Abstract
Development of lung adenocarcinoma (LADC), the most frequent histological type of lung cancer, depends in many cases on the activation of “driver” oncogenes such as KRAS, epidermal growth factor receptor (EGFR), and anaplastic lymphoma kinase (ALK). Inhibitors that target the EGFR and ALK tyrosine kinases show therapeutic effects against LADCs containing EGFR gene mutations and ALK gene fusions, respectively. Recently, we and others identified the RET fusion gene as a new targetable driver gene in LADC. The RET fusions occur in 1–2% of LADCs. Existing US Food and Drug Administration‐approved inhibitors of RET tyrosine kinase show promising therapeutic effects both in vitro and in vivo, as well as in a few patients. Clinical trials are underway to investigate the therapeutic effects of RET tyrosine kinase inhibitors, such as vandetanib (ZD6474) and cabozantinib (XL184), in patients with RET fusion‐positive non‐small‐cell lung cancer.
Personalized Therapy of LADC
Lung cancer is the leading cause of cancer‐related mortality worldwide. Lung adenocarcinoma (LADC) is the most frequent type of lung cancer. LADC occurs both in smokers and non‐smokers, and its incidence is increasing.1 Genome analyses of LADC show that these tumors contain distinct genetic alterations that activate oncogenes.2, 3 Genetic alterations that result in the activation of several oncogenes are detected in a mutually exclusive manner (Fig. 1); of the hundreds of genes mutated in each case of LADC, these oncogenes are considered to be “driver genes”.4 Remarkably, molecular targeted therapy using inhibitory drugs against activated oncogene products has begun to replace conventional chemotherapy using cytotoxic drugs, even for first‐line use.2
Figure 1.
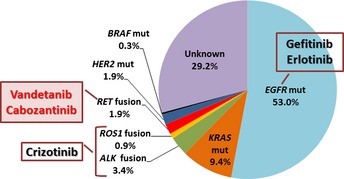
Pie chart showing the fraction of Japanese lung adenocarcinoma patients that harbor “driver” gene mutations. Surgical specimens from 319 stage I–II lung adenocarcinomas deposited in the National Cancer Center Biobank (Japan) were subjected to analysis. The EGFR,KRAS,BRAF, and HER2 mutations (mut) were examined using the high resolution melting method, whereas ALK,ROS1 and RET fusions were examined by RT‐PCR.12, 31 The protocol for this research project has been approved by the institutional review board of the National Cancer Center.
The epidermal growth factor receptor (EGFR) gene is activated by single amino acid substitution mutations or in‐frame amino acid deletion mutations in 10–20% of LADC cases in the USA and in 30–40% of cases in East Asia.2 Tumors harboring these EGFR mutations respond to EGFR tyrosine kinase inhibitors (TKIs) such as erlotinib and gefitinib, thereby improving progression‐free survival and quality of life.5, 6 In addition, 3–5% of LADC harbor fusions that result in the activation of the anaplastic lymphoma kinase (ALK) gene; such mutations are mutually exclusive with EGFR mutations. Inhibitors, such as crizotinib, that target ALK tyrosine kinase show marked therapeutic effects against ALK fusion‐positive LADCs.7, 8, 9 These results indicate that personalized therapy for LADC using TKIs selected on the basis of somatic genetic alterations has been realized already; indeed, 20% of USA/European and 40% of Asian LADC patients benefit from such therapies.
Discovery of the RET Fusion Gene as a New Targetable Driver Gene
In 2012, four studies, including one by our group, identified fusions of the RET (rearranged during transfection) oncogene10, 11, 12, 13 (Fig. 2). RET is a well‐known driver oncogene kinase for thyroid cancer, and both activating mutations and fusions of this gene have been observed.14, 15 Germline gain‐of‐function mutations in RET predispose carriers to multiple endocrine neoplasia type 2, which is characterized by medullary thyroid cancer, pheochromocytoma, and hyperparathyroidism, and also to familial medullary thyroid carcinoma syndrome. Somatic gain‐of‐function RET mutations have been observed in 30–50% of sporadic medullary thyroid cancer, and somatic RET gene fusions have been observed in 30–50% of sporadic papillary thyroid cancer. The US Food and Drug Administration (FDA) have approved two inhibitory drugs, vandetanib (ZD6474) and cabozantinib (XL184), for the treatment of advanced medullary thyroid cancer. The molecular process for generating a RET fusion is similar to the mechanism underlying ALK fusion: the most frequent RET fusion, KIF5B–RET, is generated by a pericentric inversion in chromosome 10, whereas the most frequent ALK fusion, EML4–ALK, is generated by a paracentric inversion in chromosome 2 (Fig. 2).
Figure 2.
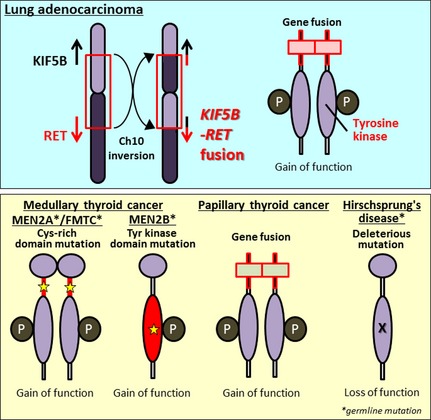
Involvement of the RET gene in lung and thyroid carcinogenesis and in a developmental disorder. Upper panel, somatic inversion in chromosome 10 results in KIF5B–RET fusions. The RET fusion protein has constitutive tyrosine (Tyr) kinase activity, representing a gain‐of‐function alteration. Lower panel, RET alterations in other diseases. A germline gain‐of‐function mutation of RET drives thyroid carcinogenesis in patients with multiple endocrine neoplasia type 2 (MEN2). Somatic gain‐of‐function mutation and translocation of RET cause medullary and papillary thyroid cancers, respectively. Germline loss‐of‐function RET mutations cause Hirschsprung's disease, a hereditary disorder characterized by the absence of enteric ganglia in variable segments of intestine. FMTC, familial medullary thyroid carcinoma; P, phosphorylation; X, inactivating mutation.
Four different strategies resulted in the discovery of the same RET fusion gene (Table 1, Fig. 3). We carried out whole‐transcriptome sequencing using RNA from 30 snap‐frozen surgical LDAC specimens to identify novel fusion‐gene transcripts.12 Ju et al.13 analyzed the whole genome and transcriptome of a single young (33‐year‐old) LADC patient. Lipson et al.11 carried out targeted‐capture sequencing of 145 cancer‐relevant genes from genomic DNA obtained from 24 formalin‐fixed paraffin‐embedded tumor samples to identify genes mutated or fused in LADC. Takeuchi et al.10 carried out a FISH‐based screen against known fusion kinase and partner genes to detect rearrangement of oncogenes in >1500 LADC cases.
Table 1.
Prevalence of RET gene fusion in non‐small‐cell lung cancer (NSCLC)
| Institution | No. of cases examined | No. of RET fusion (+) cases | RET fusion% | Fusion type | Ref. |
|---|---|---|---|---|---|
| NSCLC/lung adenocarcinoma | |||||
| National Cancer Center, Japan | 704/433 | 7/7 | 1.0/1.6 | KIF5B–RET: 7 | 12 |
| Japan Foundation for Cancer Research, Japan | 1482/1119 | 13/13 | 0.9/1.2 | KIF5B–RET: 12 | 10 |
| CCDC6–RET: 1 | |||||
| Foundation Med, USA | 643/561 | 12/12 | 1.8/2.1 | KIF5B–RET: 12 | 11 |
| Seoul National University, Korea | 21/21 | 3/3 | 14/14 | KIF5B–RET: 3 | 13 |
| (Driver mutation −) | |||||
| Chinese Academy of Sciences, China | 202/202 | 2/2 | 1.0/1.0 | CCDC6–RET: 2 | 24 |
| (Driver mutation −) | |||||
| Nagoya City University, Japan | 371/270 | 3/3 | 0.8/1.1 | KIF5B–RET: 3 | 23 |
| Memorial Sloan‐Kettering Cancer Center, USA | 69/69 | 1/1 | 1.4/1.4 | KIF5B–RET: 1 | 21 |
| (Driver mutation −) | |||||
| Fudan University Shanghai Cancer Center, China | 936/633 | 13/11 | 1.4/1.7 | KIF5B–RET: 9 | 20 |
| CCDC6–RET: 3 | |||||
| NCOA4–RET: 1 | |||||
| Tongji University School of Medicine, China | 392/231 | 6/4 | 1.5/1.7 | KIF5B–RET: 6 | 19 |
| Korea Research Institute of Bioscience and Biotechnology, Korea | 6/6 | 1/1 | 17/17 | CCDC6–RET: 1 | 22 |
| (Female non‐smoker) | |||||
| Memorial Sloan‐Kettering Cancer Center, USA | 31/31 | 5/5 | 16/16 | KIF5B–RET: 2 | 16 |
| (Driver mutation −) | TRIM33–RET: 1 | ||||
| (Unknown: 2) | |||||
| Total | 4857/3576 | 66/62 | 1.4/1.8 | KIF5B–RET: 55 | |
| CCDC6–RET: 7 | |||||
| NCOA4–RET: 1 | |||||
| TRIM33–RET: 1 | |||||
Figure 3.
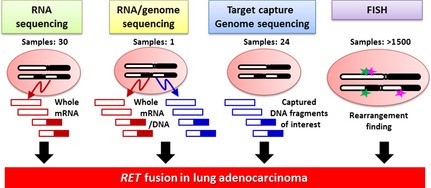
Strategies used to identify RET fusion in lung adenocarcinoma. Four different methods were used to identify novel oncogenic fusions in lung adenocarcinomas.10, 11, 12, 13
To date, RET fusions have been identified that involve four fusion partners comprising nine subtypes of fusion variants: KIF5B, CCDC6/PTC/H4, NCO4/PTC3/ELE1, and TRIM33/PTC7.16 The latter three partners are also fused to RET in thyroid cancer, whereas KIF5B is not. The deduced features of the proteins encoded by all types of RET fusion gene are similar to those of ALK: coiled‐coil domains in the N‐terminal fusion partners cause the RET domains to dimerize, resulting in activation of RET tyrosine kinase in the absence of ligands (Fig. 2). The ligand‐independent dimerization and constitutive activation of RET protein are also caused by gain‐of‐function mutations and translocations of RET, which have been detected in sporadic and hereditary thyroid cancers.15 In fact, autophosphorylation of the KIF5B–RET fusion protein, representing RET protein activation, was observed in LADC tissues harboring the corresponding RET fusion gene,12 as well as in cells cultured in the absence of serum. The transforming and signal‐addictive activities of KIF5B–RET fusion proteins are suppressed by FDA‐approved drugs (e.g., vandetanib, sorafenib, and sunitinib), which themselves suppress RET kinase.10, 11, 12 In addition, the LADC cell line, LC‐2/ad, which harbors a CCDC6–RET fusion, is sensitive to these drugs both in vitro and in vivo.17, 18 Unfortunately, these drugs are not approved for use as treatments for lung cancer; however, the existing data led us to investigate their therapeutic effects in clinical trials, as described below.
Prevalence and Characteristics of RET Fusion‐Positive LADC
Several studies have validated the presence of RET fusion in a small subset of non‐small‐cell lung cancers (NSCLCs).16, 19, 20, 21, 22, 23, 24 The total number of examined cases has reached approximately 5000 (Table 1). Most of the positive cases are LADC, but several cases involve other histological types of NSCLC, such as adenosquamous carcinoma.19, 20 The RET fusions are present in 1–2% of NSCLC/ADC of patients of both Asian and European descent. Several studies indicate that RET fusion occurs preferentially in young, never‐smoker, and light‐smoker patients.10, 12, 20
The LADCs harboring KIF5B–RET fusions are well or moderately differentiated, similar to LADCs harboring EGFR mutations. This is in contrast to EML4–ALK fusion‐positive LADCs, which tend to show signet‐ring and mucinous cribriform patterns.10 Those LADCs harboring CCDC6–RET fusions show such histological features.10, 18
In our previous study, we did not detect RET fusions in a screen of 234 squamous cell, 17 large cell, and 20 small‐cell lung cancers.12 Adenocarcinomas of other organs, such as colon (n = 200) and ovary (n = 100), were also negative for RET fusion. To date, whole‐transcriptome analysis of other organs has not identified RET fusions in cancers outside the lung. Therefore, RET fusion may occur mainly in LADC and papillary thyroid cancer.
Therapeutic Effects of RET TKIs in Patients with RET Fusion‐Positive NSCLC
In clinical trials, the ALK TKI, crizotinib, showed a dramatic therapeutic effect against NSCLCs harboring ALK gene fusions. Crizotinib was approved for use in the USA in August 2011 and for use in Japan in March 2012.8 Considering that the ALK gene fusion was first identified in NSCLC in 2007, approval has been achieved extremely rapidly. Consequently, the discovery of the RET fusion has raised expectations that patients with NSCLCs harboring RET fusions will soon benefit from targeted therapy using existing RET TKIs.
Several commercially available multikinase inhibitors, such as vandetanib (ZD6474), cabozantinib (XL184), sorafenib, sunitinib, lenvatinib (E7080), and ponatinib (AP24534), have activity against the RET kinase; however, no selective RET inhibitors have yet been developed for clinical use. Several phase II clinical trials have been initiated to investigate the therapeutic effects of such multikinase inhibitors in patients with advanced RET fusion‐positive NSCLC (Table 2). As for previous clinical trials of ALK TKIs, all of these trials have open‐label and single‐arm designs, with response rate as the primary endpoint. One study, carried out by Drilon et al. at the Memorial Sloan‐Kettering Cancer Center (NCT01639508), is testing cabozantinib, a drug recently approved by the FDA for the treatment of thyroid cancer. The therapeutic responses of the first three patients to be treated with cabozantinib were reported to be promising (Table 3).16
Table 2.
Ongoing phase II clinical trials of RET tyrosine kinase inhibitors in patients with RET fusion‐positive non‐small‐cell lung carcinoma
| Trial number† | Drug (pharmaceutical company) | Study design | Primary end‐point | Enrolment no. | Study start |
|---|---|---|---|---|---|
| NCT01639508 | Cabozantinib/XL184 (Exelixis) | 25 | July 2012 | ||
| UMIN000010095 | Vandetanib/ZD6474 (AstraZeneca) | 17 | Feb 2013 | ||
| NCT01823068 | Vandetanib/ZD6474 (AstraZeneca) | Open‐label, single arm | Response rate | 17 | April 2013 |
| NCT01877083 | Lenvatinib/E7080 (Eisai) | 20 | April 2013 | ||
| NCT01813734 | Ponatinib/AP24534 (ARIAD) | 20 | June 2013 |
†Detailed information is available at http://clinicaltrials.gov/ or https://upload.umin.ac.jp.
Table 3.
Response of lung adenocarcinoma patients to RET tyrosine kinase inhibitors
| Patient | RET fusion gene | Inhibitor | Ethnicity | Sex | Age, years | Pathological diagnosis | Smoking history (pack‐year) | Response (% decrease) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| 1 | TRIM33–RET | Cabozantinib | Caucasian | Female | 41 | Papillary adenocarcinoma | Never‐smoker | Partial response (66) | 16 |
| 2 | KIF5B–RET | Cabozantinib | African‐American | Female | 75 | Poorly differentiated adenocarcinoma | Never‐smoker | Partial response (32) | 16 |
| 3 | KIF5B–RET | Cabozantinib | Caucasian | Female | 68 | Mixed subtype adenocarcinoma | Never‐smoker | Stable disease | 16 |
| 4 | KIF5B–RET | Vandetanib | Caucasian | Male | 58 | Poorly differentiated adenocarcinoma | Former smoker (5) | Decrease in size | 26 |
The other phase II clinical trial was initiated by our own group in Japan (UMIN00001009). This trial, designated LURET (Lung Cancer with RET rearrangement study), is investigating the therapeutic effects of vandetanib in 17 patients with RET fusion‐positive NSCLC (Table 2). Because vandetanib is a multikinase inhibitor that is effective against EGFR and vascular endothelial growth factor, this drug was previously examined for its therapeutic efficacy in advanced NSCLC patients in several “all‐comer” clinical trials.25 Those trials were carried out without considering gene alterations in determining eligibility, and the trials did not show significantly greater therapeutic effects than pre‐existing therapeutic regimens. Therefore, only RET fusion‐positive cases, which represent 1–2% of all NSCLCs, are eligible for the LURET study.
To evaluate eligibility for this study, we established a diagnostic method for detecting RET fusions using a combination of RT‐PCR and FISH (Fig. 4). In this study, RNAs from frozen biopsy tissue or pleural effusion from patients with non‐squamous NSCLCs without EGFR mutations are subjected to RT‐PCR; this method enables us to detect all seven KIF5B–RET and CCDC6–RET variants identified to date.16 The positive cases are then subjected to break‐apart and fusion FISH to validate the RT‐PCR results. Cases positive by both RT‐PCR and FISH are eligible for the LURET study. The RT‐PCR screening is being carried out in >100 hospitals throughout Japan by a consortium designated LC‐SCRUM (Lung Cancer Genomic Screening Project for Individualized Medicine in Japan). The therapeutic results will be obtained within 2 years.
Figure 4.
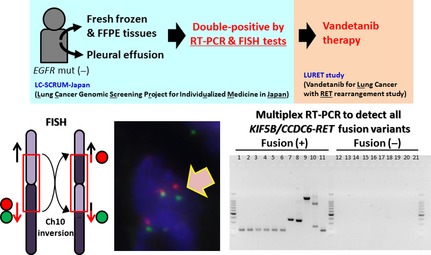
Consolidated Standards of Reporting Trials diagram of the Lung Cancer Genomic Screening Project for Individualized Medicine in Japan (LC‐SCRUM) and the Lung Cancer with RET rearrangement (LURET) study in Japan. The LC‐SCRUM screen identified 17 RET fusion‐positive cases from non‐squamous non‐small‐cell lung carcinoma cases without epidermal growth factor receptor (EGFR) mutations (mut). The RET fusion‐positive cases are defined as being positive in both RT‐PCR and subsequent FISH tests. Representative pictures of these tests are shown. Fusion‐positive cases were treated with vandetanib in the LURET study. Ch10, chromosome 10; FFPE, formalin‐fixed paraffin‐embedded.
Notably, a recent study reported that one patient with LADC harboring a KIF5B–RET fusion responded to vandetanib (Table 3). The patient was Caucasian male and a former smoker. Tumor shrinkage was observed starting in the first week, and continued for 4 weeks.26
Perspective
The RET gene is predicted to be an additional therapeutic target for therapy against LADC. Three other oncogene kinases, HER2 (activated by inflame insertion mutations), BRAF (activated by point mutation), and ROS1 (activated by gene fusion) are also promising targets for personalized therapy in addition to EGFR and ALK (Fig. 1). In fact, inhibition of these kinases has yielded therapeutic effects in several lung cancer patients. The LADCs harboring HER2 mutations responded to therapy with anti‐HER2 antibodies and HER2 TKIs.27 One LADC case harboring a BRAF mutation responded to therapy with vemurafenib, an FDA‐approved drug for the treatment of melanoma.28 The ALK TKI, crizotinib, suppresses the activity of the ROS1 tyrosine kinase due to the high structural similarity between the ALK and ROS1 tyrosine kinase domains. Consistent with this, a significant portion of the LADC patients with ROS1 fusions that were enrolled in a clinical trial responded to crizotinib.29 Therefore, developing therapies that target RET and other kinases means that increasing numbers of LADC patients will benefit from personalized therapy (Fig. 1). Thus, LADC represents a type of cancer in which “precision cancer medicine”30 based on somatic gene alterations will be realized.
Acquisition of drug resistance is a serious problem for therapies based on TKIs. The LADCs harboring ALK fusions become resistant to crizotinib by acquiring second‐site mutations in the gatekeeper region of ALK tyrosine kinase.7 Those LADCs harboring ROS1 fusions also become resistant to crizotinib, in this case through second‐site mutations in the gatekeeper region of ROS1.29 Therefore, RET fusion‐positive LADCs might also acquire resistance to RET TKIs through the same mechanism. Clinical trials of RET TKIs as a treatment for fusion‐positive NSCLCs should be carried out carefully, and focus both on efficacy and the acquisition of resistance.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
The authors thank all the collaborators in the National Cancer Center and the LC‐SCRUM/LURET studies. This work was supported in part by: the Program for Promotion of Fundamental Studies in Health Sciences from the National Institute of Biomedical Innovation; Grants‐in‐Aid from the Ministry of Health, Labor, and Welfare for the Third‐term Comprehensive 10‐year Strategy for Cancer Control and for Research on New Drug and Medical Device Development; and the National Cancer Center Research and Development Fund. The National Cancer Center Biobank is supported by the National Cancer Center Research and Development Fund, Japan.
(Cancer Sci 2013; 104: 1396–1400)
References
- 1. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010. (Sep–Oct); 60: 277–300. [DOI] [PubMed] [Google Scholar]
- 2. Oxnard GR, Binder A, Janne PA. New targetable oncogenes in non‐small‐cell lung cancer. J Clin Oncol 2013. (Mar); 31: 1097–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nat Med 2012. (Mar); 18: 349–51. [DOI] [PubMed] [Google Scholar]
- 4. Imielinski M. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012; 150: 1107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maemondo M, Inoue A, Kobayashi K et al Gefitinib or chemotherapy for non‐small‐cell lung cancer with mutated EGFR. N Engl J Med 2010. (Jun 24); 362: 2380–8. [DOI] [PubMed] [Google Scholar]
- 6. Oizumi S, Kobayashi K, Inoue A et al Quality of life with gefitinib in patients with EGFR‐mutated non‐small cell lung cancer: quality of life analysis of North East Japan Study Group 002 Trial. Oncologist 2012; 17: 863–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shaw AT, Engelman JA. ALK in lung cancer: past, present, and future. J Clin Oncol 2013. (Mar 10); 31: 1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mano H. ALKoma: a cancer subtype with a shared target. Cancer Discov 2012. (Jun); 2: 495–502. [DOI] [PubMed] [Google Scholar]
- 9. Sakamoto H, Tsukaguchi T, Hiroshima S et al CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell 2011. (May 17); 19: 679–90. [DOI] [PubMed] [Google Scholar]
- 10. Takeuchi K, Soda M, Togashi Y et al RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012. (Mar); 18: 378–81. [DOI] [PubMed] [Google Scholar]
- 11. Lipson D, Capelletti M, Yelensky R et al Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012. (Mar); 18: 382–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kohno T, Ichikawa H, Totoki Y et al KIF5B‐RET fusions in lung adenocarcinoma. Nat Med 2012. (Mar); 18: 375–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ju YS, Lee WC, Shin JY et al A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole‐genome and transcriptome sequencing. Genome Res 2012. (Mar); 22: 436–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gild ML, Bullock M, Robinson BG, Clifton‐Bligh R. Multikinase inhibitors: a new option for the treatment of thyroid cancer. Nat Rev Endocrinol 2011. (Oct); 7: 617–24. [DOI] [PubMed] [Google Scholar]
- 15. Borrello MG, Ardini E, Locati LD, Greco A, Licitra L, Pierotti MA. RET inhibition: implications in cancer therapy. Expert Opin Ther Targets 2013. (Apr); 17: 403–19. [DOI] [PubMed] [Google Scholar]
- 16. Drilon A, Wang L, Hasanovic A et al Response to cabozantinib in patients with RET fusion‐positive lung adenocarcinomas. Cancer Discov 2013. (Jun); 3: 630–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suzuki M, Makinoshima H, Matsumoto S et al Identification of a lung adenocarcinoma cell line with CCDC6‐RET fusion gene and the effect of RET inhibitors in vitro and in vivo. Cancer Sci 2013. (Apr 11); 104: 896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsubara D, Kanai Y, Ishikawa S et al Identification of CCDC6‐RET fusion in the human lung adenocarcinoma cell line, LC‐2/ad. J Thorac Oncol 2012. (Dec); 7: 1872–6. [DOI] [PubMed] [Google Scholar]
- 19. Cai W, Su C, Li X et al KIF5B‐RET fusions in Chinese patients with non‐small cell lung cancer. Cancer 2013. (Apr 15); 119: 1486–94. [DOI] [PubMed] [Google Scholar]
- 20. Wang R, Hu H, Pan Y et al RET fusions define a unique molecular and clinicopathologic subtype of non‐small‐cell lung cancer. J Clin Oncol 2012. (Dec 10); 30: 4352–9. [DOI] [PubMed] [Google Scholar]
- 21. Suehara Y, Arcila M, Wang L et al Identification of KIF5B‐RET and GOPC‐ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA‐based screen for tyrosine kinase fusions. Clin Cancer Res 2012. (Dec 15); 18: 6599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim SC, Jung Y, Park J et al A high‐dimensional, deep‐sequencing study of lung adenocarcinoma in female never‐smokers. PLoS ONE 2013; 8: e55596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yokota K, Sasaki H, Okuda K et al KIF5B/RET fusion gene in surgically‐treated adenocarcinoma of the lung. Oncol Rep 2012. (Oct); 28: 1187–92. [DOI] [PubMed] [Google Scholar]
- 24. Li F, Feng Y, Fang R et al Identification of RET gene fusion by exon array analyses in “pan‐negative” lung cancer from never smokers. Cell Res 2012. (May); 22: 928–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chu CT, Sada YH, Kim ES. Vandetanib for the treatment of lung cancer. Expert Opin Investig Drugs 2012. (Aug); 21: 1211–21. [DOI] [PubMed] [Google Scholar]
- 26. Gautschi O, Zander T, Keller FA et al A patient with lung adenocarcinoma and RET fusion treated with vandetanib. J Thorac Oncol 2013. (May); 8(5): e43–4. [DOI] [PubMed] [Google Scholar]
- 27. Mazieres J, Peters S, Lepage B et al Lung cancer that Harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol 2013. (Jun 1); 31: 1997–2003. [DOI] [PubMed] [Google Scholar]
- 28. Gautschi O, Pauli C, Strobel K et al A patient with BRAF V600E lung adenocarcinoma responding to vemurafenib. J Thorac Oncol 2012. (Oct); 7(10): e23–4. [DOI] [PubMed] [Google Scholar]
- 29. Awad MM, Katayama R, McTigue M et al Acquired resistance to crizotinib from a mutation in CD74‐ROS1. N Engl J Med 2013. (Jun 1); 368: 2395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mendelsohn J. Personalizing oncology: perspectives and prospects. J Clin Oncol 2013. (May 20); 31: 1904–11. [DOI] [PubMed] [Google Scholar]
- 31. Yoshida A, Kohno T, Tsuta K et al ROS1‐rearranged lung cancer: a clinicopathologic and molecular study of 15 surgical cases. Am J Surg Pathol 2013. (Apr); 37: 554–62. [DOI] [PubMed] [Google Scholar]


