Alcohol has pleiotropic effects across multiple organ systems, including brain, cardiovascular, endocrine, immune, musculoskeletal, and gastrointestinal systems. Moreover, some effects, such as intoxication, can be brief, but others, such as the development of Alcohol Use Disorders (AUDs), cardiovascular disease, and liver damage, can persist for a lifetime. Effects resulting in encoding of endocrine and other dysfunctions can also persist across generations. This complexity creates a barrier to the creation of therapeutics and discovery of biomarkers. However, we know that environmental factors, which can include drugs of abuse such as alcohol, can have short- and long-term effects on gene expression through epigenetic mechanisms (Holliday, 2006; Shukla et al., 2008). Epigenetic mechanisms affect the transcription and translation of many genes simultaneously. Therefore, by understanding the mechanics of these epigenetic changes, we will have the ability to craft powerful new therapeutics to offset negative effects of alcohol exposure.
Epigenetic modifications commonly occur through three mechanisms: methylation of DNA, histone post-translational modifications, and the interactions of non-coding RNA with transcriptional and translational cellular machinery (Fig. 1). Modifications that open the tightly wound chromatin structure are thought to increase gene expression, while modifications that condense chromatin structure are thought to inhibit gene expression. DNA methylation, which usually occurs at groupings of cytosine and guanine nucleotides referred to as “CpG islands”, represses gene transcription. This repression occurs with the binding of methyl-CpG binding domain proteins, the subsequent recruitment transcription inhibitory complexes, and chromatin condensation (Cedar & Bergman, 2012). Post-translational modifications of histones, by covalent addition of functional groups to histone tails, can also regulate chromatin access. These modifications include commonly studied acetylation and methylation, as well as many other modifications, including ubiquitinylation, phosphorylation, and ADP ribosylation, to name a few (Bannister & Kouzarides, 2011; Kouzarides, 2007; Strahl & Allis, 2000). Histone acetylation relaxes chromatin, which facilitates gene transcription. Histone methylation can bidirectionallly affect gene expression depending on the amino acid location on the histone tail and quantity of methylation, i.e., whether singly, di-, or tri-methylated (Zhou, Goren, & Bernstein, 2011). Non-coding RNAs (ncRNAs) can affect both gene transcription and translation. For example, long non-coding RNAs (IncRNAs, >200 nucleotides in length) can interact with transcription machinery to trigger chromatin compaction at commonly imprinted regions, including the H19 locus and X chromosome (Nagano & Fraser, 2011; Rinn & Guttman, 2014). MicroRNAs (miRNAs, ∼21 nucleotides in length) affect translation by binding the 3′-untranslated region (3′-UTR) of mRNA transcripts and, along with the miRNA-induced silencing complex, inhibit translation and enhance mRNA degradation (Fabian & Sonenberg, 2012). miRNAs can bind to many related transcripts and regulate the expression of gene networks as a whole (reviewed in Miranda, 2014).
Fig. 1. Commonly Researched Epigenetic Modifications.
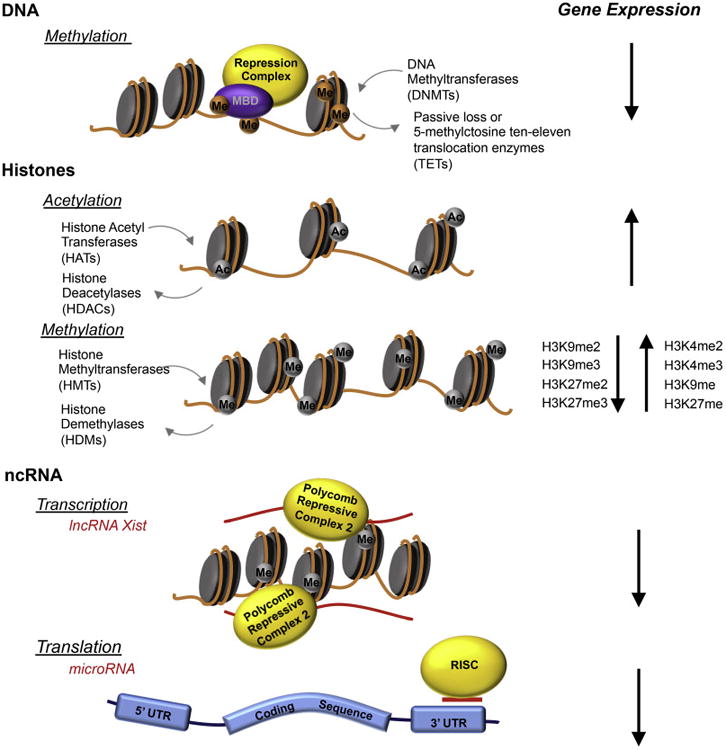
Commonly researched epigenetic modifications include covalently bonded groups attached to DNA and histones, as well as interactions with non-coding RNAs (ncRNAs). DNA methylation, via methyl-CpG binding domain proteins (MBDs), recruits repressor complexes to inhibit transcription and to encourage chromatin condensation. Histone acetylation opens chromatin to allow access to transcription machinery, while histone methylation can either condense or open chromatin, depending on the localization and degree of methyl group attachment. ncRNAs can interact at the chromatin level (e.g., long non-coding RNAs that act during imprinting to silence an allele) or at the translation level (e.g., microRNAs that bind to the 3′-UTR of mRNAs), along with the RNA-lnduced Silencing Complex (RISC), to trigger mRNA degradation.
DNA methylation, histone modification, and ncRNAs interact with each other to coordinate and regulate overall gene expression. The aim of this special edition of Alcohol is not only to provide background on what we currently know about epigenetic modifications in response to alcohol exposure, but also to further examine the impact of these changes throughout the lifespan and across generations. Additionally, we highlight, by targeting epigenetic mediators, how we can create novel therapeutics for both AUDs in adult populations and for Fetal Alcohol Spectrum Disorders (FASD). These studies provide evidence of the importance of continued research on the epigenomic alterations that occur in response to alcohol exposure, to combat the complex changes that occur across the body and across generations.
To facilitate navigation of the articles in this special issue of Alcohol, we have assembled a table that summarizes the epigenetic modifications and the timing of the alcohol exposures that are addressed in each paper (see Table 1).
Table 1. Summary of papers included in this special issue of Alcohol.
| Authors | Epigenetic modification | Exposure period | Longevity of Effects | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Histone | DNA | ncRNA | Δ Gene Expression | Adult | Adolescent | Preconception (✘) Prenatal (✓) | Exposure Period/Effects in | Cross-Generational | ||
| Reviews | ||||||||||
| Palmisano and Pandey | ✓ | ✓ | ✓ | ✓ | ✓ | Adolesence/Adulthood | ||||
| Tulisiak et al. | ✓ | ✓ | ✓ | ✓ | ||||||
| Mayfield | ✓ | ✓ | ||||||||
| Chater-Diehl et al. | ✓ | ✓ | ✓ | Prenatal/Adulthood | ||||||
| Chastain and Sarkar | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Laufer et al. | ✓ | ✓ | ✘ ✓ | Preconception Prenatal/Adulthood | ✓ | |||||
| Research – Adult Use | ||||||||||
| Restrepo et al. | ✓ | ✓ | ✓ | |||||||
| Hashimoto et al. | ✓ | ✓ | ✓ | ✓ | ||||||
| Ponomarev et al. | ✓ | ✓ | ✓ | ✓ | ||||||
| Cervera-Juanes et al. | ✓ | ✓ | ✓ | ✓ | ||||||
| Hitzemann et al. | ✓ | ✓ | ||||||||
| Research – Prenatal and Preconception Exposure/Cross-generational Effects | ||||||||||
| Veazey et al. | ✓ | ✓ | ✓ | |||||||
| Öztürk et al. | ✓ | ✓ | ||||||||
| Burrowes et al. | ✓ | ✓ | ✓ | |||||||
| Balaraman et al. | ✓ | ✓ | Developmental/Pre-pubertal | |||||||
| Rompala et al. | ✓ | ✓ | ✘ | Adult/F1 offspring | ✓ | |||||
| Asimes et al. | ✓ | ✓ | ✓ | ✘ | Adult/F1 offspring | ✓ | ||||
| Popoola et al. | ✓ | ✘✓ | Prenatal/F1, F2 offspring | ✓ | ||||||
Alcohol-Use Disorders – Adulthood
In the adult, drinking behavior can become classified as an AUD when there is evidence of dependence or withdrawal (American Psychiatric Association, 2013). It is estimated that about 30% of adults will have an AUD during their lifetime (Grant et al., 2015). Previous research has shown the presence of epigenetic modifications in alcoholic patients, including: DNA hypermethylation at specific genetic loci in blood cells, which has been associated with alcohol craving and preference (Bonsch, Lenz, Kornhuber, & Bleich, 2005; Bonsch, Lenz, Reulbach, Kornhuber, & Bleich, 2004; Hillemacher et al., 2009; Muschler et al., 2010; reviewed in Tulisiak, Harris, & Ponomarev, this issue); tri-methylation at histone 3 lysine 4 (H3K4me3), which has been associated with increased gene expression in postmortem tissue (Ponomarev, Wang, Zhang, Harris, & Mayfield, 2012); and upregulation of miRNAs, which has been associated with the downregulation of mRNA transcripts in the prefrontal cortex of postmortem samples (Lewohl et al., 2011; Nunez & Mayfield, 2012; reviewed in Mayfield, this issue).
To treat and prevent AUDs, we need to understand how the differences seen in both postmortem human tissue as well as the peripheral tissues of patients may contribute to the development of heavy, addictive drinking. This can be further understood by examining how the epigenome changes across the three components of AUDs: intoxication, withdrawal, and craving (Koob & Volkow, 2010). In this special issue on epigenetics, Cervera-Juanes, Wilhelm, Park, Grant, & Ferguson (this issue) show that within the nucleus accumbens core, which is involved in neurocircuitry for intoxication and withdrawal, chronic low level drinking results in DNA hypomethylation, and high levels of drinking result in hypermethylation. In the prefrontal cortex, which is involved in neurocircuitry for craving, Hashimoto, Gavin, Wiren, Crabbe, and Guizzetti (this issue) show altered gene expression for proteins needed for histone and DNA epigenetic modification creation and maintenance, and they show that ethanol withdrawal has a larger effect on gene expression than ethanol exposure or abstinence. Interestingly, Hashimoto et al. show that covalent modifications may act in opposition, with altered transcript expression of epigenetic modifiers that lead to open and closed chromatin conformations.
The complexity of these epigenetic changes leads to the question – What is the outcome for gene expression? Previous research has shown that changes in gene expression are not readily predictable (Ponomarev et al., 2012; Veazey, Carnahan, Muller, Miranda, & Golding, 2013; Veazey, Parnell, Miranda, & Golding, 2015). In a model system of mice predisposed to binge drinking, Hitzemann et al. (this issue) compared gene expression in the dorsal striatum, which contributes to habitual intoxication, of animals who drank to a high blood ethanol content (BEC), compared to those who drank to a lower BEC. They found a larger number of genes were expressed with a positive correlation to BEC levels than genes with a negative correlation to BEC. This effect is unexpected if there is an increased rate of DNA methylation in the dorsal striatum, as has been shown in other brain regions. Yet, these data corroborate evidence of potential maladaptive hypomethylation, specifically of Grin2b, in the dorsal striatum, contributing to increased alcohol consumption (Wang et al., 2010; Wong, Tauck, Fong, & Kendig, 1998). Additionally, epigenetic markers have been shown to vary between brain regions (Finegersh et al., 2015) and can be further complicated by comorbid conditions, including stress (Meyer, Long, Fanselow, & Spigelman, 2013; Moonat & Pandey, 2012; reviewed in Palmisano & Pandey, this issue).
While understanding how epigenetic modifications affect central control of alcohol use is critical to combating AUDs, the identification of peripheral biomarkers is also an invaluable tool and could shed light on the progression of AUDs along with associated disorders, such as alcoholic liver diseases. Epigenetic markers can serve as peripheral biomarkers of alcohol exposure, and while methylation is commonly examined (Andersen, Dogan, Beach, & Philibert, 2015), microRNAs have been indicated as biomarkers for prenatal alcohol exposure (Balaraman et al., 2014, 2016), and histone modifications have been explored as biomarkers in other fields, including cancer biology (Chervona & Costa, 2012). Restrepo, Lim, Korthuis, and Shukla (this issue) show that histone acetylation at histone 3 lysine 9, and upregulation of the protein PNPLA3, may be involved in fatty liver disease development, which can predispose patients to alcoholic liver disease. Previous studies have also suggested that the histone post-translational modification of H3K9me2 may also serve as a biomarker in FASD (Veazey et al., 2015). Further investigation into these potential peripheral markers could assist in the identification and treatment of alcohol-linked pathophysiology associated with AUDs, including the progression of alcoholic liver disease.
Prenatal and Preconception Alcohol Exposure
Drinking behavior can impact multiple generations. Alcohol is a known teratogen. As such, prenatal exposure can lead to an array of perturbations to neurological and behavioral development, resulting in Fetal Alcohol Spectrum Disorders (FASD, Hoyme et al., 2005; Jones, 2011). Prenatal alcohol exposure has been shown to affect DNA methylation, histone modification, and miRNA components (Miranda, 2012; also reviewed in Laufer, Chater-Diehl, Kapalanga, and Singh, this issue and Chater-Diehl, Laufer, & Singh, this issue). Direct fetal exposure is not the only mechanism of cross-generational effects of alcohol exposure. Recent evidence has shown that paternal drinking during the preconception period affects behavior, development, and gene expression in the following generations (Finegersh & Homanics, 2014; Jabbar et al., 2016; Knezovich & Ramsay, 2012). This cross-generational transmission likely occurs through epigenetic modifications encoded in the germline (Bohacek & Mansuy, 2015; reviewed in Chastain & Sarkar, this issue).
Ethanol exposure during the prenatal period, while toxic for neural progenitors and more developed neurons, does not kill neural stem cells (Cheema, West, & Miranda, 2000; Prock & Miranda, 2007). Neural stem cells, in response to ethanol exposure, increase proliferation and undergo premature differentiation, thus depleting the neural stem cell pool (Camarillo & Miranda, 2008; Santillano et al., 2005). These effects on neural stem cells and developing cortical neurons likely contribute to thinning of the cerebral cortex, a hallmark of FASD (Zhou, Lebel, et al., 2011). Moreover, correct neuronal and cortical development is dependent on specific DNA methylation programs (Zhou, 2012). Öztürk, Resendiz, Öztürk, and Zhou (this issue) detail the alterations to cortical development and DNA methylation within the developing cortex after prenatal alcohol exposure. miRNAs, particularly miR-9, also regulate cortical development and neurogenesis (Coolen, Katz, & Bally-Cuif, 2013). miR-9 expression is decreased in neural stem cells after ethanol exposure, and miR-9 knockdown mimics the effects of ethanol on craniofacial development in a zebrafish model (Pappalardo-Carter et al., 2013; Sathyan, Golden, & Miranda, 2007). Burrowes et al. (this issue) provide evidence that developmental chromatin remodeling, due to the maturation of the BAF (Brg/brm-associated factors) complex, can be adaptive and protective in response to ethanol exposure and act to preserve miR-9 expression. Prenatal alcohol exposure can also affect histone post-translational modifications in a dose-dependent manner in embryonic stem cells (Veazey et al., this issue). Veazey and colleagues found no correlations between histone modifications and expression levels of their genes of interest. This disconnect between epigenetic modifications and gene expression was also seen in AUDs, and shows that more work needs to be done to map the relationship between epigenetic modifications and gene expression, particularly since, as seen with miR-9 in neural stem cells, some adaptions may be protective.
The effects of ethanol are not limited to exposure during in utero development, but can also occur from alcohol exposure prior to conception. Work on the proopiomelanocortin (POMC) gene has shown that not only is gene expression downregulated in rats which were prenatally exposed to alcohol, but that the effect of lowered POMC expression can be transmitted through the paternal line to the F2 and F3 generations (Govorko, Bekdash, Zhang, & Sarkar, 2012). Studies have found additional genes with altered expression across generations (Asimes et al., this issue; Popoola, Nizhnikov, & Cameron, this issue; Przybycien-Szymanska, Rao, Prins, & Pak, 2014), as well as altered alcohol drinking-associated behaviors (Finegersh & Homanics, 2014; Nizhnikov, Popoola, & Cameron, 2016), and methylation programming (Asimes et al., this issue; Finegersh & Homanics, 2014). The cross-generational effects on behavior can be dependent on genetic background, as Popoola et al. show using rat strains, or conserved across strains, as Rompala, Finegersh, Slater, and Homanics show in mouse models (both in this issue). Cross-generational inheritance may also vary by maternal and paternal lineage. Asimes et al. (this issue) show that DNA methylation patterns in alcohol-naïve offspring can widely vary by which parent (or if both parents) had been exposed to binge-like drinking. More research needs to be done to understand the effects of genetic background, to better understand the contribution of maternal lineage verses paternal lineage, and how inheritance of these changes in gene expression and behavior varies based on the sex of the offspring (Finegersh & Homanics, 2014; Vassoler, Byrnes, & Pierce, 2014).
Epigenetic Therapeutics
There is an important question that remains – Can alterations to the epigenome serve as sites for therapeutic intervention? Previous research has shown that inhibition of DNA methylation (Barbier et al., 2015; Ponomarev et al., this issue; Warnault, Darcq, Levine, Barak, & Ron, 2013) and histone deacetylation (Sanchis-Segura, Lopez-Atalaya, & Barco, 2009; Simon-O'Brien et al., 2015; Warnault et al., 2013) can decrease drinking behavior. Interestingly, Ponomarev et al. (this issue) show that the efficacy of histone deacetylation inhibition may be dependent on the drinking model examined, indicating that further study is needed to determine the most efficacious use of these inhibitors. During prenatal exposure to alcohol, DNMT inhibition mimics alcohol's negative effects on adult neural stem cell dynamics (Zhou, Balaraman, et al., 2011), and methyl supplementation through diet can ameliorate some effects of prenatal alcohol exposure (Downing et al., 2011). Choline, which can act as a methyl donor (Otero, Thomas, Saski, Xia, & Kelly, 2012), has neurological benefits within the hippocampus after third-trimester equivalent alcohol exposure (Monk, Leslie, & Thomas, 2012; Thomas, Garrison, & O'Neill, 2004). The benefits of choline may be due to a combination of actions, including the ability to restore and augment miRNA expression within the hippocampus after prenatal alcohol exposure (Balaraman, Idrus, Miranda, & Thomas, this issue).
Future Directions
We are just beginning to understand the effects of alcohol on the epigenome. The studies outlined here show how alcohol exposure can affect the epigenome, from AUDs, to prenatal alcohol exposure, to cross-generational effects. More work is needed to understand the interaction of all the methods of epigenetic modifications, including less commonly explored factors such as histone phosphorylation, arginine methylation (see Hashimoto et al., this issue), ubiquitination, sumoylation, citrullination, ADP-ribosolyation, as well as the dynamics of DNA methylation removal through 5-hydroxymethyl cysteine. Additionally, the interactions of long non-coding RNAs in normal neuronal development in disease are just beginning to be discovered and will provide an extra layer of complexity to gene expression regulation (Fatica & Bozzoni, 2014; Wapinski & Chang, 2011). With further investigation, it may be possible to develop novel therapeutics and discover new biomarkers to combat the negative effects of alcohol exposure throughout the lifespan.
Highlights.
Epigenetic modifications change global gene expression in response to environmental factors.
Alcohol consumption can affect the epigenome across the lifespan and generations.
Further research into epigenetic changes may allow for the identification of biomarkers and the creation of novel therapeutics.
Acknowledgments
Supported by NIH grants AA024659 (RCM), HD086765 (RCM), AA10422 (GEH), and AA020889 (GEH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 5th. 1–0. American Psychiatric Association; 2013. Substance-Related and Addictive Disorders. Retrieved from http://dsm.psychiatryonline.org.ezproxy.library.tamu.edu/doi/full/10.1176/appi.books.9780890425596.dsm16. [Google Scholar]
- Andersen AM, Dogan MV, Beach SRH, Philibert RA. Current and Future Prospects for Epigenetic Biomarkers of Substance Use Disorders. Genes. 2015;6(4):991. doi: 10.3390/genes6040991. https://doi.org/10.3390/genes6040991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asimes A, Torcaso A, Pinceti E, Kim CK, Zeleznik-Le NJ, Pak TR. Adolescent binge-pattern alcohol exposure alters genome-wide DNA methylation patterns in the hypothalamus of alcohol-naïve male offspring. Alcohol. 2017 doi: 10.1016/j.alcohol.2016.10.010. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaraman S, Idrus NM, Miranda RC, Thomas JD. Postnatal choline supplementation selectively attenuates hippocampal microRNA alterations associated with developmental alcohol exposure. Alcohol. 2017 doi: 10.1016/j.alcohol.2016.12.006. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaraman S, Lunde ER, Sawant O, Cudd TA, Washburn SE, Miranda RC. Maternal and Neonatal Plasma MicroRNA Biomarkers for Fetal Alcohol Exposure in an Ovine Model. Alcoholism: Clinical and Experimental Research. 2014;38(5):1390–1400. doi: 10.1111/acer.12378. https://doi.org/10.1111/acer.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaraman S, Schafer JJ, Tseng AM, Wertelecki W, Yevtushok L, Zymak-Zakutnya N, et al. Plasma miRNA Profiles in Pregnant Women Predict Infant Outcomes following Prenatal Alcohol Exposure. PLOS ONE. 2016;11(11):e0165081. doi: 10.1371/journal.pone.0165081. https://doi.org/10.1371/journal.pone.0165081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzahdes T. Regulation of chromatin by histone modifications. Cell Research. 2011;27(3):381–395. doi: 10.1038/cr.2011.22. https://doi.org/10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier E, Tapocik JD, Juergens N, Pitcairn C, Borich A, Schank JR, et al. DNA Methylation in the Medial Prefrontal Cortex Regulates Alcohol-Induced Behavior and Plasticity. Journal of Neuroscience. 2015;35(15):6153–6164. doi: 10.1523/JNEUROSCI.4571-14.2015. https://doi.org/10.1523/JNEUROSCI.4571-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohacek J, Mansuy IM. Molecular insights into transgenerational non-genetic inheritance of acquired behaviours. Nature Reviews Genetics. 2015;16(11):641–652. doi: 10.1038/nrg3964. https://doi.org/10.1038/nrg3964. [DOI] [PubMed] [Google Scholar]
- Bönsch D, Lenz B, Kornhuber J, Bleich SC. DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Neuroreport. 2005;16(2):167–170. doi: 10.1097/00001756-200502080-00020. Miscellaneous Article. [DOI] [PubMed] [Google Scholar]
- Bönsch D, Lenz B, Reulbach U, Kornhuber J, Bleich S. Homocysteine associated genomic DNA hypermethylation in patients with chronic alcoholism. Journal of Neural Transmission. 2004;111(12):1611–1616. doi: 10.1007/s00702-004-0232-x. https://doi.org/10.1007/S00702-004-0232-X. [DOI] [PubMed] [Google Scholar]
- Burrowes SG, Salem NA, Tseng AM, Balaraman S, Pinson MR, Garcia C, et al. The BAF (BRG1/BRM-Associated Factor) Chromatin-Remodeling Complex exhibits ethanol sensitivity in fetal neural progenitor cells and regulates transcription at the mir-9-2 encoding gene locus. Alcohol. 2017 doi: 10.1016/j.alcohol.2017.01.003. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarillo C, Miranda RC. Ethanol exposure during neurogenesis induces persistent effects on neural maturation: evidence from an ex vivo model of fetal cerebral cortical neuroepithelial progenitor maturation. Gene Expression. 2008;14159(3) [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Programming of DNA Methylation Patterns. 2012 Jun 4; doi: 10.1146/annurev-biochem-052610-091920. [review-article], Retrieved February 15, 2017, from http://www.annualreviews.org.lib-ezproxy.tamu.edu:2048/doi/10.1146/annurevbiochem-052610-091920. [DOI] [PubMed]
- Cervera-Juanes R, Wilhem LJ, Park B, Grant KA, Ferguson B. Genome-wide analysis of the nucleus accumbens identifies DNA methylation signals differentiating low/binge from heavy alcohol drinking. Alcohol. 2017 doi: 10.1016/j.alcohol.2016.11.003. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain LG, Sarkar DK. Alcohol effects on the epigenome in the germline: role in the inheritance of alcohol-related pathology. Alcohol. 2017 doi: 10.1016/j.alcohol.2016.12.007. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater-Diehl EJ, Laufer BI, Singh SM. Changes to histone modifications following prenatal alcohol exposure: An emerging picture. Alcohol. 2017 doi: 10.1016/j.alcohol.2017.01.005. this issue. [DOI] [PubMed] [Google Scholar]
- Cheema ZF, West JR, Miranda RC. Ethanol Induces Fas/Apo [Apoptosis]-1 mRNA and Cell Suicide in the Developing Cerebral Cortex. Alcoholism: Clinical and Experimental Research. 2000;24(4):535–543. https://doi.org/10.1111/j.1530-0277.2000.tb02022.x. [PubMed] [Google Scholar]
- Chervona Y, Costa M. Histone modifications and cancer: biomarkers of prognosis? American Journal of Cancer Research. 2012;2(5):589. [PMC free article] [PubMed] [Google Scholar]
- Coolen M, Katz S, Bally-Cuif L. miR-9: a versatile regulator of neurogenesis. Frontiers in Cellular Neuroscience. 2013;7 doi: 10.3389/fncel.2013.00220. https://doi.org/10.3389/fncel.2013.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing C, Johnson TE, Larson C, Leakey TI, Siegfried RN, Rafferty TM, et al. Subtle Decreases in DNA Methylation and Gene Expression at the Mouse Igf2 Locus Following Prenatal Alcohol Exposure: Effects of a Methyl-Supplemented Diet. Alcohol (Fayetteville, NY) 2011;45(1):65. doi: 10.1016/j.alcohol.2010.07.006. https://doi.org/10.1016/j.alcohol.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nature Structural & Molecular Biology. 2012;19(6):586–593. doi: 10.1038/nsmb.2296. https://doi.org/10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature Reviews Genetics. 2014;75(1):7–21. doi: 10.1038/nrg3606. https://doi.org/10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Finegersh A, Homanics GE. Paternal Alcohol Exposure Reduces Alcohol Drinking and Increases Behavioral Sensitivity to Alcohol Selectively in Male Offspring. PLOS ONE. 2014;9(6):e99078. doi: 10.1371/journal.pone.0099078. https://doi.org/10.1371/journal.pone.0099078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegersh A, Ferguson C, Maxwell S, Mazariegos D, Farrell D, Homanics GE. Repeated vapor ethanol exposure induces transient histone modifications in the brain that are modified by genotype and brain region. Frontiers in Molecular Neuroscience. 2015;8 doi: 10.3389/fnmol.2015.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorko D, Bekdash RA, Zhang C, Sarkar DK. Male Germline Transmits Fetal Alcohol Adverse Effect on Hypothalamic Proopiomelanocortin Gene Across Generations. Biological Psychiatry. 2012;72(5):378–388. doi: 10.1016/j.biopsych.2012.04.006. https://doi.org/10.1016/j.biopsych.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BF, Goldstein RB, Saha TD, Chou SP, Jung J, Zhang H, et al. Epidemiology of DSM-5 Alcohol Use Disorder: Results From the National Epidemiologic Survey on Alcohol and Related Conditions III. JAMA Psychiatry. 2015;72(8):757–766. doi: 10.1001/jamapsychiatry.2015.0584. https://doi.org/10.1001/jamapsychiatry.2015.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto JG, Gavin DP, Wiren KM, Crabbe JC, Guizzetti M. Prefrontal cortex expression of chromatin modifier genes in male WSP and WSR mice changes across ethanol dependence, withdrawal, and abstinence. Alcohol. 2017 doi: 10.1016/j.alcohol.2017.01.010. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillemacher T, Frieling H, Hartl T, Wilhelm J, Kornhuber J, Bleich S. Promoter specific methylation of the dopamine transporter gene is altered in alcohol dependence and associated with craving. Journal of Psychiatric Research. 2009;43(4):388–392. doi: 10.1016/j.jpsychires.2008.04.006. https://doi.org/10.1016/jjpsychires.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Oberbeck D, lancu O, Darakjian P, McWeeney S, Spence S, et al. Alignment of the transcriptome with individual variation in animals selectively bred for High Drinking-ln-the-Dark (HDID) Alcohol. 2017 doi: 10.1016/j.alcohol.2017.02.176. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. Epigenetics: A Historical Overview. Epigenetics. 2006;1(2):76–80. doi: 10.4161/epi.1.2.2762. https://doi.org/10.4161/epi.l2.2762. [DOI] [PubMed] [Google Scholar]
- Hoyme HE, May PA, Kalberg WO, Kodituwakku P, Gossage JP, Trujillo PM, et al. A practical clinical approach to diagnosis of fetal alcohol spectrum disorders: clarification of the 1996 institute of medicine criteria. Pediatrics. 2005;115(1):39–47. doi: 10.1542/peds.2004-0259. https://doi.org/10.1542/peds.2004-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbar S, Chastain LG, Gangisetty O, Cabrera MA, Sochacki K, Sarkar DK. Preconception Alcohol Increases Offspring Vulnerability to Stress. Neuropsychopharmacology. 2016;41(11):2782–2793. doi: 10.1038/npp.2016.92. https://doi.org/10.1038/npp.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KL. The effects of alcohol on fetal development. Birth Defects Research Part C: Embryo Today: Reviews. 2011;93(1):3–11. doi: 10.1002/bdrc.20200. https://doi.org/10.1002/bdrc.20200. [DOI] [PubMed] [Google Scholar]
- Knezovich JG, Ramsay M. The Effect of Preconception Paternal Alcohol Exposure on Epigenetic Remodeling of the H19 and Rasgrf1 Imprinting Control Regions in Mouse Offspring. Frontiers in Genetics. 2012;3 doi: 10.3389/fgene.2012.00010. https://doi.org/10.3389/fgene.2012.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of Addiction. Neuropsychopharmacology. 2010;35(1):217–238. doi: 10.1038/npp.2009.110. https://doi.org/10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin Modifications and Their Function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. https://doi.org/10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Laufer BI, Chater-Diehl EJ, Kapalanga J, Singh SM. Long-term alterations to DNA methylation as a biomarker of prenatal alcohol exposure: From mouse models to human children with fetal alcohol spectrum disorders. Alcohol. 2017 doi: 10.1016/j.alcohol.2016.11.009. this issue. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, Mayfield RD. Up-Regulation of MicroRNAs in Brain of Human Alcoholics. Alcoholism: Clinical and Experimental Research. 2011;35(11):1928–1937. doi: 10.1111/j.1530-0277.2011.01544.x. https://doi.org/10.1111/j.1530-0277.2011.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield RD. Emerging roles for ncRNAs in alcohol use disorders. Alcohol. 2017 doi: 10.1016/j.alcohol.2017.01.004. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EM, Long V, Fanselow MS, Spigelman I. Stress Increases Voluntary Alcohol Intake, but Does not Alter Established Drinking Habits in a Rat Model of Posttraumatic Stress Disorder. Alcoholism: Clinical and Experimental Research. 2013;37(4):566–574. doi: 10.1111/acer.12012. https://doi.org/10.1111/acer.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RC. MicroRNAs and Fetal Brain Development: Implications for Ethanol Teratology during the Second Trimester Period of Neurogenesis. Frontiers in Genetics. 2012;3 doi: 10.3389/fgene.2012.00077. https://doi.org/10.3389/fgene.2012.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RC. Chapter Seven - MicroRNAs and Ethanol Toxicity. In: Pandey SC, editor. International Review of Neurobiology. Vol. 115. Academic Press; 2014. pp. 245–284. Retrieved from https://www.sciencedirect.com/science/article/pii/B978012801311300007X. [DOI] [PubMed] [Google Scholar]
- Monk BR, Leslie FM, Thomas JD. The effects of perinatal choline supplementation on hippocampal cholinergic development in rats exposed to alcohol during the brain growth spurt. Hippocampus. 2012;22(8):1750–1757. doi: 10.1002/hipo.22009. https://doi.org/10.1002/hipo.22009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonat S, Pandey SC. Stress, Epigenetics, and Alcoholism. Alcohol Research: Current Reviews. 2012;34(4):495. doi: 10.35946/arcr.v34.4.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschler M, Hillemacher T, Kraus C, Kornhuber J, Bleich S, Frieling H. DNA methylation of the POMC gene promoter is associated with craving in alcohol dependence. Journal of Neural Transmission. 2010;117(4):513–519. doi: 10.1007/s00702-010-0378-7. https://doi.org/10.1007/s00702-010-0378-7. [DOI] [PubMed] [Google Scholar]
- Nagano T, Fraser P. No-Nonsense Functions for Long Noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. https://doi.org/10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Nizhnikov ME, Popoola DO, Cameron NM. Transgenerational Transmission of the Effect of Gestational Ethanol Exposure on Ethanol Use-Related Behavior. Alcoholism: Clinical and Experimental Research. 2016;40(3):497–506. doi: 10.1111/acer.12978. https://doi.org/10.1111/acer.12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez YO, Mayfield RD. Understanding Alcoholism Through microRNA Signatures in Brains of Human Alcoholics. Frontiers in Genetics. 2012;3 doi: 10.3389/fgene.2012.00043. https://doi.org/10.3389/fgene.2012.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero NKH, Thomas JD, Saski CA, Xia X, Kelly SJ. Choline Supplementation and DNA Methylation in the Hippocampus and Prefrontal Cortex of Rats Exposed to Alcohol During Development. Alcoholism: Clinical and Experimental Research. 2012;36(10):1701–1709. doi: 10.1111/j.1530-0277.2012.01784.x. https://doi.org/10.1111/J.1530-0277.2012.01784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OÖztürk NC, Resendiz M, Öztürk H, Zhou FC. DNA methylation program in normal and alcohol-induced thinning cortex. Alcohol. 2017 doi: 10.1016/j.alcohol.2017.01.006. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmisano M, Pandey SC. Epigenetic mechanisms of alcoholism and stress disorders. Alcohol. 2017 doi: 10.1016/j.alcohol.2017.01.001. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappalardo-Carter DL, Balaraman S, Sathyan P, Carter ES, Chen WJA, Miranda RC. Suppression and Epigenetic Regulation of MiR-9 Contributes to Ethanol Teratology: Evidence from Zebrafish and Murine Fetal Neural Stem Cell Models. Alcoholism: Clinical and Experimental Research. 2013;37(10):1657–1667. doi: 10.1111/acer.12139. https://doi.org/10.1111/acer.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Stelly CE, Morikawa H, Blednov YA, Mayfield RD, Harris RA. Mechanistic insights into epigenetic modulation of ethanol consumption. Alcohol. 2017 doi: 10.1016/j.alcohol.2017.01.016. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene Coexpression Networks in Human Brain Identify Epigenetic Modifications in Alcohol Dependence. Journal of Neuroscience. 2012;32(5):1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. https://doi.org/10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoola DO, Nizhnikov ME, Cameron NM. Strain-specific programming of prenatal ethanol exposure across generations. Alcohol. 2017 doi: 10.1016/j.alcohol.2017.01.002. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prock TL, Miranda RC. Embryonic Cerebral Cortical Progenitors Are Resistant to Apoptosis, but Increase Expression of Suicide Receptor DISC-Complex Genes and Suppress Autophagy Following Ethanol Exposure. Alcoholism: Clinical and Experimental Research. 2007;31(4):694–703. doi: 10.1111/j.1530-0277.2007.00354.x. https://doi.org/10.1111/j.1530-0277.2007.00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybycien-Szymanska MM, Rao YS, Prins SA, Pak TR. Parental Binge Alcohol Abuse Alters F1 Generation Hypothalamic Gene Expression in the Absence of Direct Fetal Alcohol Exposure. PLoS ONE. 2014;9(2):1–13. doi: 10.1371/journal.pone.0089320. https://doi.org/10.1371/journal.pone.0089320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo RJ, Lim RW, Korthuis RJ, Shukla SD. Binge alcohol alters PNPLA3 levels in liver through epigenetic mechanism involving histone H3 acetylation. Alcohol. 2017 doi: 10.1016/j.alcohol.2017.01.009. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J, Guttman M. RNA and dynamic nuclear organization. Science. 2014;345(6202):1240–1241. doi: 10.1126/science.1252966. https://doi.org/10.1126/science.1252966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompala GR, Finegersh A, Slater M, Homanics GE. Paternal preconception alcohol exposure imparts intergenerational alcohol-related behaviors to male offspring on a pure C57BL/6J background. Alcohol. 2017 doi: 10.1016/j.alcohol.2016.11.001. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchis-Segura C, Lopez-Atalaya JP, Barco A. Selective Boosting of Transcriptional and Behavioral Responses to Drugs of Abuse by Histone Deacetylase Inhibition. Neuropsychopharmacology. 2009;34(13):2642–2654. doi: 10.1038/npp.2009.125. https://doi.org/10.1038/npp.2009.125. [DOI] [PubMed] [Google Scholar]
- Santillano DR, Kumar LS, Prock TL, Camarillo C, Tingling JD, Miranda RC. Ethanol induces cell-cycle activity and reduces stem cell diversity to alter both regenerative capacity and differentiation potential of cerebral cortical neuroepithelial precursors. BMC Neuroscience. 2005;6:59–17. doi: 10.1186/1471-2202-6-59. https://doi.org/10.1186/1471-2202-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyan P, Golden HB, Miranda RC. Competing Interactions between Micro-RNAs Determine Neural Progenitor Survival and Proliferation after Ethanol Exposure: Evidence from an Ex Vivo Model of the Fetal Cerebral Cortical Neuroepithelium. The Journal of Neuroscience. 2007;27(32):8546–8557. doi: 10.1523/JNEUROSCI.1269-07.2007. https://doi.org/10.1523/JNEUROSCI.1269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging Role of Epigenetics in the Actions of Alcohol. Alcoholism: Clinical and Experimental Research. 2008;32(9):1525–1534. doi: 10.1111/j.1530-0277.2008.00729.x. https://doi.org/10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- Simon-O'Brien E, Alaux-Cantin S, Warnault V, Buttolo R, Naassila M, Vilpoux C. The histone deacetylase inhibitor sodium butyrate decreases excessive ethanol intake in dependent animals. Addiction Biology. 2015;20(4):676–689. doi: 10.1111/adb.12161. https://doi.org/10.1111/adb.12161. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–5. doi: 10.1038/47412. https://doi.org/10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Garrison M, O'Neill TM. Perinatal choline supplementation attenuates behavioral alterations associated with neonatal alcohol exposure in rats. Neurotoxicology and Teratology. 2004;26(1):35–45. doi: 10.1016/j.ntt.2003.10.002. https://doi.org/10.1016/j.ntt.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Tulisiak CT, Harris RA, Ponomarev I. DNA Modifications in Models of Alcohol Use Disorders. Alcohol. 2017 doi: 10.1016/j.alcohol.2016.11.004. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassoler FM, Byrnes EM, Pierce RC. The impact of exposure to addictive drugs on future generations: Physiological and behavioral effects. Neuropharmacology. 2014;76(Part B):269–275. doi: 10.1016/j.neuropharm.2013.06.016. https://doi.Org/10.1016/j.neuropharm.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey KJ, Carnahan MN, Muller D, Miranda RC, Golding MC. Alcohol-Induced Epigenetic Alterations to Developmentally Crucial Genes Regulating Neural Sternness and Differentiation. Alcoholism: Clinical and Experimental Research. 2013;37(7):1111–1122. doi: 10.1111/acer.12080. https://doi.org/10.1111/acer.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey KJ, Parnell SE, Miranda RC, Golding MC. Dose-dependent alcohol-induced alterations in chromatin structure persist beyond the window of exposure and correlate with fetal alcohol syndrome birth defects. Epigenetics & Chromatin. 2015;8 doi: 10.1186/s13072-015-0031-7. https://doi.org/10.1186/s13072-015-0031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey KJ, Wang H, Behdi YS, Skiles WM, Chang RCA, Golding MC. Disconnect between alcohol-induced alterations in chromatin structure and gene transcription in a mouse embryonic stem cell model of exposure. Alcohol. 2017 doi: 10.1016/j.alcohol.2017.01.007. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lanfranco MF, Gibb SL, Yowell QV, Carnicella S, Ron D. Long-Lasting Adaptations of the NR2B-Containing NMDA Receptors in the Dorsomedial Striatum Play a Crucial Role in Alcohol Consumption and Relapse. Journal of Neuroscience. 2010;30(30):10187–10198. doi: 10.1523/JNEUROSCI.2268-10.2010. https://doi.org/10.1523/JNEUROSCI.2268-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends in Cell Biology. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. https://doi.Org/10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Warnault V, Darcq E, Levine A, Barak S, Ron D. Chromatin remodeling – a novel strategy to control excessive alcohol drinking. Translational Psychiatry. 2013;3(2):e231. doi: 10.1038/tp.2013.4. https://doi.org/10.1038/tp.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SME, Tauck DL, Fong EG, Kendig JJ. Glutamate Receptor-Mediated Hyperexcitability after Ethanol Exposure in Isolated Neonatal Rat Spinal Cord. Journal of Pharmacology and Experimental Therapeutics. 1998;285(1):201–207. [PubMed] [Google Scholar]
- Zhou D, Lebel C, Lepage C, Rasmussen C, Evans A, Wyper K, et al. Developmental cortical thinning in fetal alcohol spectrum disorders. Neurolmage. 2011;58(1):16–25. doi: 10.1016/j.neuroimage.2011.06.026. https://doi.Org/10.1016/j.neuroimage.2011.06.026. [DOI] [PubMed] [Google Scholar]
- Zhou FC. DNA methylation program during development. Frontiers in Biology. 2012;7(6):485–494. doi: 10.1007/s11515-012-9246-1. https://doi.org/10.1007/s11515-012-9246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Balaraman Y, Teng M, Liu Y, Singh R, Nephew KP. Alcohol alters DNA Methylation Patterns and Inhibits Neural Stem Cell Differentiation. Alcoholism, Clinical and Experimental Research. 2011;35(4):735. doi: 10.1111/j.1530-0277.2010.01391.x. https://doi.org/10.1111/j.1530-0277.2010.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature Reviews Genetics. 2011;12(1):7–18. doi: 10.1038/nrg2905. https://doi.org/10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]


