Abstract
The serpinopathies are human pathologies caused by mutations that promote polymerisation and intracellular deposition of proteins of the serpin superfamily, leading to a poorly understood cell toxicity. The dementia FENIB is caused by polymerisation of the neuronal serpin neuroserpin (NS) within the endoplasmic reticulum (ER) of neurons. With the aim of understanding the toxicity due to intracellular accumulation of neuroserpin polymers, we have generated transgenic neural progenitor cell (NPC) cultures from mouse foetal cerebral cortex, stably expressing the control protein GFP (green fluorescent protein), or human wild type, G392E or delta NS. We have characterised these cell lines in the proliferative state and after differentiation to neurons. Our results show that G392E NS formed polymers that were mostly retained within the ER, while wild type NS was correctly secreted as a monomeric protein into the culture medium. Delta NS was absent at steady state due to its rapid degradation, but it was easily detected upon proteasomal block. Looking at their intracellular distribution, wild type NS was found in partial co-localisation with ER and Golgi markers, while G392E NS was localised within the ER only. Furthermore, polymers of NS were detected by ELISA and immunofluorescence in neurons expressing the mutant but not the wild type protein. We used control GFP and G392E NPCs differentiated to neurons to investigate which cellular pathways were modulated by intracellular polymers by performing RNA sequencing. We identified 747 genes with a significant upregulation (623) or downregulation (124) in G392E NS-expressing cells, and we focused our attention on several genes involved in the defence against oxidative stress that were up-regulated in cells expressing G392E NS (Aldh1b1, Apoe, Gpx1, Gstm1, Prdx6, Scara3, Sod2). Inhibition of intracellular anti-oxidants by specific pharmacological reagents uncovered the damaging effects of NS polymers. Our results support a role for oxidative stress in the cellular toxicity underlying the neurodegenerative dementia FENIB.
Abbreviations: A1AT, alpha1 antitrypsin; Aldh, aldehyde dehydrogenase; ApoE, apolipoprotein E; ER, endoplasmic reticulum; FENIB, familial encephalopathy with neuroserpin inclusion bodies; GPx, glutathione peroxidase; GST, glutathione transferase; HRP, horseradish peroxidase; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; NS, neuroserpin; NFκB, nuclear factor κB; Prdx6, peroxiredoxin 6; Scara, scavenger receptor class A; SOD, superoxide dismutase; unfolded protein response, UPR; wild type, WT
Keywords: Neuroserpin, Serpin polymers, Familial encephalopathy with neuroserpin inclusion bodies, Protein conformational disease, Neurodegeneration, Neural progenitor cells, Neurons, Oxidative stress
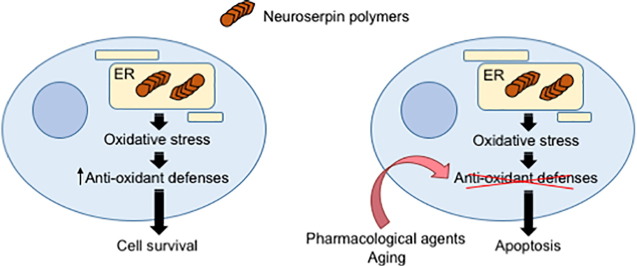
Graphical abstract
Highlights
-
•
A cell model system for FENIB was generated using mouse neural progenitor cells.
-
•
Cells expressing polymerogenic neuroserpin upregulated several anti-oxidant genes.
-
•
Inhibition of the anti-oxidant defences led to apoptosis of these cells.
-
•
Oxidative stress and lipid oxidation may contribute to neurodegeneration in FENIB.
1. Introduction
The serpinopathies are protein conformational diseases characterised by the polymerisation and intracellular deposition of mutant variants of the serpins (serine protease inhibitors), within the endoplasmic reticulum (ER) of the cells that synthesise the protein. This common pathological mechanism results in liver disease upon polymerogenic mutations in alpha-1 antitrypsin (A1AT), and a rare but fatal neurodegenerative dementia caused by mutations in neuroserpin (NS) [reviewed in Roussel et al., 2011]. The latter is an autosomal dominant condition known as FENIB (familial encephalopathy with neuroserpin inclusion bodies) (Davis et al., 1999), described in patients carrying one of six different mutations in the NS gene: S49P, S52R (Davis et al., 1999); H338R, G392E (Davis et al., 2002); G392R (Coutelier et al., 2008) and L47P (Hagen et al., 2011). In all cases, mutant NS was found to accumulate within affected neurons forming periodic acid-Schiff (PAS)-positive and diastase-resistant inclusion bodies (Collins bodies), most abundant in the cerebral cortex but also present in other regions of the central nervous system (Davis et al., 1999)(Hagen et al., 2011). For the first four mutations, a strong correlation was found between the predicted tendency to polymer formation by each mutant variant of NS and the number of Collins bodies and severity of FENIB (Davis et al., 2002), suggesting a toxic gain-of-function for NS polymers. Polymer formation by mutant NS and its correlation with the disease phenotype was later confirmed in COS-7 and PC12 cell models of FENIB, where overexpression of each mutant variant lead to intracellular accumulation of polymeric NS within the ER to a degree that was proportional to the severity of FENIB in patients (Miranda et al., 2004)(Miranda et al., 2008)(Moriconi et al., 2015).
The toxicity of NS polymers is supported by the phenotypes observed in two animal models of FENIB. Transgenic mice overexpressing S49P and S52R NS showed the formation of abundant intraneuronal Collins bodies, neuronal loss in the cerebral cortex and hippocampus, and pathological phenotypes reminiscent of FENIB during late adulthood (Galliciotti et al., 2007). Overexpression of human S49P, S52R, H338R and G392E NS in Drosophila melanogaster led to a decrease in locomotor activity, with decreasing mobility correlating to increased polymer content in the brain (Miranda et al., 2008). Despite these results, the mechanism of toxicity of NS polymers in cell models of disease has been elusive so far. Accumulation of NS polymers within the ER fails to induce a classical unfolded protein response (UPR), contrarily to a truncated version of NS (delta NS) lacking the last third of the aminoacidic sequence, which does not fold properly, does not polymerise and activates the UPR (Davies et al., 2009). Instead, NS polymers activate the ER overload response through a poorly understood signalling pathway that depends on Ca2 + and leads to the activation of nuclear factor κB (NFκB) (Davies et al., 2009). Nevertheless, three different cell model systems, transiently transfected COS-7 cells, stable inducible PC12 cells and stable inducible HeLa cells, failed to show clear signs of cell malfunction and death upon NS polymer accumulation (Miranda et al., 2004)(Miranda et al., 2008)(Roussel et al., 2013), precluding a detailed investigation of the cellular mechanisms underlying NS polymer toxicity. This lack of a toxic phenotype could be related to the proliferative nature of these cell lines, but a neuronal model with stable overexpression of NS has not been developed to date. Mouse neural progenitor cells can be isolated from several regions of the mouse foetal brain, propagated in vitro as primary cells and differentiated to mature, non-dividing neurons through a well-defined protocol (Conti et al., 2005)(Soldati et al., 2012). They can also be stably transfected for expression of heterologous proteins, making them a suitable system for cellular studies on neuronal function.
Oxidative stress, the imbalance between generation and disposal of reactive oxygen species (ROS), is an important factor in several neurodegenerative disorders including Alzheimer's disease, Parkinson's disease, Huntington's disease and amyotrophic lateral sclerosis [reviewed in Cobb and Cole, 2015]. Under physiological conditions, ROS have important roles in signalling and immune defence, and their levels are kept under check by several antioxidant defence systems, including enzymatic (mainly superoxide dismutase, glutathione peroxidase, catalase and thioredoxin reductase) and non-enzymatic (specially glutathione, GSH) mechanisms, which can either scavenge ROS or decrease their formation [reviewed in Li et al., 2013]. Neurons are particularly vulnerable to oxidative stress due to their high energy requirements, to a decrease in antioxidant defences with age and to their terminally differentiated nature, and so oxidative stress is a key player in neurodegenerative diseases, although it is not clear whether oxidative stress is a cause, a consequence or both in these pathologies [reviewed in Gandhi and Abramov, 2012]. The ER, where NS polymer formation takes place, provides an oxidizing environment for correct formation of disulfide bonds during protein folding. Accumulating evidence suggests that ROS can be generated as a by-product of protein oxidation during normal ER function and also upon ER stress due to accumulation of misfolded proteins. Both ER stress and oxidative stress, through ROS generation, may increase the leak of Ca2 + from the ER lumen, as well as induce protein and lipid oxidation. High levels of ROS generation within the mitochondria further increase Ca2 + release from the ER, generating a vicious cycle of ROS production and cellular oxidative stress [reviewed in Malhotra and Kaufman, 2007].
We report here the generation of a novel cell model system for FENIB, consisting of mouse neural progenitor cells (NPCs) derived from mouse foetal cerebral cortex, stably expressing either a control green fluorescence protein (GFP) or one of three NS variants: wild type (WT), the highly polymerogenic G392E or delta NS, a truncated version of NS lacking the last 134 aminoacids and used as a misfolding protein control (Davies et al., 2009). By RNA sequencing comparison of control GFP and polymer-containing G392E NS cells differentiated to neurons, we have identified several genes related to oxidative stress as relevant for the toxicity of NS polymers in FENIB. The relevance of this pathway was confirmed by an increase in apoptosis of cells expressing G392E NS upon pharmacological inhibition of their anti-oxidant defences.
2. Material and methods
2.1. Reagents and antibodies
Unless stated otherwise, reagents, buffers, culture media and serum for cell cultures were purchased from Sigma-Aldrich (Milan, Italy). Custom-made rabbit polyclonal anti-NS antibody and rabbit polyclonal anti-GAPDH antibody were from Abcam (Cambridge, UK). The mouse monoclonal anti-NS antibodies were made in-house as reported before (Miranda et al., 2008). Anti-KDEL was from Enzo Life Sciences (through 3VChimica S.r.l., Italy), anti-GM130 from BD Transduction Laboratories and anti-catalase from Merk Millipore (both through SIAL S.r.l., Italy). Goat polyclonal anti-rabbit-HRP (horseradish peroxidase) and rabbit anti-mouse-HRP are from Sigma-Aldrich (Milan, Italy). Goat anti-mouse IgG-Alexa Fluor® 488 and -Alexa Fluor® 594, and goat anti-rabbit IgG-Alexa Fluor® 594 were from Life Technologies (Milan, Italy).
2.2. Generation and culture of neural progenitor cell cultures with stable neuroserpin expression
The open reading frame of human NS was amplified by PCR and subcloned into the pTP6 expression vector. This vector contains the CAGG promoter followed by the NS gene, an internal ribosome entry site (IRES) and the gene for puromycin resistance, allowing selection for simultaneous expression of both genes. NPCs derived from the mouse cerebral cortex at embryonic day 13.5 and protocols to culture them in vitro were previously described (Soldati et al., 2012). Briefly, NPCs were maintained in expansion medium consisting of basal medium [DMEM/F12 (LifeTechnologies, Milan, Italy), 1% penicillin/streptomycin, 0.1 M l-glutamine (LifeTechnologies, Milan, Italy), 1 mM Hepes, 7.5% NaHCO3, 0.6% glucose] supplemented with 20 ng/ml of human recombinant epidermal growth factor (EGF; R&D System, through Aurogene, Rome, Italy), 10 ng/ml of human recombinant basic fibroblast growth factor (bFGF; R&D System, through Aurogene, Rome, Italy), and 1% N2 supplement (LifeTechnologies, Milan, Italy). NPCs were routinely cultured at 37 °C in a 5% CO2 atmosphere within flasks previously coated with 10 μg/ml poly-ornithine (for 4 h) and 5 μg/ml laminin (for 12 h), seeding them at a cell density of 0.5–1 × 104 cells/cm2 and passaging them every 3–5 days using Accutase. NPCs expanded for up to 20 passages in vitro since their original derivation were used for this work. Early passage NPCs were transfected with pTP6-NS in three different versions: wild type, G392E and delta NS. The transfection was performed with Amaxa Nucleofector device and mouse Neural Stem Cell Nucleofector kit (Lonza, Cologne AG, Germany) according to manufacturer instructions. Two days after transfection, NPCs with stable integration of the pTP6-NS transgene were selected with puromycin (1.6 μg/ml), giving rise to cell lines consisting in mixed populations of cells with random stable insertion of the transfected plasmid DNA. A NPC line expressing pTP6-hrGFP (Vallier et al., 2004)(Pratt et al., 2000) was used as a negative control for non-specific effects due to the overexpression approach. For neuronal differentiation, NPCs were plated in expansion medium (20.000 cells/cm2) and 24 h later this was replaced with neuronal differentiation medium [basal medium containing 10 ng/ml of bFGF, 1% N2, 2% B27, and 0.5 μM DAPT (Tocris Bioscience, Bristol, UK)]. Three days later this medium was partially replaced with fresh medium. After 6 days under these conditions, NPCs differentiated into a mixed culture of neurons (around 60–70%) and glial or residual progenitor cells [as reported in Sinno et al., 2013].
2.3. SDS and non-denaturating PAGE and western blot analysis
Each cell pellet from proliferative cells was obtained from a T25 flask adding 200 μl of lysis buffer (150 mM NaCl, 50 mM Tris-Cl, pH 7.5, 1% v/v Nonidet P-40, plus 1 mM protease inhibitor cocktail). Differentiated cells were lysed as for proliferative cells but adding 50 μl of complete lysis buffer directly into the flask (T25) and using a cell scraper (Corning Inc., through Sigma-Aldrich, Milan, Italy) for cell detachment and lysis. The soluble fraction was collected by centrifugation at 17.000 g, 4 °C for 15 min, and proteins in the insoluble pellet were extracted by heating at 95 °C in loading buffer containing 10% v/v beta-mercaptoethanol and 4% w/v SDS. 50 μg of total protein from each lysate for proliferative cells and 65 μl (corresponding to one flask) for differentiated cells, and the equivalent volume of each culture medium, were mixed with loading buffer without or with beta-mercaptoethanol and SDS as above and analysed by non-denaturing 7.5% w/v acrylamide PAGE (Miranda et al., 2008) or 10% w/v acrylamide SDS-PAGE (Miranda et al., 2008) as indicated. For enzymatic digestions, 20 μg of protein and the equivalent volume of culture medium were incubated with 1000 U of endoglycosidase H (endoH) for 3 h at 37 °C. Proteins were then separated by SDS-PAGE and analysed by western blot as described above. The HRP signal was developed using the LiteAblot PLUS and TURBO extra sensitive chemoluminescent substrates (Euroclone, Milan, Italy) and exposed to film or visualised on a ChemiDoc system (BioRad, Italy).
2.4. Sandwich ELISA
Quantification of NS was performed by sandwich ELISA with anti-NS antibodies made in house as described previously (Miranda et al., 2008). Briefly, 96-well plates (Costar 3590, Corning Inc., New York, US) were coated with antigen-purified rabbit polyclonal anti-NS antibody (2 μg/ml), washed (0.9% w/v NaCl, 0.05% v/v Tween20) and blocked with blocking buffer (PBS, 0.25% w/v bovine serum albumin, 0.05% v/v Tween20, 0.025% w/v sodium azide). Standards (recombinant purified monomeric or polymeric NS) and samples were diluted in blocking buffer and incubated for 2 h. After washing, wells were incubated with either a pool of monoclonal antibodies (1A10 and 10B8, 0,5 μg/ml each) or with an anti-NS polymer monoclonal antibody (7C6, 1 μg/ml). Rabbit anti-mouse IgG-HRP labeled antibody was used for detection with TMB substrate solution, and HRP activity was measured in a GloMax plate reader (Promega) at 450 nm. The concentration of NS in each sample was calculated against the appropriate standard curve (monomeric protein for WT NS, polymers for G392E NS), normalised to the total protein concentration of the sample (Bradford assay), and graphed in arbitrary units (A.U.).
2.5. Immunofluorescence staining and confocal microscopy
Cells for immunocytochemistry were grown on coverslips pre-treated with poly-ornithine and laminin, in 24-well plates. Proliferating cells (around 60% confluence) or cells differentiated for six-days in vitro were washed, fixed in ice-cold 4% (w/v) paraformaldehyde for 30 min at room temperature and washed again. Cells were then treated with permeabilisation solution (0.5% saponin, 1% BSA, 0.5% EDTA pH 8 in PBS) for 10 min, then incubated for 30 min in blocking buffer (PBS plus 0.1% saponin, 0.5% BSA, 2% normal goat serum, 5 mM glycine, 0.1% sodium azide), and immunostained with anti-NS rabbit polyclonal antibody or the anti-NS polymer monoclonal antibody 7C6, anti-KDEL and anti-GM130 antibodies, and the corresponding secondary antibodies (goat anti-mouse IgG-Alexa Fluor® 488 and -Alexa Fluor® 594, and goat anti-rabbit IgG-Alexa Fluor® 594). Nuclear DNA was counter-stained with DRAQ5® (Abcam, Cambridge, UK). Coverslips were mounted with FluorSave (Calbiochem, VWR International, Milan, Italy) plus 2% DABCO. Imaging was performed on a Zeiss 780 confocal microscope.
2.6. MTT assay
The culture medium was replaced by a solution of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, 0.5 mg/ml] in culture medium. After 3 h of incubation at 37 °C, this solution was removed and the produced formazan was solubilized in 250 μl of isopropanol plus 0.1% v/v HCl and 0.1% v/v NP-40. The optical density of the blue formazan was measured in a GloMax plate reader (Promega) at 560 nm.
2.7. RNA extraction and real-time RT-PCR
Proliferating NPCs plated in 6-well plates were lysed in 300 μl/well of RLT buffer (Qiagen RNeasy Mini kit). Total RNA was purified out using Qiagen RNeasy mini kit and eluted in 50 μl of RNase-free water. RNA was quantified using a Thermo Scientific NanoDrop 2000 spectrophotometer. NPC cultures differentiated for 6 days in T25 flasks were lysed in 400 μl of RLT buffer/flask; 20 μl of RNase-free water were used for RNA elution. For real-time reverse-transcriptase polymerase chain reaction (real-time RT-PCR), RNA was reverse-transcribed using the Qiagen QuantiTect reverse transcription kit and amplified on a Rotor-Gene Q (Qiagen), using Qiagen QuantiFast SYBR Green PCR kit. Primers for real-time RT-PCR were designed using Primer3 (http://bioinfo.ut.ee/primer3/) and purchased from Sigma-Aldrich Co (Milan, Italy). Primer sequences used for this study are listed in Supplementary Table 2. Melt curve analysis was performed at the end of each real-time RT-PCR run. All primer pairs used for this study showed a single amplification peak, which was absent in the corresponding no-template control reaction, indicating primer specificity. Relative gene expression levels in different samples were determined with the built-in comparative quantitation method using Eef1a1 or Rpl19 as reference genes, with similar results. Statistical analysis of experimental data was performed with Microsoft Excel software.
2.8. RNA-sequencing
Total RNA was extracted and quantified as described above. RNA-Seq library preparation and sequencing was performed by the IGA Technology Services (Udine, Italy), using the Illumina-TruSeq RNA Sample Preparation Kit V2 (Illumina, San Diego, USA) according to manufacturer's instructions. The final libraries for single-read sequencing of 50 base pairs were carried out on an Illumina HiSeq2000. Each sample produced about 20 millions of reads. Read quality was evaluated using FastQC (version 0.11.2, Babraham Institute, Cambridge, UK) tool. Reads were mapped to the mouse Ensembl GRCm38 build reference genome using Tophat (version 2.0.12) (Trapnell et al., 2012) using default settings plus the “—no-novel-juncs” option. Gene structure annotations corresponding to the Ensembl annotation release 75 were used to build a transcriptome index and provided to Tophat during the alignment step using the “-G” parameter. The same gene annotations were used to quantify the gene-level read counts using HTSeq-count (version 0.6.1) script. Subsequently differential gene expression analyses were performed using Bioconductor (Gentleman et al., 2004) R (version 3.2.2) (R Core Team, 2015) package DESeq2 (version 1.4.5) (Love et al., 2014). Genes significantly up- or down-regulated in NS-expressing samples (q-value < 0.1) were clustered by enrichment pathway analysis using Bioconductor R packages ClusterProfiler (Yu et al., 2012) and Pathview (Luo and Brouwer, 2013), Gene Ontology Database (Ashburner et al., 2000) and Reactome Pathway Database (Croft et al., 2014). All RNA-seq raw and processed data files were deposited at NCBI in the Gene Expression Omnibus (GEO) repository with accession number GSE96096.
2.9. Neurotoxicity experiments
NPCs were cultured in differentiation medium for 6 days in 24-well plates. At day 5 of differentiation, cells were incubated with 100 μM of diethyl maleate (DEM) or 500 μM of 3-amino-1,2,4 triazole (ATZ) for 1 h, or with 100 μM H2O2 for 30 min, at 37 °C, then cultured in fresh differentiation medium for 24 h at 37 °C. For DEM plus H2O2 treatment, cells were incubated always at 37 °C, first with 100 μM DEM for 1 h, then with fresh differentiation medium for 18 h, next with 100 μM H2O2 for 30 min, and finally with fresh differentiation medium for 6 h, for a total of 24 h. At the end of the treatments, cells were washed in PBS and fixed in 4% (v/v) paraformaldehyde for 15 min, stained for cell counting with 3 μg/ml of the nuclear staining DAPI, and analysed with a 40 × objective, counting at least 150 cells distributed in 15 areas per sample.
2.10. TUNEL assay
TUNEL staining was performed by using the in situ Cell Death Detection Kit, Fluorescein (Roche, 11,684,795,910). Briefly, cells were cultured for 6 days in differentiation medium and were pre-treated or not with 100 μM of DEM. Twenty-four hours after DEM treatment, cells were fixed in 4% paraformaldehyde for 15 min at RT. Cells were then washed with PBS for three times. Cell permeabilisation and staining were performed following the manufacturer's instructions. Briefly, cells were permeabilised in a 0.1% Triton X-100 in 0.1% sodium citrate solution for 2 min and then washed again in PBS. Finally, cells were incubated with labelling solution at 37 °C for 1 h.
3. Results
3.1. Expression and handling of wild type and G392E neuroserpin in proliferating NPCs
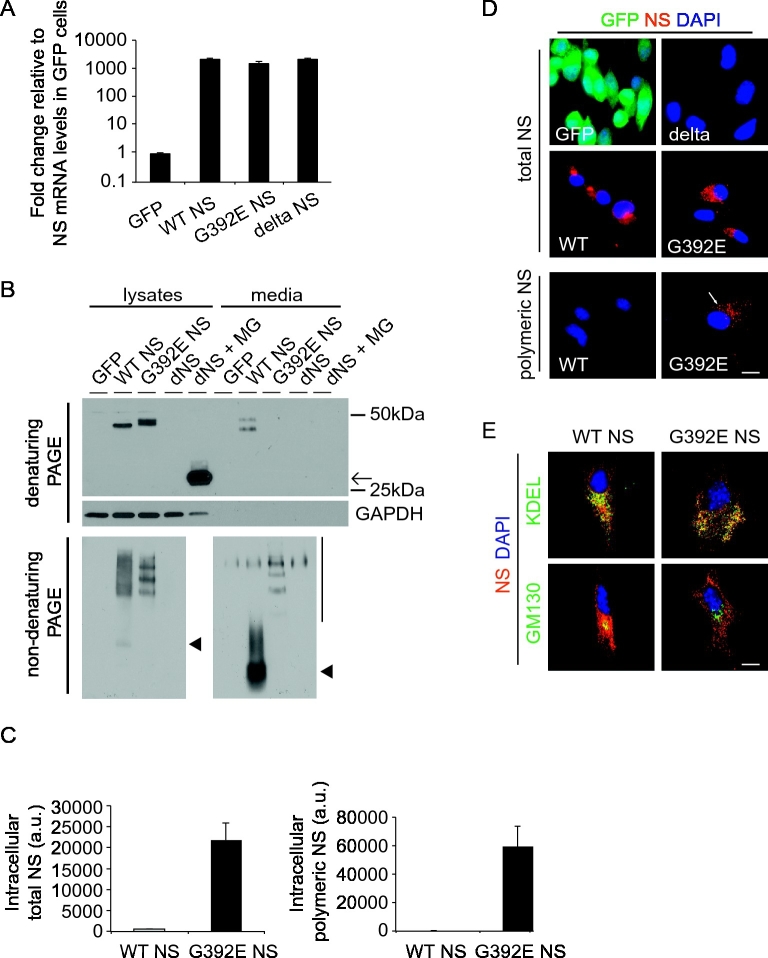
We aimed to create a neuronal model to study the cellular phenotype of the dementia FENIB using non-transformed cell cultures. We cloned the wild type (WT), G392E and delta variants of human NS in the bicistronic expression vector pTP6, which is transcribed into a single mRNA encoding for both the gene of interest and the antibiotic resistance gene (puromycin). Previously described NPCs (Soldati et al., 2012) derived from E13.5 mouse cerebral cortex were transfected with the plasmids containing the different versions of NS. Three stable NPC-NS lines were established by selection with puromycin at 1.6 μg/ml, giving rise to mixed populations of cells with random integration for each transgene. Additionally, we used NPC cultures expressing GFP from the same plasmid, as a negative control. We first investigated the expression levels of NS in NPCs cultured in proliferating conditions by real-time RT-PCR analysis. Using primer pairs targeting both the endogenous mouse NS gene and the human NS transgenes, we found that the mRNA levels of NS were almost one thousand times higher in the transgenic cultures expressing human NS relative to the expression levels of the endogenous mouse gene as detected in GFP cultures. WT, G392E and delta NS transgenes were all expressed at very similar levels (Fig. 1A). We next characterised the protein levels and conformation of NS in our proliferative NPC cultures by performing SDS and non-denaturing PAGE followed by western blot analysis of the cell lysates and culture media. The results presented in Fig. 1B show that WT NS was found as a single band of approximately 50 kDa in the cell lysate, and as a double band in the culture medium (Fig. 1B, upper panel), in a monomeric conformation (Fig. 1B, lower panels, arrowheads). In contrast, G392E NS was detected by SDS-PAGE in the cell lysate as two close bands due to differential glycosylation (Moriconi et al., 2015) and no signal was found in the culture medium (Fig. 1B, upper panel). After separation by non-denaturing PAGE, all G392E NS was found in a polymeric conformation (Fig. 1B, lower panels). Delta NS was detectable only in cell lysates after proteasomal block with the MG132 inhibitor, as expected for this truncated version of NS (Fig. 1B, upper panel). No signal for NS was detected in the samples from GFP expressing cells used as negative control. The intracellular accumulation of NS polymers was confirmed in lysates of G392E NS cells by sandwich ELISA using several monoclonal antibodies against NS, including the anti-NS polymer 7C6 monoclonal antibody, as described before (Miranda et al., 2008) (Fig. 1C).
Fig. 1.
A neural progenitor cell model of FENIB. A. Real-time RT-PCR quantification of NS gene expression in NPCs cultured in proliferating conditions, using primer pairs targeting both the exogenous human NS transgene and the endogenous mouse NS gene. Results are shown as the mean +/− SEM of the ratio between NS and GFP samples of three independent repeats. B. Cell lysates and culture media of stably transfected NPC cultured in proliferative conditions expressing a control protein (GFP), WT, G392E or delta NS (dNS, black arrow, treated or not with the reversible proteasomal inhibitor MG132 at 2 μM for 12 h) were resolved by 10% w/v acrylamide SDS (top and middle panels) or 7,5% w/v acrylamide non-denaturing (bottom panel) PAGE, followed by western blot. Arrowheads: WT NS monomer; vertical line: NS polymers. C. Quantification of intracellular total NS and NS polymers in proliferative NPCs by sandwich ELISA using monoclonal antibodies 1A10 + 10B8 (left graph) or 7C6 (right graph) respectively. The values correspond to NS normalised to total protein and are the mean +/− SEM of three independent repeats. D. Immunofluorescence analysis of WT, G392E and delta NS, as well as negative control GFP proliferative NPCs, with a rabbit anti-NS polyclonal antibody (upper and middle panels) and with monoclonal antibody 7C6 for detection of NS polymers (lower panels, arrow). E. Immunofluorescence colocalisation of WT and G392E NS with resident proteins of the secretory pathway in proliferative NPCs. Cells were costained with a rabbit anti-NS polyclonal antibody and antibodies against KDEL (ER) and GM130 (Golgi apparatus). The colour corresponding to each antibody is indicated for each set of panels, and only the merged images are shown. The nucleus appears blue due to DNA staining with DAPI. Scale bar: 10 μm.
Next, the intracellular distribution and conformation of NS in proliferative cells were analysed by immunofluorescence and confocal microscopy. Total NS was detected with a rabbit anti-NS polyclonal antibody, while NS polymers were detected with the 7C6 monoclonal antibody (Fig. 1D). No signal for NS was found in GFP and delta NS expressing cells (Fig. 1D, upper panels). WT NS showed a dispersed pattern compatible with its presence in the ER, with a higher intensity in a perinuclear location suggestive of its localization in the Golgi apparatus (Fig. 1D, middle left panel). G392E NS presented a spotty pattern throughout the cell (Fig. 1D, middle right panel), and only cells expressing G392E NS showed positive staining for polymeric NS (Fig. 1D, lower right panel). We confirmed the presence of WT NS within the ER and Golgi compartments by co-localisation with resident proteins (KDEL-tagged proteins and GM130, respectively), while G392E NS co-localised with the ER only, supporting its reduced secretion (Fig. 1E).
Taken together, these results show that WT NS is correctly processed and secreted in proliferative NPCs, while G392E NS forms polymers that accumulate within the ER, as seen in previous cell models and in FENIB patients (Davis et al., 2002)(Miranda et al., 2008).
3.2. Expression and handling of wild type and G392E neuroserpin in differentiated NPCs
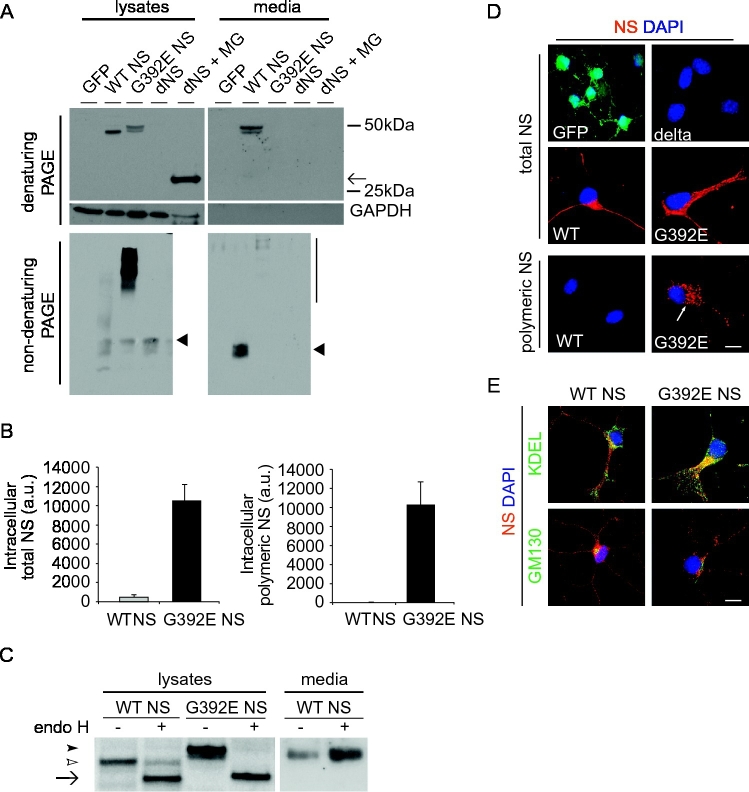
Since FENIB is a neurodegenerative condition that affects mature neurons, we next evaluated the accumulation of G392E NS polymers in our transgenic NPCs after differentiating them to neurons using previously published protocols (Sinno et al., 2013). Western blot after SDS-PAGE showed that WT NS was normally processed and secreted as a monomer in the extracellular medium, while G392E NS was found mainly in the cell lysates as a double band (Fig. 2A, upper panels), as seen above in proliferative conditions (Fig. 1B, upper panel). Delta NS was recognisable in cell lysates only after proteosomal blocking with the MG132 inhibitor (Fig. 2A, upper left panel). The results from non-denaturating PAGE and western blot analysis showed that WT NS was found as a monomeric protein, mostly in the culture medium, while G392E NS was present in the polymeric conformation, with most of the signal corresponding to the cell lysate (Fig. 2A, lower panels). Quantification of intracellular NS by sandwich ELISA showed that the amount of G392E NS in differentiated NPCs was lower when compared to proliferative conditions, but the intracellular levels of G392E NS were always higher than those of WT NS, and the presence of NS polymers was confirmed with the 7C6 monoclonal antibody in cell lysates of G392E cells (Fig. 2B), as seen above for proliferating cells.
Fig. 2.
Polymers of G392E NS accumulate in the ER of NPCs differentiated to neurons. A. Cell lysates and culture media of NPCs differentiated to neurons and expressing control GFP, WT, G392E or delta NS (dNS, black arrow, treated or not with MG132 as in Fig. 1), were resolved by 10% w/v acrylamide SDS (top and middle panels) and 7.5% w/v acrylamide non-denaturing (bottom panel) PAGE and analysed by western blot. Arrowheads: WT NS monomer; vertical line: NS polymers. B. Quantification of intracellular total NS and NS polymers by sandwich ELISA as described in Fig. 1. The values correspond to NS normalised to total protein and are expressed as the mean +/− SEM of three independent repeats. C. Endoglycosidase-H (endo H) digestion of cell lysates and culture media of differentiated NPCs cells expressing WT or G392E NS. Samples were analysed by 10% SDS-PAGE and western blot. Black arrow: non-glycosylated NS; white arrowhead: NS with two glycan chains; black arrowhead: additional slower migrating band only detectable in G392E samples. D. Immunofluorescence analysis of NPCs expressing WT, G392E and delta NS, as well as the negative control protein GFP, after 6 days of differentiation, using a rabbit anti-NS polyclonal antibody (upper and middle panels) or the 7C6 monoclonal antibody for polymer detection (arrow, lower panels). E. Immunofluorescence colocalisation of WT and G392E NS with resident proteins of the secretory pathway. Cells were costained with a rabbit anti-NS polyclonal antibody and antibodies against KDEL (ER) and GM130 (Golgi apparatus). The colour corresponding to each antibody is indicated for each set of panels, and only the merged images are shown. The nucleus appears blue due to DNA staining with DAPI. Scale bar: 10 μm.
We next assessed the intracellular localisation and secretory processing of WT and G392E NS in differentiated NPCs by treating cell lysates and culture medium with the deglycosylating enzyme endoglycosidase H (endoH), which only removes N-glycans that have not been modified in the Golgi apparatus. We found that intracellular WT and G392E NS were sensitive to endoH, supporting their pre-Golgi nature, while WT NS in the culture medium was endoH resistant, in agreement with its correct Golgi processing and secretion (Fig. 2C). The double band observed in cell lysates of G392E NS cells collapsed to a single one after endoH treatment, supporting that the difference in migration was due to different glycosylation patterns, as described before (Moriconi et al., 2015).
The intracellular distribution of NS in differentiated NPC cultures was investigated by immunofluorescence. WT and G392E NS were broadly distributed within the cell soma and along the neurites, with G392E NS showing a punctuated appearance (Fig. 2D). No NS signal was found in differentiated GFP and delta NS expressing cells. When using the anti-NS polymer monoclonal antibody 7C6, only the cells expressing G392E NS were positive, with a clearly punctuated pattern reminiscent of NS inclusions (Fig. 2D, lower panels). We looked into the trafficking of WT and G392E NS by comparing their distribution with that of the ER localisation signal (KDEL) and the Golgi resident protein GM130. WT NS was found in partial co-localisation with both the ER and Golgi markers, indicative of correct synthesis and transport of the protein, while G392E NS co-localised almost completely with KDEL and showed no co-localisation with GM130 in the Golgi, in agreement with its accumulation within the ER (Fig. 2E).
These results confirm that the behaviour of WT and the polymerogenic variant G392E NS seen in previous models of FENIB is recapitulated in our novel model of differentiated progenitor cells from mouse foetal cerebral cortex.
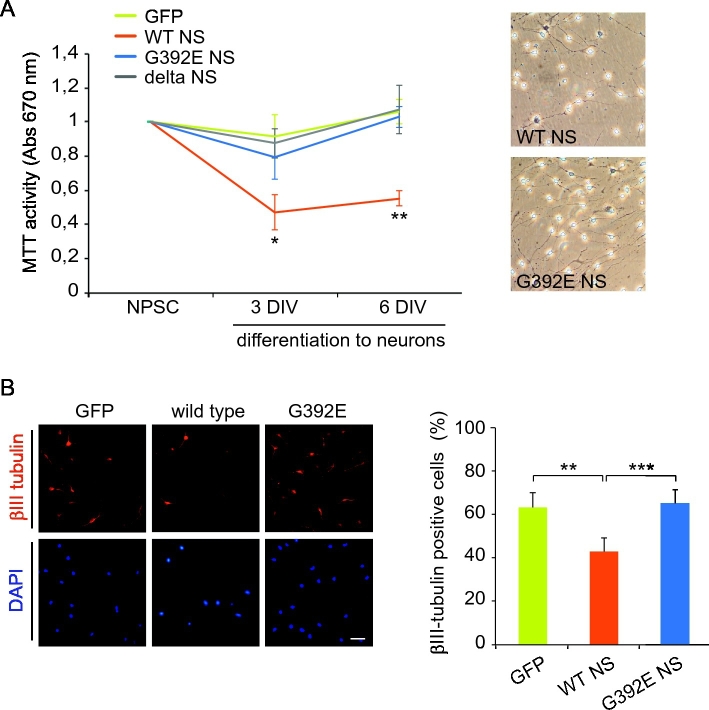
3.3. The expression of WT NS is toxic during differentiation of NPCs
In order to look at the effects of NS expression on cellular viability, we first performed MTT assays in our cell lines during differentiation in vitro for six days. As shown in Fig. 3A, we compared the vitality of cells at time 0 (proliferative conditions), and 3 and 6 days after starting their differentiation. We found that MTT activity in WT NS cells was strongly decreased after 3 days, reaching around 50% of activity at day 6. The expression of GFP, delta or G392E NS did not affect MTT activity at any of the time points analysed. We next analysed the percentage of neuronal cells generated after differentiation by immunofluorescence detection of the specific neuronal protein ßIII-tubulin, a microtubule protein almost exclusive of mature neurons (Fig. 3B). In control cells expressing GFP and in cells expressing G392E NS, the differentiation protocol induced a neuronal phenotype in over 60% of the cells, while overexpression of WT NS lead to a reduced percentage of neurons of about 40% of the surviving cells. These results suggest that WT NS overexpression impairs neuron generation in our cell culture system.
Fig. 3.
Overexpression of WT NS is toxic for NPCs upon neuronal differentiation. A. Time course of cell viability during neuronal differentiation analysed by MTT assay. NPCs were cultured and differentiated to neurons and cell viability was analysed at times 0 (proliferative conditions), 3 and 6 days after starting differentiation. Data are indicated as mean +/− SEM of three independent repeats, t-test: *p ≤ 0.05; ** p ≤ 0.01. Micrographs: bright field images of NPCs overexpressing WT and G392E NS at the end of the 6 days differentiation treatment. B. Percentage of ßIII-tubulin positive cells after differentiation of GFP, WT and G392E NS expressing NPCs for 6 days. Nuclei are stained with DAPI. Scale bar: 30 μm. Data are indicated as mean +/− SEM of three independent repeats (200 to 500 cells counted per condition in each repeat), t-test: **p ≤ 0.01; ***p ≤ 0.005.
3.4. Cells overexpressing G392E NS show higher levels of several genes involved in the anti-oxidant defence
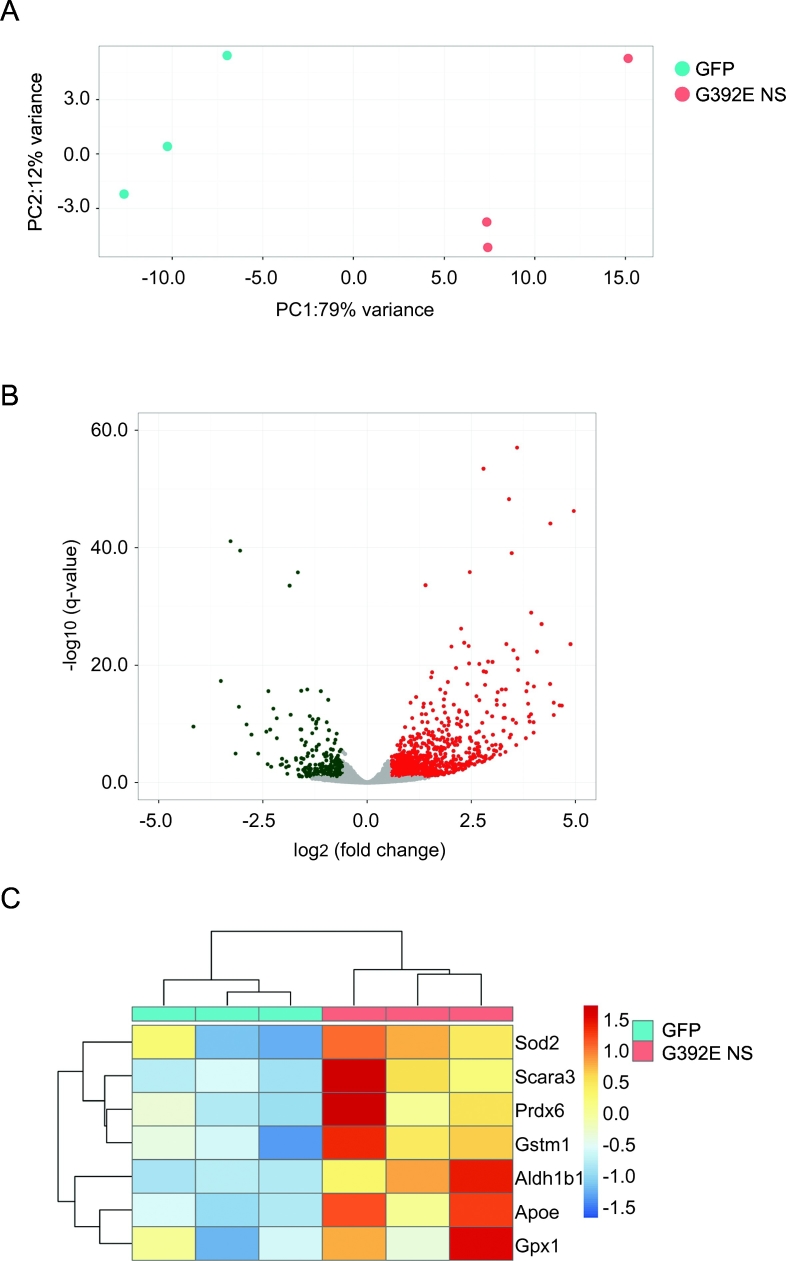
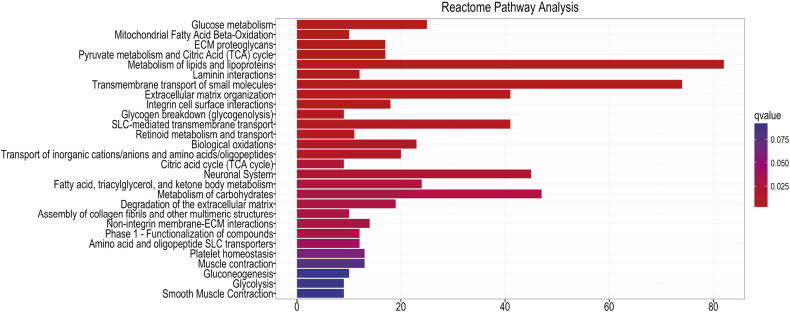
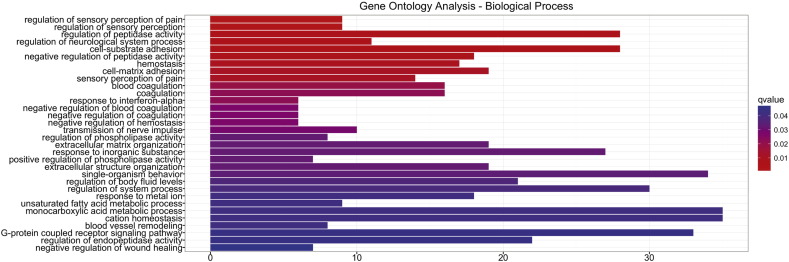
Since no evident cell toxicity was found in neurons upon accumulation of G392E NS polymers, we decided to perform an unbiased RNA sequencing (RNAseq) analysis to compare the entire transcriptome of differentiated NPCs expressing GFP or G392E NS. We decided to use the GFP expressing cells rather than the WT NS expressing cells as reference sample, due to the toxic phenotype observed for the latter. In the principal component analysis of the RNAseq results the three independent samples of GFP or G392E NS differentiated cells showed good reproducibility and tended, respectively, to cluster together, while samples from GFP and G392E NS cells were well separated due to their very different RNA profiles (Fig. 4A). Indeed, the RNAseq analysis resulted in the identification of 1960 genes whose expression was significantly modulated in G392E NS cells in comparison to the GFP samples, with 623 genes overexpressed at least 2-fold (log2 > 1) and 124 genes underexpressed at least 2-fold (log2 > − 1) (Fig. 4B). These genes were selected for further analysis by Gene Ontology (GO) Reactome and Reactome Pathway Database, which highlighted the alteration of several cellular pathways potentially due to the presence of NS polymers within the ER (Supplementary Fig. S1, Supplementary Fig. S2). Among them we found several pathways involved in lipid and carbohydrate metabolism and biological oxidation (Fig. S1). Since oxidative stress plays a major role in the aetiology of various neurodegenerative disorders, we were interested to see that genes overexpressed in differentiated G392E NS cells included a few that, according to our GO analysis or previously published work, are potentially involved in the response to oxidative stress (Fig. 4C and Supplementary Table 1). In particular, Prdx6 (coding for peroxiredoxin 6), Aldh1b1 (coding for aldehyde dehydrogenase 1b1), Scara3 (coding for scavenger receptor class A member 3), Gstm1 (coding for glutathione transferase M1) and Apoe (coding for apolipoprotein E) are all implicated in biological processes related to oxidative stress according to GO analysis and previous publications (Supplementary Table 1, (Han et al., 1998)(Brown et al., 2013)), and they were all significantly up-regulated in G293E NS cells (Supplementary Table 1, Fig. 4C).
Fig. 4.
RNA sequencing analysis of differentiated NPCs overexpressing GFP and G392E NS. NPCs stably expressing GFP or G392E NS were differentiated for 6 days in vitro, and their total RNA recovered and analysed by next generation sequencing. A. Results of principal component analysis on RNA sequencing data from three GFP (red dots) and three G392E NS (blue dots) independent samples. The plot shows the first two principal components (PC1, PC2). GFP and G392E samples showed high similarity between replicates and clustered according to their respective genotypes. B. Volcano plot of log2 fold-change (x-axis) versus − log10 q-value (y-axis, representing the probability that the gene is differentially expressed) of the results of RNA sequencing analysis of GFP and G392E NS samples. 1960 genes were significantly downregulated (green dots) or upregulated (red dots) in G392E NS cells in comparison with GFP cells. C. Heat-map of the regularised-logarithm transformed scaled counts (Love et al., 2014) of the results of RNA sequencing analysis for the indicated genes involved in oxidative metabolism. Scara3, Prdx6, Gstm1, Aldh1b1 and Apoe showed consistent upregulation in G392E NS samples, while higher variability across experimental replicates was detectable for Sod2 and Gpx1. Condition tree (top) and gene tree (left side) are represented.
Supplementary Fig. S1.
Bar plot representation of the results of Reactome Pathway analysis of RNA sequencing data from GFP and G392E NS samples, showing the categories of metabolic pathways most enriched in genes upregulated or downregulated in G392E NS differentiated NPCs.
Supplementary Fig. S2.
Bar plot representation of the results of Gene Ontology analysis of RNA sequencing data from GFP and G392E NS samples, showing the categories of biological processes most enriched in genes upregulated or downregulated in G392E NS differentiated NPCs.
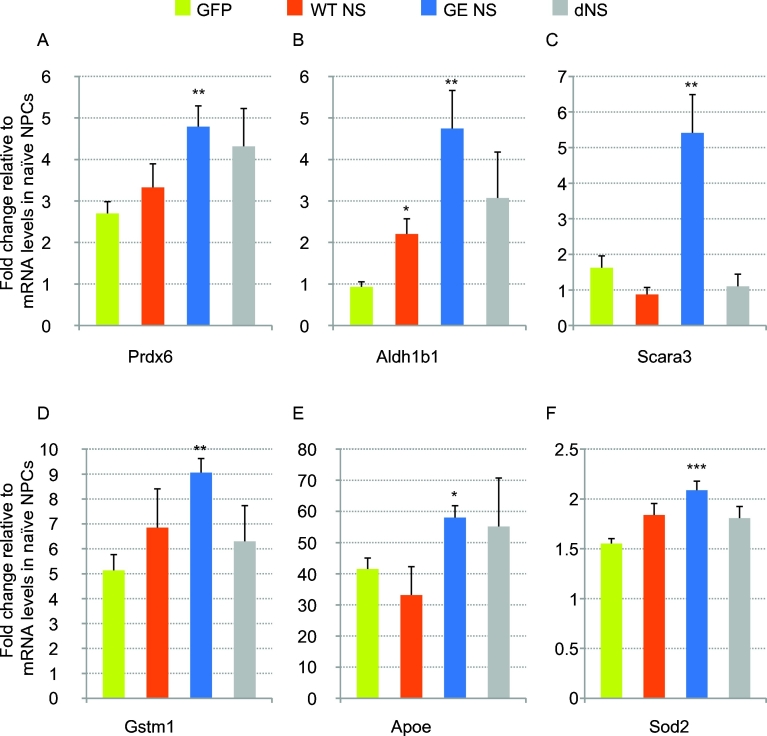
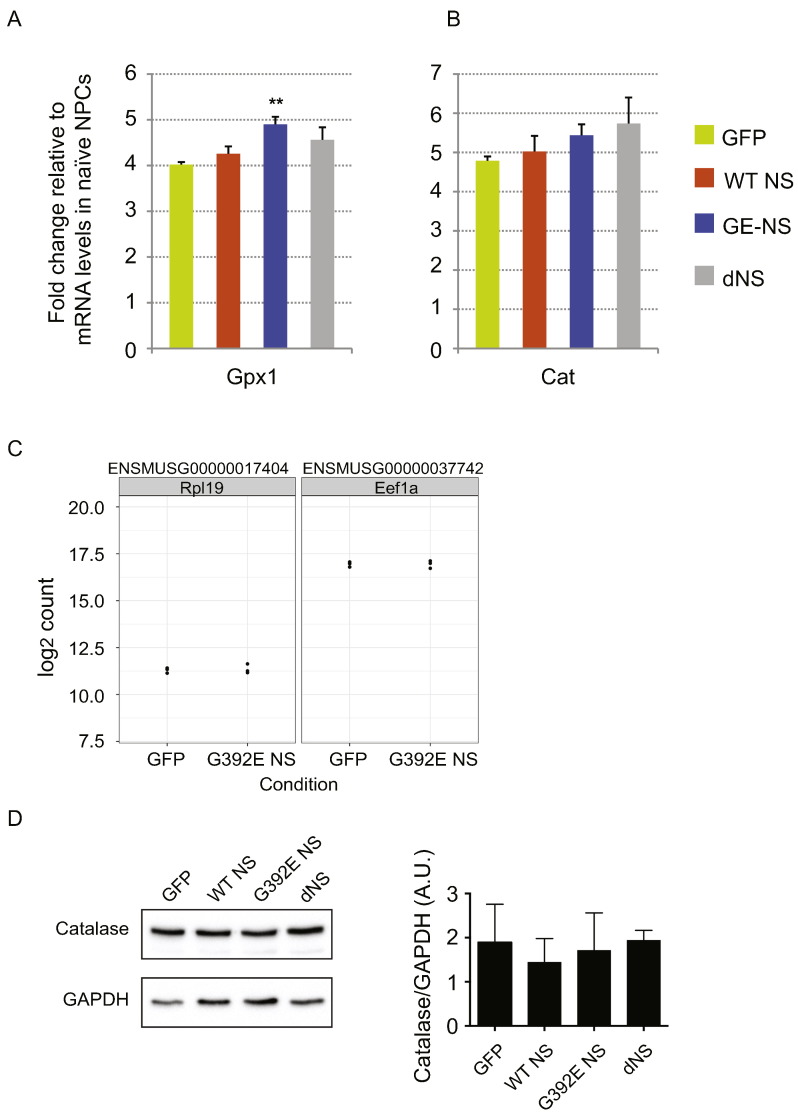
To confirm that up-regulation of these genes was specifically due to the presence of polymerogenic G392E NS, we analysed their expression by real-time RT-PCR in differentiated NPCs expressing WT NS or delta NS, in addition to cultures expressing GFP or G392E NS (Fig. 5). In this analysis, we included three additional genes involved in oxidative stress, namely Gpx1 (coding for glutathione peroxidase 1), Sod2 (coding for superoxide dismutase 2) and Cat (coding for catalase), which showed a trend towards up-regulation in G392E NS cells in our RNA sequencing dataset, although not statistically significant due to high variability in the three replicates used for RNA sequencing analysis (Supplementary Table 1, Fig. 4C, Fig. S3 and data not shown). Our results confirmed that mRNA levels of these genes were significantly higher in cells expressing G392E NS when compared to control cells expressing GFP, with the exception of catalase (Fig. 5 and Fig. S3). Moreover, there were no significant differences between GFP cells and cells expressing WT or delta NS, except for Aldh1B1 that showed a moderate increase in WT cells (Fig. 5B).
Fig. 5.
Real-time RT-PCR analysis of selected genes in differentiated NPCs. NPCs stably overexpressing GFP or wild type (WT), G392E or delta NS (dNS) were differentiated for 6 days in vitro and their total RNA recovered for gene expression analysis by real-time RT-PCR using primer pairs targeting the indicated genes (see Supplementary Table 2). Data are shown as mean +/− SEM of the ratio between transgenic cultures and control (non-transfected, proliferating) NPCs of five independent repeats. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001, according to Student's t-test performed between GFP and NS cultures.
Real-time RT-PCR analysis of selected genes in differentiated NPCs. NPCs stably overexpressing GFP or wild type (WT), G392E or delta NS (dNS) were differentiated for 6 days in vitro and their total RNA recovered for gene expression analysis by real-time RT-PCR using primer pairs targeting the indicated genes (see Supplementary Table 2). Data are shown as mean +/− SEM of the ratio between transgenic cultures and control (non-transfected, proliferating) NPCs of five independent repeats. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001, according to Student's t-test performed between GFP and NS cultures.
Supplementary Fig. S3.
NPSC stably overexpressing GFP or wild type (WT), G392E or delta NS (dNS) were differentiated for 6 days in vitro and their total RNA recovered for gene expression analysis by real-time RT-PCR using primer pairs targeting the indicated genes. Data are shown as mean +/− SEM of the ratio between transgenic cultures and control (non-transfected) NPCs of five independent repeats. **p ≤ 0.01, according to Student's t-test performed between GFP and NS cultures. C. Expression levels in our RNA sequencing analysis of Eef1a and Rpl19, used in this study as reference genes for real-time RT-PCR experiments. Data are shown as normalised counts of reads for genes across the two cell lines. D. Catalase protein levels were determined in cell lysates of NPCs differentiated for six days by 10% SDS-PAGE followed by western blot with an anti-catalase specific antibody (upper panel). GAPDH (lower panel) in the same membrane was used as a loading control. Four independent repeats were used for densitometry analysis (histogram) showing no significant differences by non-parametric ANOVA (p = 0.561).
These results suggest that NS polymers activate cellular anti-oxidant defences in differentiated NPCs by up-regulation of several genes involved in oxidative stress.
3.5. Differentiated G392E NS cells are resistant to an oxidative insult but show enhanced toxicity upon inhibition of the anti-oxidant defences
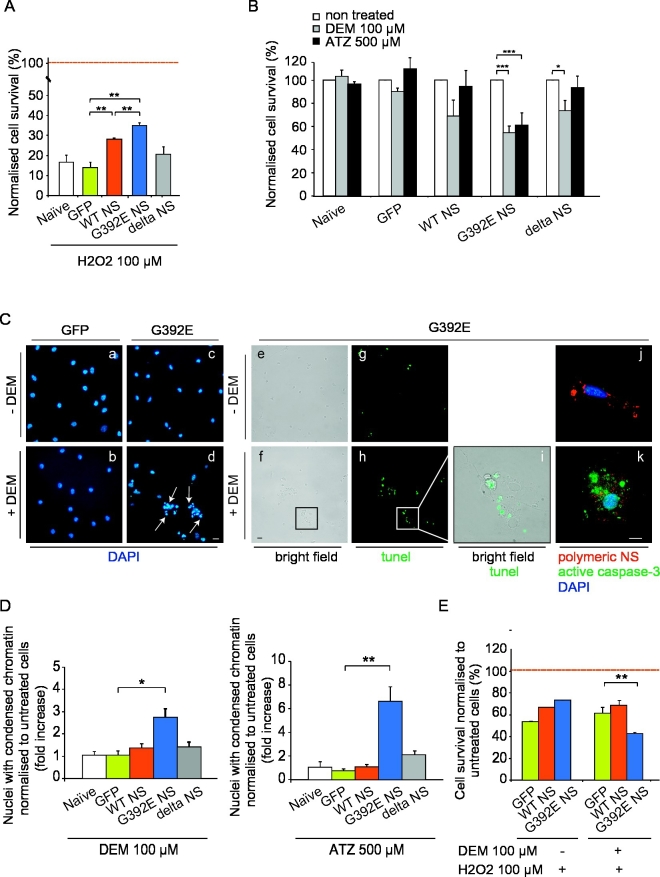
In order to investigate oxidative stress in our differentiated NPC cultures, we first decided to assess their sensitivity to an exogenous oxidative insult such as H2O2. We measured cell survival after exposure to 100 μM H2O2 and found that G392E NS cells, and to a lesser extent WT NS cells, were more resistant to the oxidative insult when compared to naïve (non-transgenic), GFP and delta NS cells (Fig. 6A), suggesting that cells expressing WT and specially G392E NS are better adapted to cope with oxidative stress.
Fig. 6.
Polymers of G392E NS cause oxidative stress in differentiated NPCs. A. NPCs were differentiated for 6 days and each cell line was incubated in the absence or presence of H2O2 (100 μM) for 30 min. Cell survival was quantified 24 h later by counting DAPI-positive cells. Results are expressed as the percentage of surviving neurons compared with control cultures not treated with H2O2 (100% survival reference line), and are the mean +/− SEM of three independent repeats, t-test: **p ≤ 0.01. B. Differentiated NPCs were incubated for 1 h in control conditions (non treated) or with either the glutathione depletor DEM (100 μM) or the catalase inhibitor ATZ (500 μM). Neuronal survival was quantified 24 h later by counting DAPI-positive stained cells. Results are expressed as the percentage of surviving neurons compared to control cultures, and are the mean +/− SEM of three independent repeats, t-test: *p ≤ 0.05; ***p ≤ 0.005. C. Differentiated NPCs expressing GFP or G392E NS were treated or not with 100 μM DEM and stained with DAPI (a–d), TUNEL assay (e–i) or anti-cleaved caspase-3 antibody (green) and anti-NS polymers (7C6 antibody, red) (j–k), to assess the nature of nuclei showing chromatin condensation. Arrows: nuclei with condensed chromatin. D. Differentiated cells were incubated for 1 h with either 100 μM DEM or 500 μM ATZ and their nuclear morphology was analysed 24 h later. The toxic effect of DEM and ATZ was quantified by counting DAPI-stained nuclei with condensed chromatin. Results are expressed as the ratio between treated and non-treated cells, and data are expressed as the mean +/− SEM of three independent repeats, t-test: *p ≤ 0.05; **p ≤ 0.01. E. Differentiated NPCs were pre-incubated for 1 h without or with 100 μM DEM and then further incubated for 30 min with 100 μM H2O2 in the presence or absence of DEM. Neuronal survival was quantified 24 h later by counting DAPI-stained cells. Results are expressed as the percentage of surviving neurons compared with control non-treated cultures, and data are the mean +/− SEM of three independent repeats, t-test: **p ≤ 0.01. In panels A, B, D and E, 15 different fields (to a total of 150 to 350 cells) were counted per condition in each independent repeat.
We next decided to investigate the effect of pharmacological inhibition of the cellular defences against oxidative stress. Glutathione (GSH) is an important antioxidant metabolite that prevents cell damage caused by the presence of reactive oxygen species (ROS), lipid oxidases and free radicals, and is involved in the mechanism of action of several ROS-detoxifying enzymes [reviewed in Schulz et al., 2000]. Cellular levels of GSH can be depleted by treating the cells with diethyl maleate (DEM) (Mitchell et al., 1983). Catalases are strong cellular scavengers of H2O2 [reviewed in Dringen et al., 2005], and can be inhibited with 3-amino-1,2,4 triazole (ATZ) (Margoliash et al., 1960). Since mRNA and protein levels of catalase were similar in all differentiated NPC lines (Fig. S3, panels B and D), the inhibitory effect of ATZ should be similar for all of them. The biological effects of catalase and GSH inhibitors were examined in differentiated NPCs expressing WT, G392E or delta NS, using GFP and naïve NPCs as negative controls, by culturing the cells without or with 100 μM DEM or 500 μM ATZ for 1 h and quantifying cell survival 24 h later by counting the surviving cells stained with DAPI. We found that the pharmacological inhibition of these antioxidant defences reduced the viability of G392E NS expressing cells when compared to GFP NPCs (Fig. 6B). These data were further confirmed by counting the number of cells with condensed chromatin, an indication of apoptosis, after GSH depletion and catalase inhibition. As shown in Fig. 6C, in differentiated cultures expressing G293E NS the impairment of these antioxidant defences caused a phenotype of chromatin condensation, which corresponded to positive staining with TUNEL (Fig. 6C, panels d, h, i). It also led to positivity to an anti-cleaved caspase-3 antibody in cells containing G392E NS polymers (Fig. 6C, panel k), confirming the apoptotic nature of this nuclear phenotype. The quantification of condensed nuclei (Fig. 6D) showed a high number of apoptotic nuclei in differentiated cells expressing G392E NS treated with DEM (100 μM) or ATZ (500 μM).
After having found that G392E NS cells showed a toxic phenotype when the antioxidant defences were blocked by DEM, we tested their response under this condition to the additional stress caused by treating them with H2O2. Differentiated cells were pre-incubated with DEM as described in the methods section and then further exposed to a mild H2O2 treatment (100 μM H2O2 for 30 min). As shown in Fig. 6E we found that, after treatment with DEM, the addition of H2O2 was more toxic to cells overexpressing G392E NS than GFP or WT NS cells, indicative of a higher sensitivity of differentiated G392E NS cells to a pro-oxidant insult when the antioxidant defences are impaired.
4. Discussion
The neurodegenerative dementia FENIB is a rare condition caused by aberrant polymerisation of NS within the ER. Studies conducted in two different animal models of FENIB, namely flies (Miranda et al., 2008) and mice (Galliciotti et al., 2007) overexpressing mutant human NS, have demonstrated that the presence of polymers is toxic to the organism, but the exact mechanism of this toxicity is still poorly understood. This is mainly due to the absence of a toxic phenotype in the cell culture models of the disease available so far (Miranda et al., 2004)(Miranda et al., 2008)(Kroeger et al., 2009)(Davies et al., 2009)(Roussel et al., 2013).
Our first aim in the present work was to create a novel cell model system closer to the conditions found in FENIB. Postmortem analysis of brains from FENIB patients has shown the presence of abundant inclusion bodies in some brain regions, mainly in the cerebral cortex and the substantia nigra (Davis et al., 1999)(Hagen et al., 2011). We thus chose to use NPCs derived from the developing mouse cerebral cortex to establish cell lines overexpressing NS and a selection marker from the same promoter, so NS expression could be maintained stably by culturing the cells in the presence of puromycin. Although these cultures consisted in mixed populations with stable random integration of the transgenes, the use of a bicistronic plasmid led to similar NS expression levels by real-time RT-PCR and homogeneous presence of GFP and NS by immunofluorescence, indicating low variability in our NPC cultures.
Since FENIB is characterised by the presence of inclusion bodies in mature neurons, we differentiated our cells in vitro using previously described protocols driving efficient neuronal differentiation of NPCs (Sinno et al., 2013). This led to a postmitotic population with approximately 60% of neurons, as assessed by morphology and expression of ßIII tubulin, and 40% of glial-like cells, as previously described (Sinno et al., 2013). Our results show that, although protein levels of NS were lower in differentiated cells, NS was handled in the same way both in proliferating and differentiated NPCs, with correct secretion of the wild type variant as a mature glycoprotein and accumulation of G392E NS polymers within the ER, thus reproducing the trafficking defects characteristic of FENIB as described in previous cell models of disease (Miranda et al., 2004)(Miranda et al., 2008)(Roussel et al., 2013)(Moriconi et al., 2015). Serpin polymers are atypical in that their accumulation in the ER does not activate the UPR, although they cause a different type of ER stress characterised by activation of NFkB signalling, the ER overload response or ordered protein response (Davies et al., 2009)(Lawless et al., 2004). In order to control for cellular effects due to classical misfolding and UPR, we also created a cell line overexpressing a truncated version of NS, delta NS, lacking the last third of the protein, which does not fold properly and activates the UPR (Davies et al., 2009). In our cell model, delta NS was efficiently degraded and was detectable only by blocking proteasomal degradation, as expected for a severely misfolded protein.
During the first three days of the differentiation protocol, we observed a strong decline in cultures of NPCs expressing WT NS. This could be due to a toxic effect of the overexpression of functionally active neuroserpin in this system. A similar toxicity was previously observed when generating a D. melanogaster line overexpressing functional WT NS in the fly brain (D. Crowther, personal communication). In contrast, cells expressing G392E NS did not show toxicity during routine culturing or differentiation, so we analysed them by global mRNA sequencing in comparison to control GFP cells. We found 747 genes whose expression levels were altered by at least two-fold, and decided to further investigate those involved in oxidative stress and lipid oxidation according to GO analysis and previously published studies. It has been shown that expression of the polymerogenic Z variant of A1AT in mouse liver leads to upregulation of genes involved in redox regulation, namely carbonyl reductase 3 (CBR3), glutathione S transferase alpha 1 + 2 (GSTA 1 + 2) and glutathione S transferase M3 (GST M3), and this correlated with higher ROS levels and oxidative damage in liver tissue from aged mice transgenic for Z A1AT (Marcus et al., 2012). The involvement of oxidative stress and lipid oxidation in FENIB has not been investigated so far. Our RNA sequencing analysis and real-time RT-PCR data confirmed that several genes of interest were upregulated in G392E NS cells, but not in cells expressing WT NS or the UPR eliciting mutant delta NS, except for a moderate upregulation of Aldh1b1 in WT NS cells. The most upregulated gene in G392E NS cells in comparison with GFP, WT NS and delta NS cells was Scara3, which has been shown to scavenge ROS (Han et al., 1998) and to be protective against cell death induced by oxidative stress in myeloma cells (Brown et al., 2013). Additional genes involved in the defence against oxidative stress and lipid oxidation were also significantly and specifically upregulated in GE392E NS cells: Prdx6 codes for an antioxidant enzyme whose levels increase in brains of patients suffering from Alzheimer's disease (Power et al., 2008) and Parkinson's disease or dementia with Lewy bodies (Power et al., 2002); Aldh1 family genes code for important enzymes that prevent the accumulation of toxic aldehydes derived from lipid oxidation during oxidative stress (Makia et al., 2011); the protein products of Gst family genes are protective against oxidative stress damage, particularly through detoxification of lipid oxidation products [reviewed in Sharma et al., 2004], and upregulation of GST M3 was detected upon expression of a polymerogenic variant of A1AT in mouse liver (Marcus et al., 2012); studies in knock-out mice lacking the lipid transporter apolipoprotein E have shown increased lipid peroxidation in the cortex (Montine et al., 1999). By performing real-time RT-PCR analysis with a higher number of samples than those used for the RNA sequencing analysis, we also found that mRNA levels of Sod2 and Gpx1, two important genes involved in detoxification of ROS [reviewed in Gandhi and Abramov, 2012, Cobb and Cole, 2015], were higher in G392E NS cells. The fact that G392E NS cells show a higher expression of these genes when compared to GFP, WT and delta NS cells supports a specific response in G392E NS cells to the accumulation of NS polymers within the ER, separate from other effects due to overexpression of a heterologous protein or a misfolding protein able to activate the UPR. It has been reported that polymers of mutant Z A1AT reduce the mobility of proteins in the ER lumen, rendering the ER more susceptible to stress (Ordóñez et al., 2013). It is possible that the presence of NS polymers alters the redox balance in the ER by a similar mechanism, although further studies are needed to support this hypothesis.
Based on these results, we hypothesised that polymers of G392E NS caused low levels of oxidative stress in our differentiated NPCs, triggering a protective response involving increased expression of proteins with anti-oxidant activities. Blocking the main anti-oxidant defences by pharmacological depletion of GSH and inhibition of catalase showed that G392E NS cells became more susceptible to oxidative stress than the other cell lines, with increased cell death through apoptosis. These results support the involvement of oxidative stress and lipid oxidation in the progression of the neurodegenerative dementia FENIB, as already demonstrated for more common forms of neurodegeneration as Alzheimer's and Parkinson's diseases [reviewed in Gandhi and Abramov, 2012, Cobb and Cole, 2015]. Furthermore, it is known that anti-oxidative defences decrease with age, which may help to understand why FENIB mutations like G392E and G392R cause higher number of inclusion bodies in the brain at an early age: the young brain, coping better with oxidative stress, would be more resistant to polymer accumulation and thus need a higher burden of polymers to develop the disease, while lower amounts of inclusion bodies in older brains suffice to start neurodegeneration with slower polymer-forming mutant variants of NS, like the S49P (Davis et al., 1999).
A recent report has proposed a protective role for extracellular WT NS against exogenous H2O2, through an increase in phosphorylation of AKT (Cheng et al., 2016). The study does not clarify how this effect is exerted and whether it depends on the inhibitory properties of NS, but it could provide an explanation for our observation that NPCs overexpressing WT NS are more resistant than control cells to addition of H2O2, although not as much as G392E NS expressing cells. The fact that NPCs overexpressing WT NS show similar expression levels to those seen in control cells for all except one of the anti-oxidant genes studied here by real-time RT-PCR, indicates that the signalling activated by WT and G392E NS in NPCs are probably diverse: an extracellular effect for WT NS, which is efficiently secreted as a monomeric protein, and an intracellular response to the intracellular accumulation of polymers in the case of G392E NS.
In conclusion, we show here for the first time that expression of a strongly polymerogenic variant of NS that causes severe dementia FENIB leads to upregulation of several anti-oxidant genes and to apoptotic neural cell death upon impairment of the oxidative stress defences, suggesting that oxidative stress and lipid oxidation may contribute to neurodegeneration in FENIB.
The following are the supplementary data related to this article.
List of genes identified in our RNA sequencing analysis or through literature search as potentially interesting with regards to oxidative stress caused by expression of mutant G392E NS in NPCs. FDR: false discovery rate.
List of primers used in this study for real-time RT-PCR analyses.
Conflict of interest statement
The authors declare no conflicts of interest.
Funding
This work was supported by the Telethon Foundation (Italy, grant GGP11057 to E.M.), the Pasteur Institute - Cenci Bolognetti Foundation (Italy, call 2010-under 40 grant to E.M.), and Sapienza University of Rome (Scientific Research grants 2013, 2014 and 2015 to E.M, and Starter grant 2013 to N.A.G.).
Acknowledgments
We are grateful to Silvana Caristi for technical support with confocal microscopy.
Contributor Information
Giuseppe Lupo, Email: giuseppe.lupo@uniroma1.it.
Elena Miranda, Email: mariaelena.mirandabanos@uniroma1.it.
References
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.O., Schibler J., Fitzgerald M.P., Singh N., Salem K., Zhan F., Goel A. Scavenger receptor class A member 3 (SCARA3) in disease progression and therapy resistance in multiple myeloma. Leuk. Res. 2013;37:963–969. doi: 10.1016/j.leukres.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Loh Y.P., Birch N.P. Neuroserpin attenuates H2O2-induced oxidative stress in hippocampal neurons via AKT and BCL-2 signaling pathways. J. Mol. Neurosci. MN. 2016 doi: 10.1007/s12031-016-0807-7. [DOI] [PubMed] [Google Scholar]
- Cobb C.A., Cole M.P. Oxidative and nitrative stress in neurodegeneration. Neurobiol. Dis. 2015;84:4–21. doi: 10.1016/j.nbd.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L., Pollard S.M., Gorba T., Reitano E., Toselli M., Biella G., Sun Y., Sanzone S., Ying Q.-L., Cattaneo E., Smith A. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutelier M., Andries S., Ghariani S., Dan B., Duyckaerts C., van Rijckevorsel K., Raftopoulos C., Deconinck N., Sonderegger P., Scaravilli F., Vikkula M., Godfraind C. Neuroserpin mutation causes electrical status epilepticus of slow-wave sleep. Neurology. 2008;71:64–66. doi: 10.1212/01.wnl.0000316306.08751.28. [DOI] [PubMed] [Google Scholar]
- Croft D., Mundo A.F., Haw R., Milacic M., Weiser J., Wu G., Caudy M., Garapati P., Gillespie M., Kamdar M.R., Jassal B., Jupe S., Matthews L., May B., Palatnik S., Rothfels K., Shamovsky V., Song H., Williams M., Birney E., Hermjakob H., Stein L., D'Eustachio P. The reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M.J., Miranda E., Roussel B.D., Kaufman R.J., Marciniak S.J., Lomas D.A. Neuroserpin polymers activate NF-kappaB by a calcium signaling pathway that is independent of the unfolded protein response. J. Biol. Chem. 2009;284:18202–18209. doi: 10.1074/jbc.M109.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.L., Shrimpton A.E., Holohan P.D., Bradshaw C., Feiglin D., Collins G.H., Sonderegger P., Kinter J., Becker L.M., Lacbawan F., Krasnewich D., Muenke M., Lawrence D.A., Yerby M.S., Shaw C.M., Gooptu B., Elliott P.R., Finch J.T., Carrell R.W., Lomas D.A. Familial dementia caused by polymerization of mutant neuroserpin. Nature. 1999;401:376–379. doi: 10.1038/43894. [DOI] [PubMed] [Google Scholar]
- Davis R.L., Shrimpton A.E., Carrell R.W., Lomas D.A., Gerhard L., Baumann B., Lawrence D.A., Yepes M., Kim T.S., Ghetti B., Piccardo P., Takao M., Lacbawan F., Muenke M., Sifers R.N., Bradshaw C.B., Kent P.F., Collins G.H., Larocca D., Holohan P.D. Association between conformational mutations in neuroserpin and onset and severity of dementia. Lancet Lond. Engl. 2002;359:2242–2247. doi: 10.1016/S0140-6736(02)09293-0. [DOI] [PubMed] [Google Scholar]
- Dringen R., Pawlowski P.G., Hirrlinger J. Peroxide detoxification by brain cells. J. Neurosci. Res. 2005;79:157–165. doi: 10.1002/jnr.20280. [DOI] [PubMed] [Google Scholar]
- Galliciotti G., Glatzel M., Kinter J., Kozlov S.V., Cinelli P., Rülicke T., Sonderegger P. Accumulation of mutant neuroserpin precedes development of clinical symptoms in familial encephalopathy with neuroserpin inclusion bodies. Am. J. Pathol. 2007;170:1305–1313. doi: 10.2353/ajpath.2007.060910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S., Abramov A.Y. Mechanism of oxidative stress in neurodegeneration. Oxidative Med. Cell. Longev. 2012;2012:428010. doi: 10.1155/2012/428010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A.J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J.Y.H., Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen M.C., Murrell J.R., Delisle M.-B., Andermann E., Andermann F., Guiot M.C., Ghetti B. Encephalopathy with neuroserpin inclusion bodies presenting as progressive myoclonus epilepsy and associated with a novel mutation in the proteinase inhibitor 12 gene. Brain Pathol. 2011;21:575–582. doi: 10.1111/j.1750-3639.2011.00481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H.J., Tokino T., Nakamura Y. CSR, a scavenger receptor-like protein with a protective role against cellular damage causedby UV irradiation and oxidative stress. Hum. Mol. Genet. 1998;7:1039–1046. doi: 10.1093/hmg/7.6.1039. [DOI] [PubMed] [Google Scholar]
- Kroeger H., Miranda E., MacLeod I., Pérez J., Crowther D.C., Marciniak S.J., Lomas D.A. Endoplasmic reticulum-associated degradation (ERAD) and autophagy cooperate to degrade polymerogenic mutant serpins. J. Biol. Chem. 2009;284:22793–22802. doi: 10.1074/jbc.M109.027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawless M.W., Greene C.M., Mulgrew A., Taggart C.C., O'Neill S.J., McElvaney N.G. Activation of endoplasmic reticulum-specific stress responses associated with the conformational disease Z alpha 1-antitrypsin deficiency. J. Immunol. 2004;1950(172):5722–5726. doi: 10.4049/jimmunol.172.9.5722. [DOI] [PubMed] [Google Scholar]
- Li J.O.W., Li W., Jiang Z.-G., Ghanbari H.A. Oxidative stress and neurodegenerative disorders. Int. J. Mol. Sci. 2013;14:24438–24475. doi: 10.3390/ijms141224438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinforma. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makia N.L., Bojang P., Falkner K.C., Conklin D.J., Prough R.A. Murine hepatic aldehyde dehydrogenase 1a1 is a major contributor to oxidation of aldehydes formed by lipid peroxidation. Chem. Biol. Interact. 2011;191:278–287. doi: 10.1016/j.cbi.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra J.D., Kaufman R.J. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid. Redox Signal. 2007;9:2277–2293. doi: 10.1089/ars.2007.1782. [DOI] [PubMed] [Google Scholar]
- Marcus N.Y., Blomenkamp K., Ahmad M., Teckman J.H. Oxidative stress contributes to liver damage in a murine model of alpha-1-antitrypsin deficiency. Exp. Biol. Med. 2012;237:1163–1172. doi: 10.1258/ebm.2012.012106. (Maywood NJ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margoliash E., Novogrodsky A., Schejter A. Irreversible reaction of 3-amino-1:2:4-triazole and related inhibitors with the protein of catalase. Biochem. J. 1960;74:339–348. doi: 10.1042/bj0740339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda E., Römisch K., Lomas D.A. Mutants of neuroserpin that cause dementia accumulate as polymers within the endoplasmic reticulum. J. Biol. Chem. 2004;279:28283–28291. doi: 10.1074/jbc.M313166200. [DOI] [PubMed] [Google Scholar]
- Miranda E., MacLeod I., Davies M.J., Pérez J., Römisch K., Crowther D.C., Lomas D.A. The intracellular accumulation of polymeric neuroserpin explains the severity of the dementia FENIB. Hum. Mol. Genet. 2008;17:1527–1539. doi: 10.1093/hmg/ddn041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.B., Russo A., Biaglow J.E., McPherson S. Cellular glutathione depletion by diethyl maleate or buthionine sulfoximine: no effect of glutathione depletion on the oxygen enhancement ratio. Radiat. Res. 1983;96:422–428. [PubMed] [Google Scholar]
- Montine T.J., Montine K.S., Olson S.J., Graham D.G., Roberts L.J., Morrow J.D., Linton M.F., Fazio S., Swift L.L. Increased cerebral cortical lipid peroxidation and abnormal phospholipids in aged homozygous apoE-deficient C57BL/6J mice. Exp. Neurol. 1999;158:234–241. doi: 10.1006/exnr.1999.7067. [DOI] [PubMed] [Google Scholar]
- Moriconi C., Ordoñez A., Lupo G., Gooptu B., Irving J.A., Noto R., Martorana V., Manno M., Timpano V., Guadagno N.A., Dalton L., Marciniak S.J., Lomas D.A., Miranda E. Interactions between N-linked glycosylation and polymerisation of neuroserpin within the endoplasmic reticulum. FEBS J. 2015;282:4565–4579. doi: 10.1111/febs.13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordóñez A., Snapp E.L., Tan L., Miranda E., Marciniak S.J., Lomas D.A. Endoplasmic reticulum polymers impair luminal protein mobility and sensitize to cellular stress in alpha1-antitrypsin deficiency. Hepatol. Baltim. Md. 2013;57:2049–2060. doi: 10.1002/hep.26173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.H.T., Shannon J.M., Blumbergs P.C., Gai W.-P. Nonselenium glutathione peroxidase in human brain: elevated levels in Parkinson's disease and dementia with lewy bodies. Am. J. Pathol. 2002;161:885–894. doi: 10.1016/S0002-9440(10)64249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.H.T., Asad S., Chataway T.K., Chegini F., Manavis J., Temlett J.A., Jensen P.H., Blumbergs P.C., Gai W.-P. Peroxiredoxin 6 in human brain: molecular forms, cellular distribution and association with Alzheimer's disease pathology. Acta Neuropathol. 2008;115:611–622. doi: 10.1007/s00401-008-0373-3. (Berl.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt T., Sharp L., Nichols J., Price D.J., Mason J.O. Embryonic stem cells and transgenic mice ubiquitously expressing a tau-tagged green fluorescent protein. Dev. Biol. 2000;228:19–28. doi: 10.1006/dbio.2000.9935. [DOI] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2015. R: A language and environment for statistical computing. URL https://www.R-project.org/ [Google Scholar]
- Roussel B.D., Irving J.A., Ekeowa U.I., Belorgey D., Haq I., Ordóñez A., Kruppa A.J., Duvoix A., Rashid S.T., Crowther D.C., Marciniak S.J., Lomas D.A. Unravelling the twists and turns of the serpinopathies. FEBS J. 2011;278:3859–3867. doi: 10.1111/j.1742-4658.2011.08201.x. [DOI] [PubMed] [Google Scholar]
- Roussel B.D., Newton T.M., Malzer E., Simecek N., Haq I., Thomas S.E., Burr M.L., Lehner P.J., Crowther D.C., Marciniak S.J., Lomas D.A. Sterol metabolism regulates neuroserpin polymer degradation in the absence of the unfolded protein response in the dementia FENIB. Hum. Mol. Genet. 2013;22:4616–4626. doi: 10.1093/hmg/ddt310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz J.B., Lindenau J., Seyfried J., Dichgans J. Glutathione, oxidative stress and neurodegeneration. Eur. J. Biochem. 2000;267:4904–4911. doi: 10.1046/j.1432-1327.2000.01595.x. (FEBS) [DOI] [PubMed] [Google Scholar]
- Sharma R., Yang Y., Sharma A., Awasthi S., Awasthi Y.C. Antioxidant role of glutathione S-transferases: protection against oxidant toxicity and regulation of stress-mediated apoptosis. Antioxid. Redox Signal. 2004;6:289–300. doi: 10.1089/152308604322899350. [DOI] [PubMed] [Google Scholar]
- Sinno M., Biagioni S., Ajmone-Cat M.A., Pafumi I., Caramanica P., Medda V., Tonti G., Minghetti L., Mannello F., Cacci E. The matrix metalloproteinase inhibitor marimastat promotes neural progenitor cell differentiation into neurons by gelatinase-independent TIMP-2-dependent mechanisms. Stem Cells Dev. 2013;22:345–358. doi: 10.1089/scd.2012.0299. [DOI] [PubMed] [Google Scholar]
- Soldati C., Cacci E., Biagioni S., Carucci N., Lupo G., Perrone-Capano C., Saggio I., Augusti-Tocco G. Restriction of neural precursor ability to respond to Nurr1 by early regional specification. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L., Rugg-Gunn P.J., Bouhon I.A., Andersson F.K., Sadler A.J., Pedersen R.A. Enhancing and diminishing gene function in human embryonic stem cells. Stem Cells. 2004;22:2–11. doi: 10.1634/stemcells.22-1-2. (Dayt. Ohio) [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes identified in our RNA sequencing analysis or through literature search as potentially interesting with regards to oxidative stress caused by expression of mutant G392E NS in NPCs. FDR: false discovery rate.
List of primers used in this study for real-time RT-PCR analyses.