Abstract
Apoptotic cell death is critical for the early development of the nervous system, but once the nervous system is established, the apoptotic pathway becomes highly restricted in mature neurons. However, the mechanisms underlying this increased resistance to apoptosis in these mature neurons are not completely understood. We have previously found that members of the miR-29 family of microRNAs (miRNAs) are induced with neuronal maturation and that overexpression of miR-29 was sufficient to restrict apoptosis in neurons. To determine whether endogenous miR-29 alone was responsible for the inhibition of cytochrome c release in mature neurons, we examined the status of the apoptotic pathway in sympathetic neurons deficient for all three miR-29 family members. Unexpectedly, we found that the apoptotic pathway remained largely restricted in miR-29-deficient mature neurons. We therefore probed for additional mechanisms by which mature neurons resist apoptosis. We identify miR-24 as another miRNA that is upregulated in the maturing cerebellum and sympathetic neurons that can act redundantly with miR-29 by targeting a similar repertoire of pro-death BH3-only genes. These results reveal that mature neurons engage redundant brakes to restrict the apoptotic pathway and ensure their long-term survival.
Keywords: Neurons, apoptosis, miR-29, miR-24, BH3-only, maturation
Introduction
In recent years, it has become increasingly clear that the threshold to undergo apoptosis can be markedly different in different cell types. For example, primary mitotic cells are sensitive to apoptotic insults, whereas postmitotic cells such as neurons, cardiomyocytes and myotubes have acquired mechanisms for restricting apoptosis [1–14]. Such differences in the regulation of apoptosis are physiologically important because while mitotic cells are at continual risk of becoming cancerous and need to maintain their ability to die rapidly, this risk is significantly lower in terminally differentiated postmitotic cells. Indeed, the ability of organisms to maintain the long-term survival of postmitotic cells such as neurons is critical for normal physiological functions [15].
What is less appreciated is that even within the same cells, the apoptotic pathway can sometimes undergo dynamic changes during and after development. A cell type that exemplifies this phenomenon is neurons, where the apoptotic pathway becomes highly restricted as young neurons become mature [16]. Apoptosis plays an important role in the developing nervous system, where it is estimated that more than 50% of neurons that are initially produced will die by apoptosis as a part of normal neuronal development [17]. However, once the nervous system is fully formed and neurons are appropriately wired, it is physiologically important for these neurons to survive long term. Indeed, the apoptotic pathway becomes highly restricted with neuronal maturation, but the molecular details are not completely understood.
Mouse sympathetic neurons provide an excellent model system for studying the regulation of the apoptotic pathway during neuronal maturation. Young P5 (postnatal day 5) neurons can undergo apoptosis in response to multiple stimuli including nerve growth factor (NGF) withdrawal, DNA damage, and Endoplasmic Reticulum stress. In contrast, those same neurons after 4–5 weeks (P28) become remarkably resistant to the same apoptotic stimuli [7, 18, 19]. Apoptotic stimuli in neurons are known to transcriptionally upregulate multiple redundant members of the pro-apoptotic BH3-only family of proteins [20]. These proteins activate Bax, which then permeablizes the mitochondria to induce the release of cytochrome c (cyt c) into the cytosol [19, 21]. Once in the cytosol, cyt c binds to Apaf-1 (Apoptotic Activating Protease 1) and forms the apoptosome complex with procaspase-9. Autoactivation of caspase-9 on the apoptosome can then activate caspase-3 to ultimately trigger cell death [22].
We and others have previously shown that one mechanism by which mature neurons become resistant to apoptosis is via the epigenetic silencing of Apaf-1[4, 23–25]. However, mature neurons induced to undergo apoptosis with NGF deprivation fail to release cyt c, (a process that is unaffected by Apaf-1 levels), despite maintaining Bax levels [18, 19]. Interestingly, mature neurons still undergo the initial steps in the apoptotic pathway, such as c-jun phosphorylation, after NGF deprivation [18]. This suggested the presence of one or more brakes upstream of cyt c release but downstream of c-jun phosphorylation in mature neurons. Indeed, mature neurons were found to have markedly elevated levels of the microRNA (miRNA) miR-29, which targets and represses multiple redundant members of the BH3-only family of proteins [7]. A neuroprotective role for miR-29 is further supported by the results of in vivo models of neuronal insult, which have found that overexpression of miR-29 is able to reduce cell death in ischemic stroke [26–28], spinal cord injury [29], and ethanol-induced toxicity [30]. However, it is unclear at this point whether miR-29 induction is the only brake employed to inhibit cyt c release in mature neurons, or if other redundant brakes also exist.
We find that deletion of all three miR-29 family members fails to re-sensitize mature neurons to apoptosis, with mature miR-29 knockout neurons remaining significantly resistant to cyt c release induced by NGF deprivation. Here we report that another miRNA, miR-24, is also upregulated with neuronal maturation and is capable of acting redundantly with miR-29 to inhibit cyt c release by targeting a similar subset of BH3-only genes. Our results highlight the ability of mature neurons to engage multiple, redundant mechanisms to restrict the apoptotic pathway and help ensure their long-term survival.
Results
Maturing neurons simultaneously restrict apoptosis both pre- and post-mitochondria
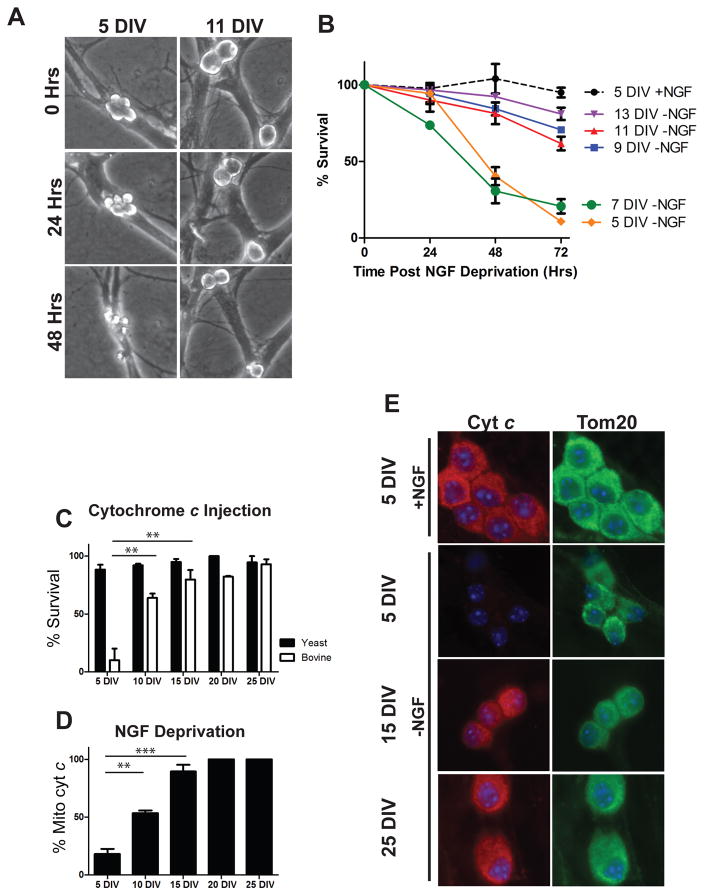
In previous studies of neuronal maturation, the mechanisms by which mature neurons become resistant to apoptosis have been investigated in post-natal day 28 neurons. At this time-point, neurons engage multiple mechanisms to restrict apoptosis both up-stream and down-stream of mitochondrial permeablization. However, the exact timing for when these brakes are initiated during the maturation process is not known. Therefore, to investigate the time course and mechanisms by which neurons become resistant to apoptosis with maturation, we matured neurons for increasing lengths of time and assessed their susceptibility to apoptotic stimuli. Sympathetic neurons were isolated from neonatal (postnatal day 0–1) mouse pups and maintained in culture for 5–25 days. Apoptosis was then induced by deprivation of Nerve Growth Factor (NGF). While neurons maintained in culture for 5–7 days remained vulnerable to NGF deprivation-induced apoptosis, marked resistance to apoptosis was observed by as early as 9 days in vitro (DIV) (Figure 1A, B).
Figure 1.
Neuronal maturation is associated with progressive resistance to neuronal apoptosis at both pre and post-mitochondrial checkpoints. A) Representative images of identical fields of 5 DIV and 11 DIV neurons imaged at 24 hour intervals after NGF deprivation. B) Quantification of sympathetic neuronal survival in response to NGF deprivation for 48 hours after maturing in culture for the indicated amount of time. Data represent mean ± SEM of 3 independent experiments. C) Neurons isolated from neonatal XIAP−/− mice and cultured for the indicated time were injected with 10 mg/mL of either bovine cyt c to induce apoptosis or yeast cyt c as a negative control and survival was quantified 24 hours post-injection. Data are displayed as mean ± SEM of 3 independent experiments for 5, 10 and 15 DIV and 2 independent experiments for 20 and 25 DIV(**=P<0.01). D) Quantification of cyt c release from neurons cultured for the indicated time and deprived of NGF for 48 hours (**=P<.01, ***=P<.001). E) Representative images of cyt c and Tom20 staining in neurons matured for the indicated amount of time and maintained (+NGF) or deprived (-NGF) for 48 hours.
Neurons have been previously shown to restrict apoptosis both before and after the mitochondrial checkpoint. To more precisely define whether these two separate brakes were engaged simultaneously or sequentially, we specifically assessed apoptosis restriction at the pre- and post-mitochondrial checkpoint. First, to assess the post-mitochondrial resistance to apoptosis, we microinjected maturing neurons with purified cyt c. These experiments were conducted in neurons from XIAP-deficient mice as XIAP (X-Linked Inhibitor of Apoptosis) is known to inhibit cyt c-induced apoptosis even in young neurons [1]. As reported earlier, neurons (XIAP−/−) maintained for 5 DIV exhibited massive cell death in response to cytosolic cyt c injection, with virtually 100% of neurons dying within 24 hours of injection. Approximately 50% of neurons became resistant to cyt c injections by 10 DIV, with greater than 80% exhibiting resistance by 15 DIV (Figure 1C).
Second, to assess pre-mitochondrial resistance to apoptosis, we subjected maturing neurons to NGF deprivation and assessed whether cyt c was maintained at the mitochondria or released into the cytosol by immunofluorescence. As mitochondrially-released cyt c is rapidly degraded in neurons, loss of cyt c signal is an established indicator of its release from mitochondria in these cells [8]. Consistent with the results of our neuronal survival experiments, neurons by 10 DIV exhibited significant resistance to NGF-deprivation induced cyt c release, with approximately 50% of neurons maintaining cyt c after 48 hrs of NGF deprivation. Nearly complete resistance at this pre-mitochondrial checkpoint was seen in 15 DIV neurons (Figure 1D, E). Together, these results show that both pre- and post-mitochondrial brakes are engaged concurrently in maturing neurons, resulting in resistance to apoptosis within 2 weeks in culture.
Loss of miR-29 is not sufficient to re-sensitize mature neurons to apoptosis
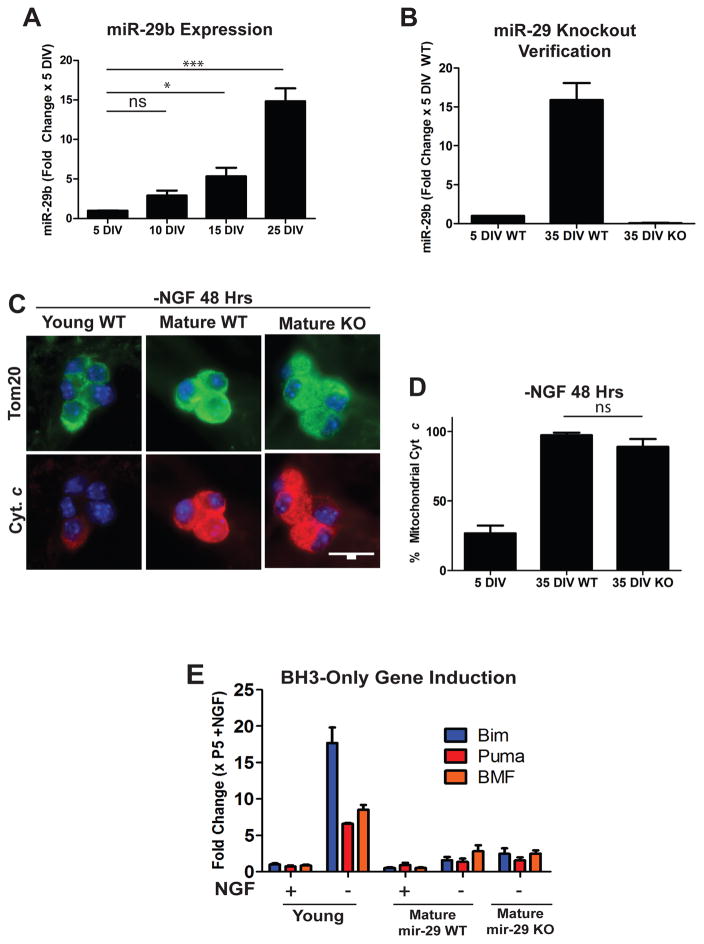
Recent studies in our lab have identified the miR-29 family as regulators of apoptosis in maturing neurons. miR-29 is induced during neuronal maturation (Figure 2A), and can inhibit the induction of multiple members of the BH3-only family of proteins to prevent Bax activation and apoptosis [7]. We hypothesized that loss of miR-29 expression may render mature neurons sensitive to apoptosis. miR-29 has three family members (miR-29a,b,c) that are expressed from two separate genomic loci. Thus, to completely delete miR-29 in sympathetic neurons, mice floxed at both miR-29 loci were crossed with a mouse line expressing Cre recombinase under a tamoxifen inducible promoter (ER-Cre). Neurons isolated from these mice were then treated with 4-hydroxy-tamoxifen in vitro to induce recombination and generate miR-29 deficient neurons. Loss of miR-29 expression was verified by RT-qPCR comparing cre-positive (hereafter referred to as miR-29 KO) and Cre-negative (hereafter referred to as miR-29 WT) littermates, with mature miR-29 KO neurons exhibiting virtually no detectable miR-29 as compared to WT controls (Figure 2B).
Figure 2.
Mature miR-29 deficient neurons remain resistant to NGF-deprivation induced cyt c release. A) Timecourse of miR-29b induction in maturing neurons. Neurons were isolated from neonatal mice and cultured for the indicated amount of time and miR-29 levels were assessed by RT-qPCR. Values are expressed as mean fold change ± SEM relative to 5 DIV neurons from 3 independent experiments (**=p<0.01, ***=p<0.001, ns=not significant). B) Verification of miR-29 deficiency in mature mir-29 knockout neurons as assessed by RT-qPCR. Values are expressed as mean fold change ± SEM relative to WT 5 DIV neurons from 3 independent experiments. C) Representative images of cyt c staining in mature WT and miR-29 KO cells deprived of NGF for 48 hours. D) Quantification of cyt c release in young WT neurons and mature WT and mature miR-29 KO neurons deprived of NGF for 48 hours. Data represent the percentage of cells with mitochondrial cyt c and are presented as mean ± SEM of 3 independent experiments. E) RT-qPCR quantification of selected BH3-only genes in young, mature WT, and mature miR-29 KO neurons deprived of NGF for 48 hours. Fold changes were normalized to P5 +NGF neurons and represent mean ± SEM of 3 independent experiments.
miR-29 WT and miR-29 KO neurons were matured in culture until 35 DIV and subjected to NGF deprivation to induce apoptosis. Since miR-29 is known to inhibit apoptosis upstream of the mitochondria (and mature neurons are known to engage an additional brake downstream of mitochondria), we specifically examined mitochondrial release of cyt c as a readout for these experiments. We were surprised to find that mature miR-29 KO neurons still exhibited negligible cyt c release in response to NGF deprivation (Figure 2C, D).
Since miR-29 is known to be able to target multiple pro-apoptotic BH3-only domain genes and inhibit their induction, we next assayed BH3-induction in young wild-type, mature wild-type, and mature miR-29 knockout neurons. Consistent with our findings that mature miR-29 knockout neurons fail to release cyt c, we found that miR-29 knockout neurons also failed to substantially induce BH3-only domain genes in response to 48 hours of NGF deprivation when compared to young neurons as measured by RT-qPCR (Figure 2E). These results indicate that loss of miR-29 expression alone is not sufficient to re-sensitize mature neurons to apoptosis, and led us to hypothesize that other brakes could be acting redundantly with miR-29 to the induction of BH3-only genes, cyt c release, and apoptosis in mature neurons.
Other microRNAs with predicted targets in the apoptotic pathway are upregulated during neuronal maturation
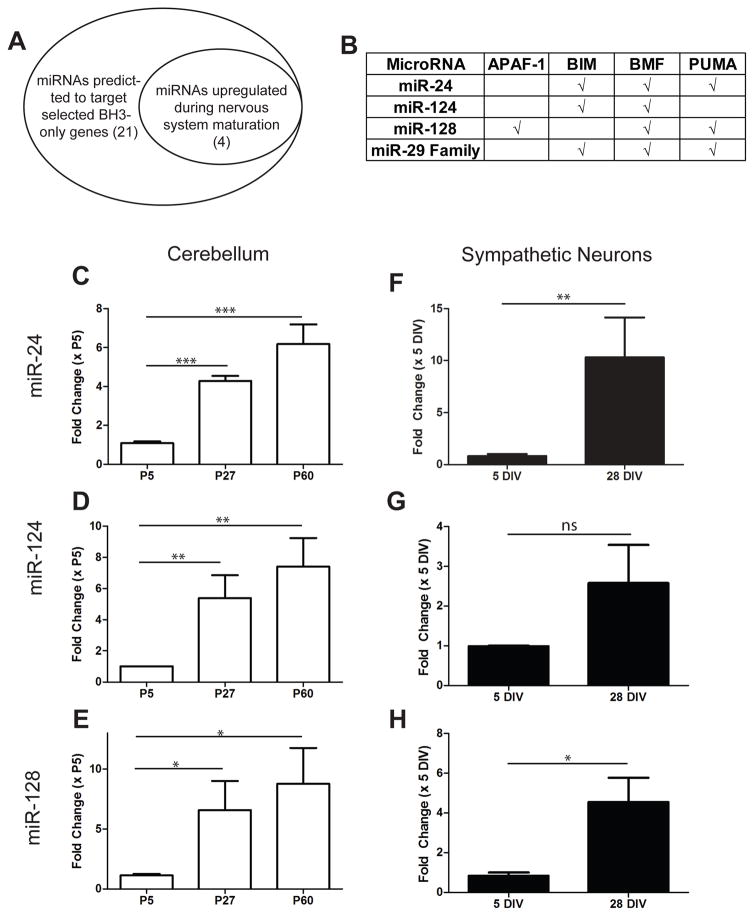
To identify other miRNAs that are induced during maturation and capable of acting redundantly with miR-29 to inhibit cyt c release and apoptosis, we utilized two approaches (Figure 3A). First, we used TargetScan software (www.targetscan.org) to identify predicted miRNAs that could target the BH3-only genes. Second, we reviewed existing literature on miRNAs that are known to increase with brain maturation, in addition to performing small-RNA-Seq on maturing cerebellum, to identify miRNAs that were upregulated in maturing neurons. While the fold increase in miR-29 was the most striking (Figure 2A, Supplementary Table S1), we identified three other miRNAs upregulated more than 5-fold in maturing neurons that were predicted to target the 3′ UTRs of multiple BH3-only family genes: miR-24, miR-124, and miR-128 (Figure 3B). We confirmed the upregulation of these miRNAs during neuronal maturation using RT-qPCR in developing cerebellum (Figure 3C–E) and sympathetic neurons (Figure 3F–H). These results identify miR-24, miR-124, and miR-128 as potential candidates that could act redundantly with miR-29 to restrict apoptosis in mature neurons.
Figure 3.
Other miRNAs predicted to regulate the apoptotic pathway are also induced with neuronal maturation. A) Schematic showing candidate pools of miRNAs that may also regulate cell death in maturing neurons. B) Table of predicted targets for candidate miRNAs by TargetScan software. C–E) Relative expression levels of candidate miRNAs in maturing cerebellum. Values are expressed relative to young (P5) cerebellum and represent mean ± SEM of 3 independent experiments. F–H) Relative expression levels of candidate miRNAs in young (5 DIV) and mature (28 DIV) sympathetic neurons measured by RT-qPCR. Values are expressed as fold-change relative to expression in young neurons and represent mean ± SEM of 3 independent experiments (*=P<0.05 **=p<0.01 ***=P<0.001, ns=not significant).
Overexpression of miR-24 in Young Neurons Inhibits Cyt c Release and Cell Death
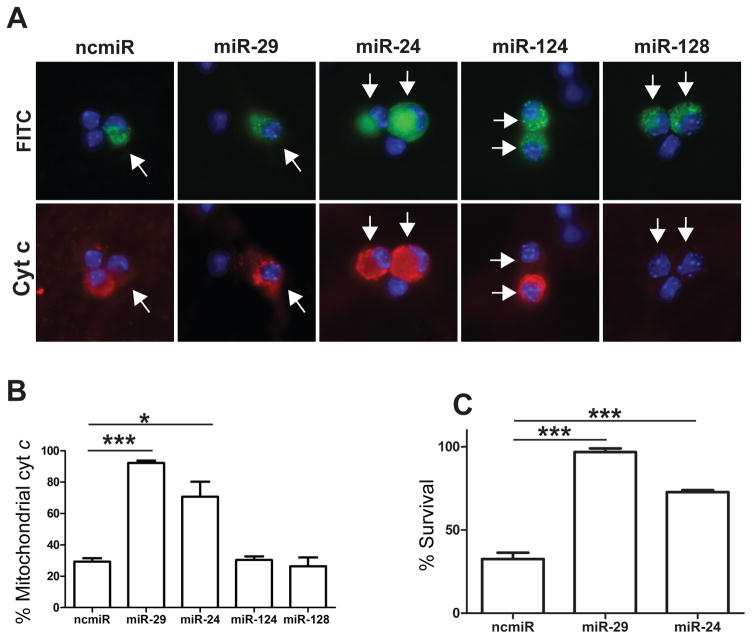
To determine whether any of the selected miRNAs were capable of inhibiting BH3-only gene induction and cell death in a manner similar to miR-29, we overexpressed mimics to the selected miRNAs in young neurons using single-cell microinjection. Neurons isolated from neonatal mice were injected at 3 DIV with selected mimics or negative control miRNA and, after 48 hours, were deprived of NGF to induce apoptosis. As a positive control, we also injected neurons with a mimic to miR-29b which, as reported previously, inhibited cyt c release in young neurons. Among the candidate miRNAs, we found only miR-24 overexpression to be capable of inhibiting cyt c release in neurons deprived of NGF (Figure 4A, B). The ability of miR-24, but not miR-124 and miR-128, to inhibit cyt c release is likely because miR-24 is predicted to target more members of the redundant BH3-only family than either miR-124 or miR-128 (Figure 3B). Importantly, we also examined whether miR-24 expression could inhibit apoptosis. Consistent with the observed effect on cyt c release, we found that miR-24 significantly inhibits apoptosis in NGF-deprived young neurons (Figure 4C).
Figure 4.
Overexpression of miR-29 or miR-24 is sufficient to inhibit cyt c release and cell death in young, NGF deprived neurons. A) Representative images of sympathetic neurons injected with mimics to candidate miRNAs or negative control mimic. Injected cells (arrows) are marked with FITC-Dextran (green) and cyt c release (red) was assessed after 48 hours of NGF deprivation. B) Quantification of cyt c release in injected cells after 48 hours of NGF deprivation. Data are represented as mean ± SEM of 3 independent experiments (*=P<0.05, ***=P<0.001). C) Quantification of neuronal survival in neurons injected with mimics to miR-29, miR-24, or a negative control (ncmiR) after 48 hours of NGF deprivation. Survival is expressed as the percentage of cells remaining compared to the number alive pre-deprivation. Data represent mean ± SEM of at least 3 independent experiments.
miR-24 Can Inhibit Bim and Puma Induction in NGF-Deprived Neurons
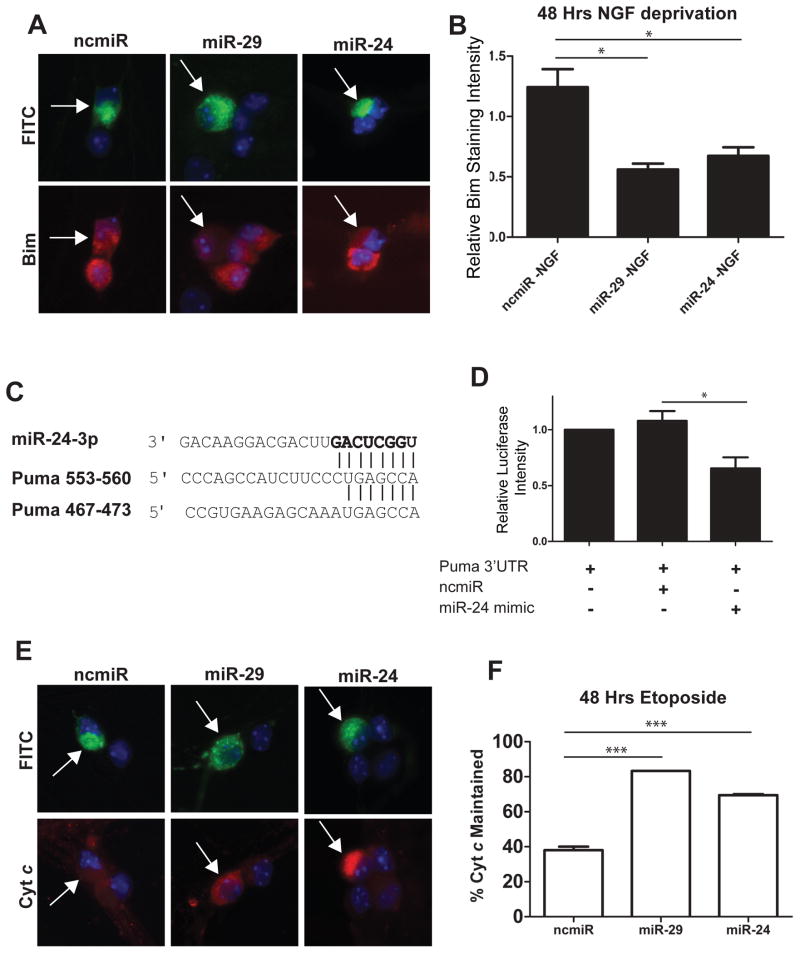
While previously published reports have validated miR-24 binding sites in the Bim 3′UTR, and miR-24 has been previously shown to downregulate Bim expression and inhibit apoptosis in cardiac tissue [31], its potential role in regulating neuronal apoptosis has not been investigated. To examine if miR-24 could inhibit Bim induction in neurons, young (P3 equivalent) sympathetic neurons were injected with mimics to miR-24, miR-29 (positive control), or a negative control miRNA, and were then subjected to NGF deprivation to trigger Bim induction. Just as seen with miR-29, miR-24 expression potently suppressed Bim induction in NGF-deprived neurons (Figure 5A, B).
Figure 5.
Overexpression of miR-29 or miR-24 is sufficient to inhibit the induction of Bim and Puma in young sympathetic neurons. A) Representative images of Bim staining in neurons. Neurons were injected at 3 DIV with mimics to miR-29, miR-24, or a negative control, along with FITC-Dextran (Green) to mark injected cells. At 5 DIV neurons were deprived of NGF and after 48 hours of NGF deprivation, neurons were fixed and stained for Bim (red). B) Quantification of normalized Bim staining intensity. Bim staining intensity was measured, and values for injected cells were normalized to NGF-deprived, mock injected neurons. Data presented as mean intensity ± SEM of 3 independent experiments (*=P>0.05). C) Sequence and alignment of miR-24 seed sequence with two putative miR-24 target sites in Puma 3′UTR. D) Luciferase activity was measured 48 hours after transfection in HEK293T cells transfected with reporter plasmids containing the Puma 3′UTR fused to a firefly luciferase gene. Plasmids were transfected either alone or with 100 nM mimics of miR-24 or negative control. Expression was normalized by measuring the ratio of firefly to renilla luciferase. Values are plotted relative to vector alone and represent mean ± SEM of 3 independent experiments (*=P<0.05). E) Representative images of cyt c staining in neurons injected with mimics to miR-29, miR-24, or negative control miR (ncmiR) and treated with 20 μM etoposide. Green indicates injected cells (arrows). F) Quantification of cyt c release in young neurons injected with mimics to miR-29, miR-24, or negative control after 48 hours of etoposide treatment. Data are plotted as mean ± SEM of 3 independent experiments (***=P<0.001).
We next sought to determine if miR-24 could also target Puma, another BH3-only family member that has been found to act redundantly with Bim to induce neuronal apoptosis [32]. Puma has two conserved miR-24 sites in its 3′UTR (Figure 5C). To validate these sites, we cloned the 3′UTR of Puma into a luciferase reporter vector. We then cotransfected this vector with either a negative control miRNA or a miR-24 mimic into HEK293T cells. We found that miR-24 was indeed able to significantly repress the expression of luciferase in cells expressing the Puma 3′UTR compared to a negative control (Figure 5D).
To test the functional ability of miR-24 expression to inhibit Puma, we took advantage of the fact that in sympathetic neurons, cyt c release and apoptosis in response to DNA damage is known to be entirely dependent on Puma expression [33]. Thus, we examined whether miR-24 expression was able to effectively inhibit cyt c release in young neurons treated with the DNA damaging agent etoposide. Consistent with our observation that miR-24 is able to target Puma, we found that the release of cyt c was inhibited in young neurons expressing miR-24 in response to etoposide treatment (Figure 5E, F). Together, these results identify miR-24 as a miRNA that is not only induced with neuronal maturation, but as seen with miR-29, also targets multiple BH3-only genes and inhibit neuronal apoptosis.
Discussion
The ability of maturing neurons to dynamically switch the apoptotic pathway from a permissive to a restrictive state is of critical importance. This allows young neurons to permit physiological apoptosis during nervous system development, but once established, to highly restrict apoptosis to promote the long term survival of mature neurons. Previous studies had identified miR-29-mediated inhibition of BH3-only genes and the epigenetic silencing of the Apaf-1 promoter as mechanisms by which apoptosis is restricted pre- and post-mitochondria in mature sympathetic neurons [4, 7]. Our results now highlight the finding that the apoptotic pathway is even more restricted in adult neurons than previously appreciated, with mature neurons engaging redundant brakes at the pre-mitochondrial checkpoint.
Our previous results brought focus on the neuroprotective capability of miR-29 in mature neurons. Not only is miR-29 markedly induced with neuronal maturation but expression of miR-29 alone is sufficient to inhibit apoptosis in young neurons by its ability to target the BH3-only gene family. Delivery of miR-29 can also confer neuroprotection in models of stroke and spinal cord injury and alcohol-induced toxicity in vivo [26–30]. Our results now show that, while overexpression of miR-29 promotes neuronal survival, loss of miR-29 failed to restore the ability of mature neurons to release cyt c in response to NGF-deprivation. Thus, endogenous miR-29 does not seem to be the only brake restricting apoptosis at the pre-mitochondrial checkpoint in mature neurons. These results prompted us to identify other brakes that function redundantly with miR-29 to effectively inhibit apoptosis in mature neurons.
Our finding that miR-24 is upregulated in maturing cerebellum and sympathetic neurons is consistent with recent findings that miR-24 expression is up-regulated in maturing cerebral cortex [34]. miR-24 is also a miRNA that has been found to be a key regulator of apoptosis in ischemic heart muscle through its ability to target Bim, a BH3-only protein also known to be important for neuronal apoptosis [35]. The observation that the upregulation correlates with an increase in resistance to apoptosis made miR-24 an attractive candidate as a molecule that may act redundantly with miR-29 to inhibit apoptosis. Indeed, our results show that expression of mir-24 in young sympathetic neurons can target the BH3-only proteins Bim and Puma, which are thought to be the two most important BH3-only proteins for neuronal cell death [32], and inhibit apoptosis at the level of cyt c release. It is also interesting to note that miR-24 is expressed in two separate clusters with two other miRNAs, miR-23a/b and miR-27a/b. Recent studies have found that miR-23a/b and miR-27a/b are also capable of inhibiting neuronal apoptosis in cases of traumatic brain injury or ischemia [34, 36]. Thus, miRNAs of this cluster may work synergistically to inhibit the apoptotic pathway in the adult brain.
Increased resistance to apoptosis with maturation has been observed in many populations of neurons in response to diverse insults. For example, the neonatal brain is far more vulnerable to hypoxia ischemia-induced apoptosis or traumatic brain injury than the adult brain [37]. Likewise, greater numbers of neurons survive nerve crush or axotomy if the injury is done on 3-week-old mice as compared to 1-week-old mice [38]. Our work provides mechanistic insight into the multiple brakes engaged by mature neurons that allow these cells to withstand diverse stresses and survive long-term.
An interesting question here is why do mature neurons not shut down all the components of the apoptotic pathway? Mature sympathetic neurons repress Apaf-1 but continue to express many apoptotic proteins including Bax, caspase-9 and caspase-3 [4]. A potential explanation for this has come from recent studies that have found these proteins to have functions outside of the canonical apoptotic pathway. For example, Bax is known to regulate mitochondrial dynamics and caspases-9 and -3 have essential roles in synaptic plasticity and axon pruning, which are events that are important for optimal neuronal function and plasticity [39]. Thus, maintaining these proteins, albeit with increased regulation, permits mature neurons to utilize these proteins while simultaneously limiting the risk of apoptosis.
Despite the mechanisms described here that provide neurons with improved capacity to survive injury and apoptosis, adult neurons can still be vulnerable and undergo cell death in situations of acute brain injury or neurodegenerative disease. It is possible that even the partial removal of these apoptotic brakes that could occur with injury or chronic neurodegeneration could increase the vulnerability of adult neurons. Indeed, consistent with a role of miR-29 in neuroprotection, its levels are reduced in Alzheimer’s disease, Huntington’s disease, and during hypoxic-ischemic brain injury [27, 40, 41]. Restoring these brakes on the apoptotic pathway via overexpression of miR-29 or miR-24 could be an effective therapeutic strategy that promotes long-term neuroprotection. A previous study reported that mice partially deleted for miR-29 (deletion of the miR-29a/b1 loci) had grossly normal brains but exhibited an ataxic phenotype [42]. Our findings that apoptosis is not increased in miR-29 knockout neurons support the hypothesis that observed phenotypes in these studies are more likely to be due to the effect of miR-29 on neuronal function rather than survival.
Together, our results identify miR-29 as sufficient but not solely responsible for the inhibition of the apoptotic pathway in mature neurons, and highlight the redundant brakes employed by mature neurons to inhibit apoptosis and promote long-term survival.
Materials and Methods
Cell Culture
For young neurons, sympathetic ganglia were isolated and cultured from neonatal CD1 mice unless otherwise indicated as described previously [1]. All animal handling and protocols were carried out in accordance with established practices as described in the National Institutes of Health Guide for Care and Use of Laboratory Animals and as approved by the Animal Care and Use Committee of the University of North Carolina (UNC).
Neurons were then maintained in culture for 5 days. For mature neurons, ganglia from P5-P12 mice were isolated and cultured as previously described until 28–35 DIV [4]. NGF deprivation experiments were performed by washing cells three times with media containing no NGF, then refeeding cells in NGF-free media containing NGF-neutralizing antibody. For DNA damage experiments, cells were treated with 20 μM Etoposide (Sigma). For luciferase assay experiments, HEK293T cells were grown in DMEM/F12 medium supplemented with 10% FBS, 100u/mL penicillin, and 100μg/mL streptomycin.
RNA Isolation and RT-qPCR
RNA was isolated from tissue or cultured cells using the Zymo Research Direct-zol RNA MiniPrep kit (Genesee) according to the manufacturer’s instructions. Mature miRNA expression was determined using hydrolysis probe based-miRNA assays (Taqman/Life Technologies). RT primers specific for miR-29a, miR-29b, miR-29c, miR-24, miR-124, miR-128 and U6 were used to amplify the indicated genes from 10 ng of isolated RNA using the Superscript III Reverse Transcriptase system (Life Technologies) according to manufacturer instructions. cDNA was amplified using TaqMan universal PCR master mix (Life Technologies) on an ABI 7500 Real-Time PCR system. Relative quantification and statistical comparisons were performed using the delta-delta-ct method. Samples were internally normalized to U6 SnoRNA expression. For non-miRNA gene expression, cDNA libraries were prepared using 50–100 ng RNA. RNA was pre-treated with RQ1 DNase (Promega) for 30 minutes at 37°C followed by 10 min incubation with DNAse Stop Solution at 65°C for 10 min. cDNA was reverse-transcribed using random hexamers (Invitrogen) and the Superscript III Reverse Transcriptase System according to manufacturer instructions. cDNA was diluted 1:20 in each qPCR reaction, along with 400 nM of forward and reverse primers and Power SYBR Green PCR master mix (Applied Biosystems). BH3-only gene and GAPDH primers have been previously published [7]. Reactions were amplified in an ABI 7500 Real-Time PCR system. Relative quantification and statistical comparisons were performed using the delta-delta CT method. Samples were internally normalized to GAPDH expression.
Small RNA sequencing library production and mapping
Libraries for Illumina sequencing were prepared using a modification of the TruSeq protocol. Briefly, 1 ug total RNA was ligated to 3 pmol of the 3′ linker using T4 RNA ligase 2. RNA size fractions corresponding to 35–70 nucleotides (insert plus linker) were gel isolated and ligated to 3 pmol of the 5′ linker. Products were reverse transcribed, PCR amplified to mid-log phase, and size isolated. Libraries were barcoded using indexed 5′ linkers. Libraries were sequenced on an Illumina HiSeq 2000. These libraries were aligned to the mm9 genome. miRNA annotations were downloaded from miRBase r18.
Immunofluorescence Staining
Immunofluorescence was carried out as previously described [1]. The primary antibodies used were as follows: anti-cyt c (BD Biosciences #556432); anti-Tom20 (Santa Cruz sc11415); anti-Bim (Cell Signaling #2189). Secondary antibodies were anti-mouse Cy3 (The Jackson Laboratory) or anti-rabbit Alexa Fluor 488 (Life Technologies). Nuclei were stained with Hoechst 33258 (Molecular Probes). For analysis of cyt c release, neurons treated with apoptotic stimuli in the presence of 25 μM Q-VD-OPH to inhibit downstream caspase activation and preserve cells for staining. Images were acquired using an ORCA-ER digital B/W charge-coupled device camera (Hamamatsu) mounted to a DMI6000 microscope (Leica) using Metamorph 7.6 software and processed using Adobe Photoshop. For fluorescence intensity measurements, average pixel intensities were measured in individual injected neuronal cell bodies using Metamorph 7.6 software and were normalized to neighboring uninjected cell bodies on the same plate.
Assessment of Neuronal Survival
Neuronal survival was assessed by the presence of intact, phase bright cell bodies at the indicated time after treatment. Survival was quantified as the percentage of healthy cells at the indicated time point compared to immediately prior to treatment. This method of assessing survival correlates well with independent methods of measuring cell death such as trypan blue exclusion and calcein AM staining [1].
Cloning of Puma 3′UTR and Luciferase Assays
A 719-bp segment of the Puma 3′UTR was amplified from mouse genomic DNA and cloned into a modified PGL3-control plasmid (Promega), in which the multiple cloning site was placed downstream of the firefly luciferase gene. For luciferase assays, 60,000 HEK293T cells were plated into each well of a 12-well plate and transfected with 1.5 μg of PGL3-3′UTR reporter construct or empty vector, 100 ng phRL renilla luciferase (Promega) and 100 nM of either miR-24-3p or cel-mir-67 MIRIDIAN mimic (GE Dharmacon) as a negative control. Transfections were performed using lipofectamine 2000 according to manufacturer’s instructions. 48 hours post transfection, cells were lysed and firefly- and renilla-luciferase intensities were measured using Promega Dual-Luciferase Reporter system on a Fluorscan Ascent Type 379 fluorescence plate reader (Thermo). Firefly luciferase intensity was normalized to renilla luciferase intensity to control for cell number and transfection efficiency.
Single Cell Microinjection
Cells were injected with Miridian mimics (GE Dharmacon) to miR-29, miR-24, miR-124, miR-128 or cel-miR-67 as a negative control (30 uM needle concentration) as previously described [7]. Briefly, mimics were dissolved in sterile RNAse-free water and mixed with microinjection buffer containing 100 mM KCl and 10 mM KPi (pH 7.4) along with 8 mg/mL lysine-fixable Fluorescein Dextran (Invitrogen) to mark injected cells. For cyt c injections, 10 mg/mL of mammalian (bovine) or yeast cyt c were injected as previously described [1] and cell survival was quantified by comparing the percentage of surviving cells immediately after injection and 24 hours post-injection.
Generation of miR-29 knockout neurons
To generate sympathetic neurons deficient for all three miR-29 family members, mice floxed at both genomic loci for miR-29 (miR-29a/b1 and miR-29b2/c)(kindly provided by Dr. He, Duke University) were crossed with ER-Cre (CAG-Cre/Esr1/ strain: 004453. Jackson Laboratories). Neurons were isolated from Cre-positive and Cre-negative littermates as previously described and treated with 500 nM 4-OH-tamoxifen (Sigma) for 10 days to induce recombination. Neurons were then matured until 28–35 DIV prior to experimental treatments. Cre-positive and negative littermates were genotyped for Cre expression using the following primers: Forward – gatggacatgttcagggatcgcc. Reverse: ctcccaccgtcagtacgtgagat. Knockout of miR-29 family expression was confirmed via RT-qPCR.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism Software version 5.0c. For comparisons between two means, unpaired student’s T-Test was used. For comparisons between more than two groups, one-way ANOVA with Newman-Keuls multiple comparisons test was used. A p-value of less than 0.05 was considered significant.
Acknowledgments
We would like to thank members of the Deshmukh lab for their critical review of the manuscript. We also thank Dr. JrGang Cheng for help with generating the luciferase construct. This work was supported by NIH grants NS042197 and NS083384 to MD.
References
- 1.Potts PR, Singh S, Knezek M, Thompson CB, Deshmukh M. Critical function of endogenous XIAP in regulating caspase activation during sympathetic neuronal apoptosis. J Cell Biol. 2003;163:789–799. doi: 10.1083/jcb.200307130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright KM, Linhoff MW, Potts PR, Deshmukh M. Decreased apoptosome activity with neuronal differentiation sets the threshold for strict IAP regulation of apoptosis. J Cell Biol. 2004;167:303–313. doi: 10.1083/jcb.200406073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Potts MB, Vaughn AE, McDonough H, Patterson C, Deshmukh M. Reduced Apaf-1 levels in cardiomyocytes engage strict regulation of apoptosis by endogenous XIAP. J Cell Biol. 2005;171:925–930. doi: 10.1083/jcb.200504082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright KM, Smith MI, Farrag L, Deshmukh M. Chromatin modification of Apaf-1 restricts the apoptotic pathway in mature neurons. J Cell Biol. 2007;179:825–832. doi: 10.1083/jcb.200708086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaughn AE, Deshmukh M. Glucose metabolism inhibits apoptosis in neurons and cancer cells by redox inactivation of cytochrome c. Nat Cell Biol. 2008;10:1477–1483. doi: 10.1038/ncb1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith MI, Huang YY, Deshmukh M. Skeletal muscle differentiation evokes endogenous XIAP to restrict the apoptotic pathway. PLoS One. 2009;4:e5097. doi: 10.1371/journal.pone.0005097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kole AJ, Swahari V, Hammond SM, Deshmukh M. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 2011;25:125–130. doi: 10.1101/gad.1975411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gama V, Swahari V, Schafer J, Kole AJ, Evans A, Huang Y, Cliffe A, Golitz B, Sciaky N, Pei X-H, Xiong Y, Deshmukh M. The E3 ligase PARC mediates the degradation of cytosolic cytochrome c to promote survival in neurons and cancer cells. Sci Signal. 2014;7:ra67. doi: 10.1126/scisignal.2005309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao R, Ferry A, Dupont-Versteegden E. Cell death-resistance of differentiated myotubes is associated with enhanced anti-apoptotic mechanisms compared to myoblasts. Apoptosis. 2011;16:221–234. doi: 10.1007/s10495-010-0566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchis D, Mayorga M, Ballester M, Comella JX. Lack of Apaf-1 expression confers resistance to cytochrome c-driven apoptosis in cardiomyocytes. Cell Death Differ. 2003;10:977–986. doi: 10.1038/sj.cdd.4401267. [DOI] [PubMed] [Google Scholar]
- 11.Mayorga M, Bahi N, Ballester M, Comella JX, Sanchis D. Bcl-2 is a key factor for cardiac fibroblast resistance to programmed cell death. J Biol Chem. 2004;279:34882–34889. doi: 10.1074/jbc.M404616200. [DOI] [PubMed] [Google Scholar]
- 12.Oláh G, Szczesny B, Brunyánszki A, López-García IA, Gerö D, Radák Z, Szabo C. Differentiation-Associated Downregulation of Poly(ADP-Ribose) Polymerase-1 Expression in Myoblasts Serves to Increase Their Resistance to Oxidative Stress. PLoS ONE. 2015;10:e0134227. doi: 10.1371/journal.pone.0134227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nam YJ, Mani K, Ashton AW, Peng CF, Krishnamurthy B, Hayakawa Y, Lee P, Korsmeyer SJ, Kitsis RN. Inhibition of both the extrinsic and intrinsic death pathways through nonhomotypic death-fold interactions. Mol Cell. 2004;15:901–912. doi: 10.1016/j.molcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Benn SC, Perrelet D, Kato AC, Scholz J, Decosterd I, Mannion RJ, Bakowska JC, Woolf CJ. Hsp27 Upregulation and Phosphorylation Is Required for Injured Sensory and Motor Neuron Survival. Neuron. 2002;36:45–56. doi: 10.1016/s0896-6273(02)00941-8. [DOI] [PubMed] [Google Scholar]
- 15.Wright KM, Deshmukh M. Restricting apoptosis for postmitotic cell survival and its relevance to cancer. Cell Cycle. 2006;5:1616–1620. doi: 10.4161/cc.5.15.3129. [DOI] [PubMed] [Google Scholar]
- 16.Kole AJ, Annis RP, Deshmukh M. Mature neurons: equipped for survival. Cell Death Dis. 2013;4:e689. doi: 10.1038/cddis.2013.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oppenheim RW. Cell death during development of the nervous system. Annu Rev Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- 18.Easton RM, Deckwerth TL, Parsadanian AS, Johnson EM., Jr Analysis of the mechanism of loss of trophic factor dependence associated with neuronal maturation: a phenotype indistinguishable from Bax deletion. J Neurosci. 1997;17:9656–9666. doi: 10.1523/JNEUROSCI.17-24-09656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Putcha GV, Deshmukh M, Johnson EM., Jr Inhibition of apoptotic signaling cascades causes loss of trophic factor dependence during neuronal maturation. J Cell Biol. 2000;149:1011–1018. doi: 10.1083/jcb.149.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristiansen M, Menghi F, Hughes R, Hubank M, Ham J. Global analysis of gene expression in NGF-deprived sympathetic neurons identifies molecular pathways associated with cell death. BMC Genomics. 2011;12:551. doi: 10.1186/1471-2164-12-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ham J, Towers E, Gilley J, Terzano S, Randall R. BH3-only proteins: key regulators of neuronal apoptosis. Cell Death Differ. 2005;12:1015–1020. doi: 10.1038/sj.cdd.4401689. [DOI] [PubMed] [Google Scholar]
- 22.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 23.Donovan M, Cotter TG. Caspase-independent photoreceptor apoptosis in vivo and differential expression of apoptotic protease activating factor-1 and caspase-3 during retinal development. Cell Death Differ. 2002;9:1220–1231. doi: 10.1038/sj.cdd.4401105. [DOI] [PubMed] [Google Scholar]
- 24.Madden SD, Donovan M, Cotter TG. Key apoptosis regulating proteins are down-regulated during postnatal tissue development. Int J Dev Biol. 2007;51:415–423. doi: 10.1387/ijdb.062263sm. [DOI] [PubMed] [Google Scholar]
- 25.Yakovlev AG, Ota K, Wang G, Movsesyan V, Bao W-L, Yoshihara K, Faden AI. Differential Expression of Apoptotic Protease-Activating Factor-1 and Caspase-3 Genes and Susceptibility to Apoptosis during Brain Development and after Traumatic Brain Injury. J Neurosci. 2001;21:7439–7446. doi: 10.1523/JNEUROSCI.21-19-07439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouyang Y-B, Xu L, Lu Y, Sun X, Yue S, Xiong X-X, Giffard RG. Astrocyte-enriched miR-29a targets PUMA and reduces neuronal vulnerability to forebrain ischemia. Glia. 2013;61:1784–1794. doi: 10.1002/glia.22556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khanna S, Rink C, Ghoorkhanian R, Gnyawali S, Heigel M, Wijesinghe DS, Chalfant CE, Chan YC, Banerjee J, Huang Y, Roy S, Sen CK. Loss of miR-29b following acute ischemic stroke contributes to neural cell death and infarct size. J Cereb Blood Flow Metab. 2013;33:1197–1206. doi: 10.1038/jcbfm.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandi G, Nakka VP, Dharap A, Roopra A, Vemuganti R. MicroRNA miR-29c Down-Regulation Leading to De-Repression of Its Target DNA Methyltransferase 3a Promotes Ischemic Brain Damage. PLoS ONE. 2013;8:e58039. doi: 10.1371/journal.pone.0058039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X-J, Zheng X-P, Zhang R, Guo Y-L, Wang J-H. Combinatorial effects of miR-20a and miR-29b on neuronal apoptosis induced by spinal cord injury. Int J Clin Exp Pathol. 2015;8:3811–3818. [PMC free article] [PubMed] [Google Scholar]
- 30.Qi Y, Zhang M, Li H, Frank JA, Dai L, Liu H, Chen G. MicroRNA-29b Regulates Ethanol-induced Neuronal Apoptosis in the Developing Cerebellum through SP1/RAX/PKR Cascade. J Biol Chem. 2014;289:10201–10210. doi: 10.1074/jbc.M113.535195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian L, Van Laake LW, Huang Y, Liu S, Wendland MF, Srivastava D. miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J Exp Med. 2011;208:549–560. doi: 10.1084/jem.20101547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren D, Tu HC, Kim H, Wang GX, Bean GR, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science. 2010;330:1390–1393. doi: 10.1126/science.1190217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyttenbach A, Tolkovsky AM. The BH3-only protein Puma is both necessary and sufficient for neuronal apoptosis induced by DNA damage in sympathetic neurons. J Neurochem. 2006;96:1213–1226. doi: 10.1111/j.1471-4159.2005.03676.x. [DOI] [PubMed] [Google Scholar]
- 34.Chen Q, Xu J, Li L, Li H, Mao S, Zhang F, Zen K, Zhang CY, Zhang Q. MicroRNA-23a/b and microRNA-27a/b suppress Apaf-1 protein and alleviate hypoxia-induced neuronal apoptosis. Cell Death Dis. 2014;5:e1132. doi: 10.1038/cddis.2014.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian L, Van Laake LW, Huang Y, Liu S, Wendland MF, Srivastava D. miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J Exp Med. 2011;208:549–560. doi: 10.1084/jem.20101547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabirzhanov B, Zhao Z, Stoica BA, Loane DJ, Wu J, Borroto C, Dorsey SG, Faden AI. Downregulation of miR-23a and miR-27a following Experimental Traumatic Brain Injury Induces Neuronal Cell Death through Activation of Proapoptotic Bcl-2 Proteins. J Neurosci. 2014;34:10055–10071. doi: 10.1523/JNEUROSCI.1260-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu BR, Liu CL, Ouyang Y, Blomgren K, Siesjo BK. Involvement of Caspase-3 in Cell Death After Hypoxia-Ischemia Declines During Brain Maturation. J Cereb Blood Flow Metab. 2000;20:1294–1300. doi: 10.1097/00004647-200009000-00003. [DOI] [PubMed] [Google Scholar]
- 38.Snider WD, Elliott JL, Yan Q. Axotomy-induced neuronal death during development. J Neurobiol. 1992;23:1231–1246. doi: 10.1002/neu.480230913. [DOI] [PubMed] [Google Scholar]
- 39.Unsain N, Barker Philip A. New Views on the Misconstrued: Executioner Caspases and Their Diverse Non-apoptotic Roles. Neuron. 2015;88:461–474. doi: 10.1016/j.neuron.2015.08.029. [DOI] [PubMed] [Google Scholar]
- 40.Shioya M, Obayashi S, Tabunoki H, Arima K, Saito Y, Ishida T, Satoh J. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol Appl Neurobiol. 2010;36:320–330. doi: 10.1111/j.1365-2990.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- 41.Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol Disease. 2008;29:438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Papadopoulou AS, Serneels L, Achsel T, Mandemakers W, Callaerts-Vegh Z, Dooley J, Lau P, Ayoubi T, Radaelli E, Spinazzi M, Neumann M, Hébert SS, Silahtaroglu A, Liston A, D’Hooge R, Glatzel M, De Strooper B. Deficiency of the miR-29a/b-1 cluster leads to ataxic features and cerebellar alterations in mice. Neurobiol Dis. 2015;73:275–288. doi: 10.1016/j.nbd.2014.10.006. [DOI] [PubMed] [Google Scholar]