of Mulago National Referral, Mengo, and Case Medical Centre Hospitals for allowing them to collect samples from their patients. The authors recognize the contribution of the research assistants: Richard Okillan, Emmanuel Oumo, John Enzama, and Christopher Muchwa for recruiting the study participants and collection of samples. Deep and sincere gratitude goes to the Management of MBN Clinical Laboratories for partial sponsorship of this research project and for the excellent laboratory services.
Conflicts of Interest
All authors declare that they have no conflicts of interest.
Authors' Contributions
The authors made the following contributions: Freddie Bwanga conceived the idea, contributed to the development of the study protocol, interpreted results, analysed data, interpreted the data, and prepared the manuscript. Denish Calmax Angol contributed to the development of the study protocol, collected samples, ran the laboratory experiments, participated in data analysis, interpreted results, and prepared the manuscript. Tess Ayazika Kirabo contributed to the development of the study protocols, collection of samples, and laboratory experiments. Alfred Okeng helped in fixing the experimental issues and ran some of the PCR tests. Irene Najjingo ran part of the PCR test. Ponsiano Ocama contributed to the development of the study protocol, was the lead clinician, and critically reviewed the manuscript.
References
- 1.Owen R. J. Helicobacter—species classification and identification. British Medical Bulletin. 1998;54(1):17–30. doi: 10.1093/oxfordjournals.bmb.a011667. [DOI] [PubMed] [Google Scholar]
- 2.Ndip R. N., Malange Takang A. E., Ojongokpoko J. E. A., et al. Helicobacter pylori isolates recovered from gastric biopsies of patients with gastro-duodenal pathologies in Cameroon: current status of antibiogram. Tropical Medicine and International Health. 2008;13(6):848–854. doi: 10.1111/j.1365-3156.2008.02062.x. [DOI] [PubMed] [Google Scholar]
- 3.Tanih N. F., Ndip L. M., Clarke A. M., Ndip R. N. An overview of pathogenesis and epidemiology of helicobacter pylori infection. African Journal of Microbiology Research. 2010;4(6):426–436. [Google Scholar]
- 4.Vu C., Ng Y. Y. Prevalence of Helicobacter pylori in Peptic Ulcer Disease in a Singapore Hospital. Singapore Medical Journal. 2000;41(10):478–481. [PubMed] [Google Scholar]
- 5.Goodwin C. S., Mendall M. M., Northfield T. C. Helicobacter pylori infection. Lancet. 1997;349(9047):265–269. doi: 10.1016/S0140-6736(96)07023-7. [DOI] [PubMed] [Google Scholar]
- 6.Go M. Natural history and epidemiology of Helicobacter pylori infection. Alimentary Pharmacology & Therapeutics. 2002;16:3–15. doi: 10.1046/j.1365-2036.2002.0160s1003.x. [DOI] [PubMed] [Google Scholar]
- 7.Kusters J. G., van Vliet A. H. M., Kuipers E. J. Pathogenesis of Helicobacter pylori infection. Clinical Microbiology Reviews. 2006;19(3):449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asrat D., Nilsson I., Mengistu Y., et al. Prevalence of Helicobacter pylori infection among adult dyspeptic patients in Ethiopia. Annals of Tropical Medicine and Parasitology. 2004;98(2):181–189. doi: 10.1179/000349804225003190. [DOI] [PubMed] [Google Scholar]
- 9.Ndip R. N., Malange A. E., Akoachere J. F. T., Mackay W. G., Titanji V. P. K., Weaver L. T. Helicobacter pylori antigens in the faeces of asymptomatic children in the Buea and Limbe health districts of Cameroon: a pilot study. Tropical Medicine and International Health. 2004;9(9):1036–1040. doi: 10.1111/j.1365-3156.2004.01299.x. [DOI] [PubMed] [Google Scholar]
- 10.Dube C., Nkosi T., Clarke A., Mkwetshana N., Green E., Ndip R. Helicobacter pylori in an asymptomatic population of Eastern Cape Province, South Africa: public health implication. Reviews on Environmental Health. 2009;24(3):249–255. doi: 10.1515/REVEH.2009.24.3.249. [DOI] [PubMed] [Google Scholar]
- 11.Holcombe C. Helicobacter pylori: the African enigma. Gut. 1992;33(4):429–431. doi: 10.1136/gut.33.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J. W., Kim N., Kim J. M., et al. Prevalence of primary and secondary antimicrobial resistance of Helicobacter pylori in Korea from 2003 through 2012. Helicobacter. 2013;18(3):206–214. doi: 10.1111/hel.12031. [DOI] [PubMed] [Google Scholar]
- 13.Kumala W., Rani A. Patterns of Helicobacter pylori isolate resistance to fluoroquinolones, amoxicillin, clarithromycin and metronidazoles. Southeast Asian Journal of Tropical Medicine and Public Health. 2006;37(5):970–974. [PubMed] [Google Scholar]
- 14.Cattoir V., Nectoux J., Lascols C., et al. Update on fluoroquinolone resistance in Helicobacter pylori: new mutations leading to resistance and first description of a gyrA polymorphism associated with hypersusceptibility. International Journal of Antimicrobial Agents. 2007;29(4):389–396. doi: 10.1016/j.ijantimicag.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Tankovic J., Lamarque D., Lascols C., Soussy C. J., Delchier J. C. Clarithromycin resistance of Helicobacter pylori has a major impact on the efficacy of the omeprazole-amoxicillin-clarithromycin therapy. Pathologie Biologie. 2001;49(7):528–533. doi: 10.1016/S0369-8114(01)00209-7. [DOI] [PubMed] [Google Scholar]
- 16.Broutet N., Guillon F., Sauty E., Lethuaire D., Mégraud F. Survey of the in vitro susceptibility of Helicobacter pylori to antibiotics in France-Preliminary results (abstr) Gastroenterology. 1998;43, supplement 2:p. A11. doi: 10.1016/S0016-5085(98)80328-4. [DOI] [Google Scholar]
- 17.Megraud F., Coenen S., Versporten A., et al. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut. 2013;62(1):34–42. doi: 10.1136/gutjnl-2012-302254. [DOI] [PubMed] [Google Scholar]
- 18.Logan R. P. H., Walker M. M. Epidemiology and diagnosis of Helicobacter pylori infection. British Medical Journal. 2001;323(7318):920–922. doi: 10.1136/bmj.323.7318.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricci C., Holton J., Vaira D. Diagnosis of Helicobacter pylori: invasive and non-invasive tests. Best Practice & Research: Clinical Gastroenterology. 2007;21(2):299–313. doi: 10.1016/j.bpg.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Smith S. M., O'Morain C., McNamara D. Antimicrobial susceptibility testing for Helicobacter pylori in times of increasing antibiotic resistance. World Journal of Gastroenterology. 2014;(29):9912–9921. doi: 10.3748/wjg.v20.i29.9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.HainLifescience. GenoType HelicoDR: Molecular Genetic Assay for Identification of Resistance to Fluoroquinolones and Clarithromycin of Helicobacter pylori from Culture and Biopsy Samples. http://www.hain-lifescience.de/en/products/microbiology/helicobacter/genotype-helicodr.html.
- 22.Cambau E., Allerheiligen V., Coulon C., et al. Evaluation of a new test, GenoType HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori. Journal of Clinical Microbiology. 2009;47(11):3600–3607. doi: 10.1128/jcm.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lottspeich C., Schwarzer A., Panthel K., Koletzko S., Rüssmann H. Evaluation of the novel Helicobacter pylori ClariRes real-time PCR assay for detection and clarithromycin susceptibility testing of H. pylori in stool specimens from symptomatic children. Journal of Clinical Microbiology. 2007;45(6):1718–1722. doi: 10.1128/jcm.00103-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.TaytecEnterprises. Helicobcter pylori stool antigen test. Ontario, Canada, 2009, http://www.taytec.ca.
- 25.Qiagen. Sample & Assay Technologies. Hilden, Germany: Qiagen; 2010. QIAamp® DNA stool handbook for DNA purification from stool samples. [Google Scholar]
- 26.Brennan D. E., Omorogbe J., Hussey M., et al. Molecular detection of Helicobacter pylori antibiotic resistance in stool vs biopsy samples. World Journal of Gastroenterology. 2016;22(41):p. 9214. doi: 10.3748/wjg.v22.i41.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schabereiter-Gurtner C., Hirschl A. M., Dragosics B., et al. Novel real-time PCR assay for detection of Helicobacter pylori infection and simultaneous clarithromycin susceptibility testing of stool and biopsy specimens. Journal of Clinical Microbiology. 2004;42(10):4512–4518. doi: 10.1128/jcm.42.10.4512-4518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gisbert J. P., De La Morena F., Abraira V. Accuracy of monoclonal stool antigen test for the diagnosis of H. pylori infection: a systematic review and meta-analysis. American Journal of Gastroenterology. 2006;101(8):1921–1930. doi: 10.1111/j.1572-0241.2006.00668.x. [DOI] [PubMed] [Google Scholar]
- 29.Makristathis A., Hirschl A. M. Detection and clarithromycin susceptibility testing of Helicobacter pylori in stool specimens by real-time PCR: how to get accurate test results. Journal of Clinical Microbiology. 2007;45(8):2756–2757. doi: 10.1128/jcm.00846-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malfertheiner P., Megraud F., O'Morain C. A., et al. Management of Helicobacter pylori infection—the Maastricht IV/ Florence consensus report. Gut. 2012;61(5):646–664. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 31.Nishizawa T., Suzuki H., Hibi T. Quinolone-based third-line therapy for Helicobacter pylori eradication. Journal of Clinical Biochemistry and Nutrition. 2009;44(2):119–124. doi: 10.3164/jcbn.08-220R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajper S., Khan E., Ahmad Z., Alam S. M. Z., Akbar A., Hasan R. Macrolide and fluoroquinolone resistance in Helicobacter pylori isolates: an experience at a tertiary care centre in Pakistan. Journal of the Pakistan Medical Association. 2012;62(11):1140–1144. [PubMed] [Google Scholar]
- 33.Mégraud F. Antibiotic resistance in Helicobacter pylori infection. British Medical Bulletin. 1998;54(1):207–216. doi: 10.1093/oxfordjournals.bmb.a011671. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi I., Murakami K., Kato M., et al. Changing antimicrobial susceptibility epidemiology of Helicobacter pylori strains in Japan between 2002 and 2005. Journal of Clinical Microbiology. 2007;45(12):4006–4010. doi: 10.1128/JCM.00740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vecsei A., Kipet A., Innerhofer A., et al. Time trends of Helicobacter pylori resistance to antibiotics in children living in Vienna, Austria. Helicobacter. 2010;15(3):214–220. doi: 10.1111/j.1523-5378.2010.00753.x. [DOI] [PubMed] [Google Scholar]
- 36.Meyer J. M., et al. Evaluation of risk factors in The Surveillance of Helicobacter pylori Antimicrobial Resistance Partnership (SHARP) in the United States from 1993–1999. Gastroenterology. 2000;118, supplement, article A677 [Google Scholar]
- 37.Solnick J. V. Editorial response: antibiotic resistance in Helicobacter pylori. Clinical Infectious Diseases. 1998;27(1):90–92. doi: 10.1086/514641. [DOI] [PubMed] [Google Scholar]
- 38.Xiong L. J., Tong Y., Wang Z., Mao M. Detection of clarithromycin-resistant helicobacter pylori by stool PCR in children: a comprehensive review of literature. Helicobacter. 2013;18(2):89–101. doi: 10.1111/hel.12016. [DOI] [PubMed] [Google Scholar]
- 39.Nishizawa T., Suzuki H. Mechanisms of Helicobacter pylori antibiotic resistance and molecular testing. Frontiers in Molecular Biosciences. 2014;1:p. 19. doi: 10.3389/fmolb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y. J., Yang J. C., Jeng Y. M., Chang M. H., Ni Y. H. Prevalence and rapid identification of clarithromycin-resistant Helicobacter pylori isolates in children. Pediatric Infectious Disease Journal. 2001;20(7):662–666. doi: 10.1097/00006454-200107000-00005. [DOI] [PubMed] [Google Scholar]
- 41.Moore R. A., Beckthold B., Wong S., Kureishi A., Bryan L. E. Nucleotide sequence of the gyrA gene and characterization of ciprofloxacin-resistant mutants of Helicobacter pylori. Antimicrobial Agents and Chemotherapy. 1995;39(1):107–111. doi: 10.1128/AAC.39.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umegaki N., Shimoyama T., Nishiya D., Suto T., Fukuda S., Munakata A. Clarithromycin-resistance and point mutations in the 23S rRNA gene in Helicobacter pylori isolates from Japan. Journal of Gastroenterology and Hepatology (Australia) 2000;15(8):906–909. doi: 10.1046/j.1440-1746.2000.02072.x. [DOI] [PubMed] [Google Scholar]
- 43.Hung K.-H., Sheu B.-S., Chang W.-L., Wu H.-M., Liu C.-C., Wu J.-J. Prevalence of primary fluoroquinolone resistance among clinical isolates of Helicobacter pylori at a University Hospital in Southern Taiwan. Helicobacter. 2009;14(1):61–65. doi: 10.1111/j.1523-5378.2009.00655.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 3 (supplementary materials) shows representative hybridization results of the GTHDT on the nitrocellulose strips as obtained in our laboratory. In strip (a) the H. pylori strain was sensitive to both FQL and CLA because of presence of probe bands in the gyrA and 23S rRNA wild type loci but no mutation bands. In strip (b) the H. pylori strain was resistant to both FQL and CLA because of presence of mutation bands in both the gyrA 91 (MUT1) and 23S (MUT3) probe loci. In strip (c) the H. pylori strain was resistant to FQL because of presence of mutation band in the gyrA 91 (MUT1) but sensitive to CLA because of presence of the 23S wildtype probe bands but no mutation bands in the 23S (MUT1-MUT3) probe loci. In strip (d) (negative control) only the conjugate and amplification control bands are seen and no other bands because only PCR water was tested.
Kit insert for the Taytec® Helicobacter pylori antigen test in stool.