SUMMARY
The basal ganglia (BG) integrate inputs from diverse sensorimotor, limbic, and associative regions to guide action-selection and goal-directed behaviors. The entopeduncular nucleus (EP) is a major BG output nucleus and has been suggested to channel signals from distinct BG nuclei to target regions involved in diverse functions. Here we use single-cell transcriptional and molecular analyses to demonstrate that the EP contains at least three classes of projection neurons–glutamate/GABA coreleasing somatostatin neurons, glutamatergic parvalbumin neurons, and GABAergic parvalbumin neurons. These classes comprise functionally and anatomically distinct output pathways that differentially affect EP-target regions such as the lateral habenula (LHb) and thalamus. Furthermore, LHb- and thalamic-projecting EP neurons are differentially innervated by subclasses of striatal and pallidal neurons. Therefore, we identify previously unknown subdivisions within the EP and reveal the existence of cascading, molecularly distinct projections through striatum and globus pallidus to EP targets within epithalamus and thalamus.
eTOC Blurb
Neuronal diversity of a basal ganglia output region is defined in Wallace et al., who find that three distinct neuronal subtypes in the entopeduncular nucleus differentially modulate downstream limbic and sensorimotor targets.
INTRODUCTION
The basal ganglia (BG) are a group of interconnected nuclei that play an essential role in voluntary movement and reinforcement learning. Disruptions in BG function have serious consequences for human health ranging from Parkinson’s disease to drug addiction (Hyman et al., 2006; Nelson and Kreitzer, 2014). Despite the central importance of the BG in human health and disease, our understanding of the cellular diversity, microcircuitry, and functional neural pathways that link BG nuclei together and to other brain regions remains incomplete.
Most circuit-level schemes of BG organization describe the entopeduncular nucleus (EP in rodents or globus pallidus internus (GPi) in primates) as a homogenous group of neurons performing the same function of providing inhibitory input to thalamic nuclei to control movement. As a result these models tend to focus on sensorimotor functions of the BG within the framework of the influential “direct and indirect pathway” model (Alexander and Crutcher, 1990). However, there also is a well-described input to the BG from limbic and associative cortical regions that may operate in parallel to process distinct inputs relevant to the emotional state of the animal (Gerfen, 1984). These sensorimotor and limbic circuits are spatially intermingled at the level of the dorsal striatum in the matrix and patch (or striosome) compartments (Gerfen, 1984; Ragsdale and Graybiel, 1981). However, the extent to which these channels remain distinct in downstream BG nuclei and the cellular identity of their targets within and outside the BG are poorly understood.
Anatomical and physiological studies indicate the existence of cellular heterogeneity within the rodent EP and primate GPi (Miyamoto and Fukuda, 2015; Parent et al., 2001; Takada et al., 1994). In mice, cells projecting to the lateral habenula (LHb), a component of the limbic system whose activity is known to have a net inhibitory effect on ventral tegmental area dopamine neurons (Ji and Shepard, 2007), express GABAergic and glutamatergic markers and corelease GABA and glutamate onto individual LHb neurons (Shabel et al., 2012, 2014). LHb-projecting EP neurons also increase their firing rates to aversive stimuli and have been proposed to have a net excitatory effect on the LHb, despite the corelease of both GABA and glutamate (Hong and Hikosaka, 2008; Shabel et al., 2014; Stephenson-Jones et al., 2016). Conversely, neurons that project to thalamic regions express GABAergic markers and are not believed to send axons to the LHb (Barroso-Chinea et al., 2008; Parent et al., 2001). However, a comprehensive description of EP cellular diversity at molecular, electrophysiological, and anatomical levels is lacking.
Here we dissect the genetic, anatomical, and physiological diversity of the EP. Parallel mRNA sequencing from hundreds of individual mouse EP neurons defined two transcriptionally distinct neuronal populations intrinsic to the EP, with a third minority EP population identified by RNA fluorescent in situ hybridization (FISH). We also find three analogous populations of neurons in the human GPi, indicating evolutionary conservation within mammals. Importantly, we define a purely glutamatergic class of LHb-projecting EP neuron, that may contribute to the net excitatory effects of EP activity on the LHb seen in studies that manipulate all LHb-projecting EP neurons together (Shabel et al., 2012; Stephenson-Jones et al., 2016). Based on our results, we genetically targeted EP subpopulations for electrophysiological characterization, anatomical tracing, and functional mapping of outputs. To place the specific subtypes of EP neurons within the larger context of BG circuitry we employed viral-genetic tracing of inputs and outputs. Together, we identify previously unknown subdivisions within the EP and reveal the existence of molecularly and functionally distinct channels through the BG to distinct targets.
RESULTS
Classification of EP neuron types using genome wide expression profiling of single neurons
The EP lies posterior to the globus pallidus externus (GPe) and anterior to the subthalamic nucleus (STN) in mice (Figure 1A and 1B). The most anterior portion of the EP begins immediately posterior to the termination of axons from indirect pathway spiny projection neurons (iSPNs), which define the borders of the GPe. To identify this region we coinjected a Cre-ON AAV encoding GFP and a Cre-OFF AAV encoding tdTom to label iSPNs and dSPNs, respectively, into the striatum of an A2A-Cre mouse (Figure 1A) (Gerfen et al., 2013). Neurons found in the anterior region of the EP express Somatostatin (Sst), whereas those in the more posterior portion of the EP express Parvalbumin (Pvalb) and these two cell types have been hypothesized to form anatomically and functionally distinct subgroups (Figure 1A, 1B and S2) (Rajakumar et al., 1994; Vincent and Brown, 1986).
Figure 1. Single cell RNA sequencing defines two neuronal populations in EP.
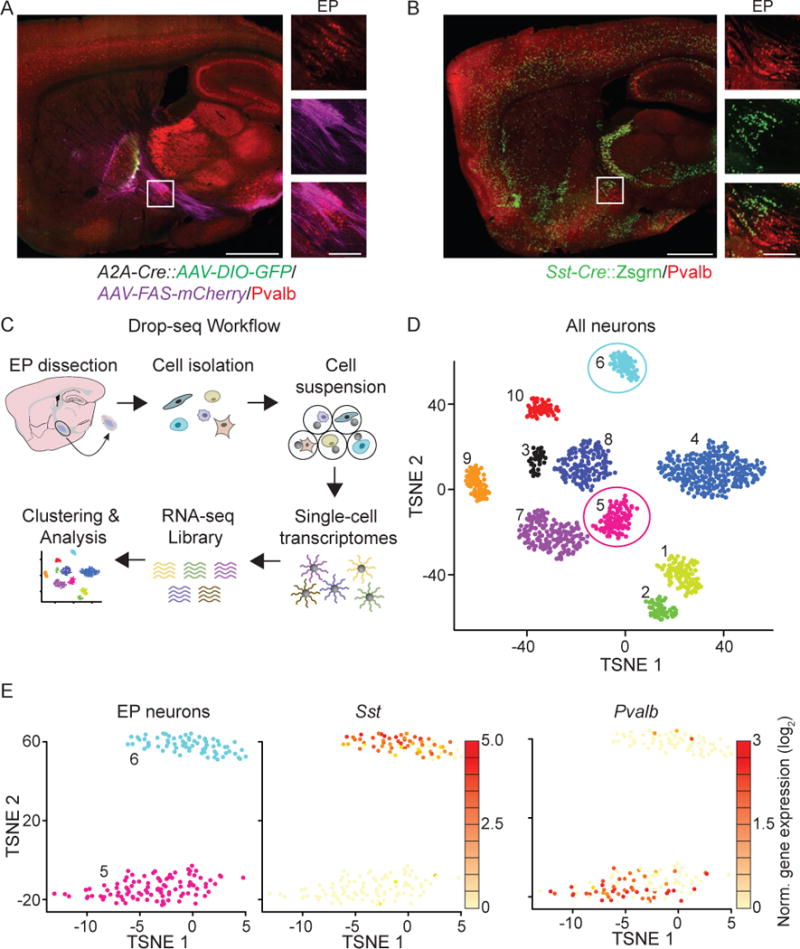
A. A sagittal section of an Adora2A-Cre (A2A-Cre) mouse (which expresses Cre in striatopallidal neurons) injected in striatum with AAV-DIO-GFP (Cre-ON) (green) and AAV-FAS-tdTom (Cre-OFF) (magenta) and immunostained for Pvalb (red). Inset shows zoom of EP.
B. A sagittal section of a Sst-cre::Zsgrn (green) mouse immunostained for Pvalb (red) depicting the Sst/Pvalb subdivisions of the EP (inset).
C. Drop-seq workflow (modified from (Macosko et al., 2015)). Cells isolated from acute slices of EP and surrounding areas are encapsulated in droplets for bead-based mRNA capture and barcoding. Thousands of single cell transcriptomes are then sequenced and analyzed.
D. TSNE plot displaying the results of clustering of the 1,615 neurons dissociated from acute microdissections. Each point represents 1 neuron and clusters are color-coded. Clusters intrinsic to EP (5 and 6) are circled in pink and blue.
E. Of the 10 clusters of neurons, clusters 6 and 5 were confirmed to be from the EP (left) and show differential expression of Sst (middle) and Pvalb (right), red/yellow denotes high and low expression, respectively. See also Figure S1, S2 and S3.
To examine neuronal heterogeneity in the EP, we performed high-throughput single-cell transcriptional profiling (“Drop-seq”) (Macosko et al., 2015). Cell suspensions from the EP and surrounding regions were generated from acute, microdissected brain slices from adult mice (Figure 1C), producing an estimated 24,000 single-cell transcriptomes attached to microparticles (STAMPs) that were sequenced at an average depth of 22,700–44,300 reads. We clustered 9,058 STAMPs (STAMPs with <400 genes were pruned) using principal components analysis and dimensionality reduction by t-Distributed Stochastic Neighbor Embedding (tSNE) (Van Der Maaten and Hinton, 2008) as described previously (Macosko et al., 2015), resulting in 10 cellular clusters (Figure 1D, see Experimental Procedures for details on sequential clustering). To determine which clusters are intrinsic to the EP, differential expression analysis (Satija et al., 2015) and digital in situ hybridization analysis (Allen Brain Atlas, (Lein et al., 2007)) identified the spatial expression patterns for differentially expressed genes (Table S1, Figure S1). Clusters 1 and 2 expressed high levels of oligodendrocyte associated genes (Olig1 and Mag) and low levels of neuronal genes (Snap25, RbFox3 and Syn1) and therefore were excluded from additional analysis (Figure S1A–C, Table S1). We were unable to define the identity of cluster 3 from its differentially expressed genes and did not analyze it further (Table S1). Two clusters had genes whose differential expression was confined to the EP (clusters 6 and 5 with 69 and 97 cells, respectively (Figure 1D and 1E)). We were able to assign other clusters (clusters 4 and 7–10) to regions surrounding the EP such as the subthalamic nucleus, zona incerta, thalamic reticular nucleus and substantia innominata (Figure S1, S2, and Table S1).
Compared to all sequenced neurons, expression of 7 genes in cluster 6 and 6 genes in cluster 5 were enriched by at least 1 log unit (2.72 fold) (p<1.28 × 10−7, Bonferroni corrected (McDavid et al., 2013); Figure 2A, 2B and Table S1). Among the genes enriched in cluster 6 were Sst, Slc17a6 (encoding the vesicular glutamate transporter Vglut2), and Tbr1, whereas cluster 5 was enriched for Kcnc3 and Lypd1 (Lynx1) (Figure 2A, B and S2). Conversely, Slc32a1 (encoding the vesicular GABA/glycine transporter Vgat), Gad1, and Gad2 (encoding, respectively, the GABA synthetic enzymes GAD67 and GAD65) were highly expressed in both clusters (Figure S2). This indicates that cluster 6 contained transcripts associated with both GABA and glutamate neurotransmission, while cluster 5 contained transcripts associated with GABA transmission only. Individual cells in cluster 6 coexpressed glutamatergic and GABAergic markers (n=59 of 69 cells), demonstrating shared GABA/glutamatergic identity at the single cell level (Figure 2A and S2). We also compared gene expression levels between clusters 5 and 6 (Figure 2C). Several other genes were enriched in this analysis such as Meis2 and Nrn1 in cluster 6 and Pvalb and Snc4b in cluster 5 (Figure 1E and 2C). Many genes encoding ion channel subunits, neurotransmitter receptors, and transcription factors were differentially expressed, suggesting molecular substrates for differences in membrane physiology and cell signaling between EP cell types (Figure 2, Table S1).
Figure 2. Genes differentially expressed across EP clusters highlight novel markers and potential functional differences.
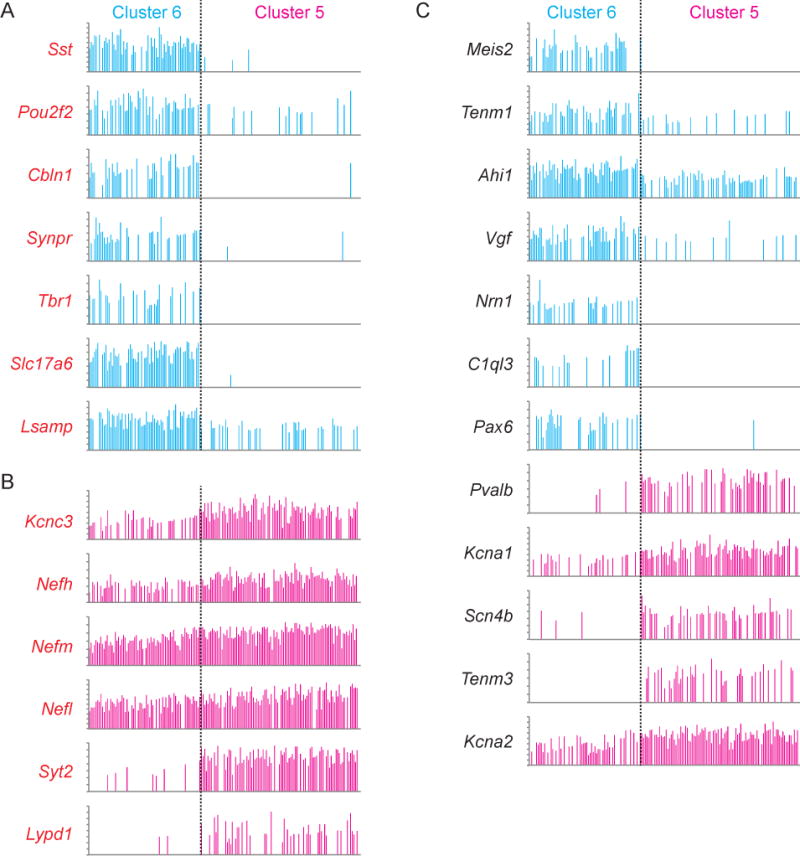
A, B. Genes enriched in cluster 6 (A, blue) or cluster 5 (B, magenta) as compared to the whole population of neurons. Each bar represents the expression level of the indicated gene for a single EP neuron (total number of cells n = 166). The y-axis is log2 normalized expression level for each gene (maxima range from 2.5–5.34). Genes in red indicate genes that are also significantly different across clusters 5 vs 6.
C. Genes not listed in panels A and B that are differentially expressed across clusters 5 and 6. Blue bars represent genes that are enriched in cluster 6 and magenta bars represent genes that are enriched in cluster 5. See also Figure S2.
In situ hybridization reveals neurotransmitter heterogeneity within EP neuron subtypes
Previous anatomical studies revealed that the EP contains markers for glutamatergic and GABAergic neurons and recent functional studies demonstrated that EP neurons are capable of coreleasing both glutamate and GABA (Kha et al., 2000; Shabel et al., 2014). The Drop-seq results suggest that the cluster of neurons expressing Sst is the putative GABA and glutamate coreleasing population and the cluster expressing Pvalb is purely GABAergic (Figure 1, 2, S1, and S2). Consistent with our Drop-Seq analysis and previously published reports Sst and Pvalb labeled largely non-overlapping cell populations in EP (Figure S3A–B) (Miyamoto and Fukuda, 2015).
To confirm the neurotransmitter systems associated with the Sst- and Pvalb-expressing neurons in EP and determine their spatial distribution, we performed three-color single-molecule FISH (Wang et al., 2012). For simplicity, probes for Gad1 and Gad2 were mixed and analyzed in a single fluorescence channel (referred to as Gad below). As expected, nearly all EP neurons in both the anterior and posterior regions expressed Gad, consistent with the GABAergic nature of the nucleus (Penney and Young, 1981). The Sst probe marked a population of anterior EP neurons (Vincent and Brown, 1986) that also expressed Slc17a6 and Gad, confirming that most Sst neurons express both GABAergic and glutamatergic markers and thus may release GABA and glutamate (Figure 3A).
Figure 3. Three neuronal classes in both mouse EP and human GPi.
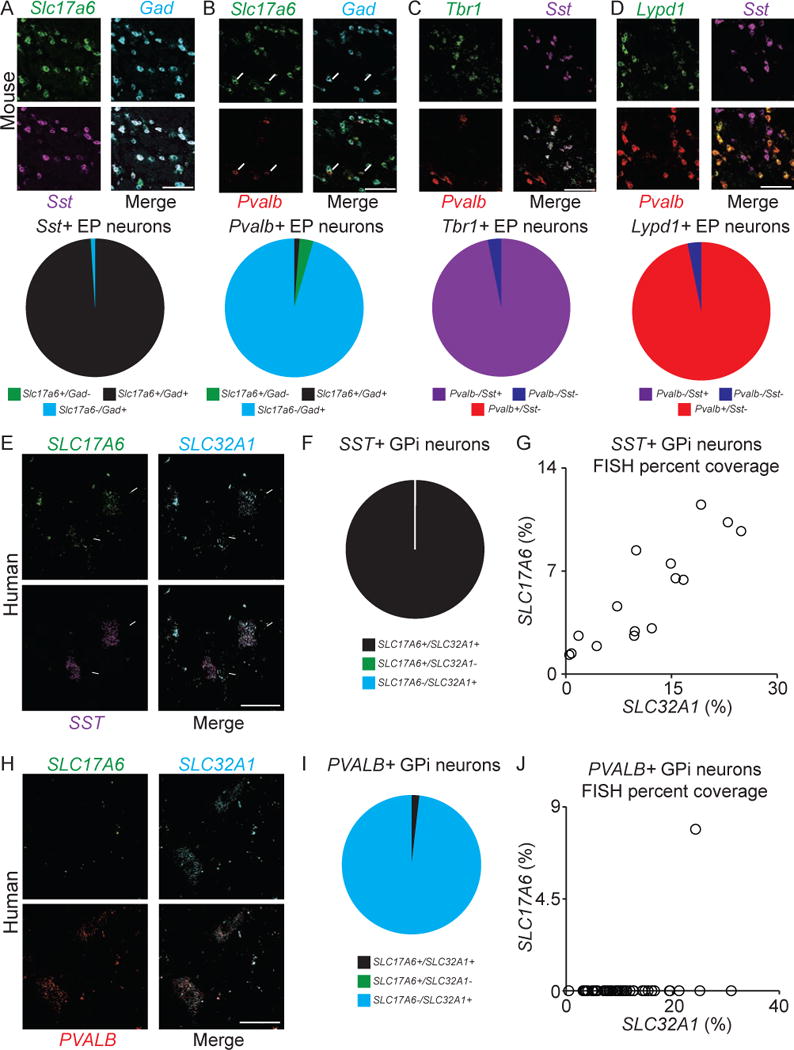
A. (Top) Sample image of a coronal section of EP probed for Sst (magenta), Slc17a6 (green), and Gad (mixed probes for Gad1 and Gad2; cyan). (Bottom) Quantification of colabeling of Sst with Slc17a6 and Gad in EP (n=959 cells, 3 mice).
B. (Top) Sample image of a coronal section of EP probed for Pvalb (red), Slc17a6 (green), and Gad (cyan). Arrows indicate Pvalb+/Slc17a6+ cells, and arrowhead indicates a Pvalb+/Gad+ cell. (Bottom) Quantification of colabeling of Pvalb with Slc17a6 and Gad in EP (n=714 cells, 3 mice).
C. (Top) Sample image of a coronal section of EP probed for Sst (magenta), Tbr1 (green), and Pvalb (red). (Bottom) Quantification of colabeling of Tbr1 with Sst and Pvalb in EP (n=223 cells, 3 mice).
D. (Top) Sample image of a coronal section of EP probed for Sst (magenta), Lypd1 (green), and Pvalb (red). (Bottom) Quantification of colabeling of Lypd1 with Sst and Pvalb in EP (n=188 cells, 3 mice).
E. Sample image of a coronal section of human GPi probed for SST (magenta), SLC17A6 (green), and SLC32A1 (cyan). Arrows indicate SST+/SLC17A6+/SLC32A1+ cells.
F. Quantification of colabeling of SST with SLC17A6 and SLC32A1 in GPi (n=14 cells).
G. Quantification of fluorescence coverage of SLC17A6 and SLC32A1 in SST+ GPi neurons.
H. Sample image of a coronal section of human GPi probed for PVALB (red), SLC17A6 (green), and SLC32A1 (cyan).
I. Quantification of colabeling of PVALB with SLC17A6 and SLC32A1 in GPi (n=56 cells).
J. Quantification of fluorescence coverage of SLC17A6 and SLC32A1 in PVALB+ GPi neurons. See also Figure S3.
Triple FISH for Pvalb, Gad, and Slc17a6 revealed additional neuronal heterogeneity within EP (Figure 3B). Most EP neurons that express Pvalb were in the posterior EP and also express Gad, but not Slc17a6, consistent with the Drop-seq results. However a small subpopulation of Pvalb neurons lacked Gad, but did express Slc17a6 (Figure 3B). Pvalb+/Slc17a6+ EP neurons were mostly found at the borders of EP and the interface between Pvalb+ and Sst+ zones. These results confirm the Drop-seq analysis demonstrating two distinct neuron-types in EP, and also reveal the existence of a minority cell type not found in the Drop-seq single-cell clustering.
Whereas Sst and Pvalb are known to mark neuronal populations in EP, the Drop-seq analysis revealed many potential additional markers for these two cell classes. We validated the expression patterns of two of those genes with FISH. Consistent with the Drop-seq findings, Tbr1 and Lypd1 exclusively and specifically labeled the Sst+ and Pvalb+/Gad+ EP cell classes, respectively (Figure 3C–D and S2G–H). Additionally, Lypd1 was not expressed in Slc17a6+/Slc32a1− EP neurons (putative Pvalb+/Slc17a6+ neurons) further distinguishing this neuronal subset from Pvalb+/Gad+ neurons (Figure S3C–E). Collectively, the FISH results indicate that there are at least three separate classes of EP neurons with distinct neurochemical properties: 1) Sst+/Tbr1+/Gad+/Slc17a6+, 2) Pvalb+/Gad+/Lypd1+, and 3) Pvalb+/Slc17a6+/Lypd1−. Thus, we will refer to these as 1) GABA/glutamate dual-transmitter; 2) purely GABAergic neurons; and 3) purely glutamatergic.
Three distinct neuronal classes in human GPi
The structure and function of the BG is believed to be well conserved throughout evolution (Grillner and Robertson, 2016). Therefore, we examined if the cell-types we described in mouse EP are also present in the analogous structure in humans, the GPi. We performed triple FISH for SST, SLC17A6, and SLC32A1, or PVALB, SLC17A6, and SLC32A1 on sections of human GPi. Similar to mouse EP, we found 1) SST+/SLC17A6+/SLC32A1+, 2) PVALB+/SLC17A6−/SLC32A1+, and 3) SLC17A6+/SLC32A1− neurons in the human GPi (Figure 3E–J and S3F–G). The first two classes appear analogous to the mouse dual-transmitter neurons and the mouse pure GABAergic neurons, such that the differential expression of somatostatin and parvalbumin are preserved across species in these cell classes. In contrast to mice, the pure glutamatergic (SLC17A6+/SLC32A1−) neurons in humans do not express significant levels of PVALB. Additionally, in the human, much like LHb-projecting GPi neurons in monkey (Saimiri sciureus) (Parent and De Bellefeuille, 1982), the majority of dual transmitter (SST+/SLC17A6+/SLC32A1+) and pure glutamatergic (SLC32A1−/SLC17A6+) neurons are located on the borders of the GPi whereas the pure GABAergic (PVALB+/SLC32A1+) cells are located in the center of the nucleus. This suggests that dual transmitter and pure glutamatergic neurons may project to the LHb, whereas pure GABAergic neurons may project to the thalamus.
Intrinsic membrane properties, AP shape, and firing rates distinguish Sst+ and Pvalb+ EP neurons
Many neurons of the BG are intrinsically active and exhibit distinct electrophysiological characteristics that govern their in vivo firing patterns and function within a circuit. To examine the intrinsic electrophysiological properties of Sst and Pvalb neurons of the EP, we performed whole-cell current clamp recordings in acute brain slices from adult Sst-IRES-Cre (Sst-Cre) (Taniguchi et al., 2011) and Pvalb-IRES-Cre (Pvalb-Cre) (Hippenmeyer et al., 2005) mice crossed to Ai6 reporter mice, identifying each neuron class by expression of green fluorophores (Figure 4A and S4). Sst-Cre+ EP neurons fired a steady train of action potentials in response to positive current injections (Figure 4B and 4C). Many Pvalb-Cre+ EP neurons, however, fired in a stuttering or bursting spike pattern which was evinced by an increase in the coefficient of variation of the interspike interval (CVISI) compared to Sst-Cre+ neurons (Figure 4B and D). The majority of Sst-Cre and Pvalb-Cre positive EP neurons also fired spikes spontaneously at rest (Figure S5A). Membrane resistance, membrane time constant and resting membrane potential were not different between Sst-Cre+ and Pvalb-Cre+ neurons, but Pvalb-Cre+ neurons had a larger capacitance suggesting that these neurons had a larger soma size (Figure 4E–F and S5D–E). The shape of the action potential also differed between groups such that Sst-Cre+ neurons had wider action potentials as measured by full-width at half-height (FWHH) and a smaller after-hyperpolarization (AHP) than the Pvalb-Cre+ neurons (Figure 4G–H). Finally, the Drop-seq data indicated that both Sst+ and Pvalb+ neurons in EP expressed the hyperpolarization-activated cyclic nucleotide-gated (HCN) channels (Table S1). These channels mediate the Ih current, and contribute to “sag” potentials (Robinson and Siegelbaum, 2003). Consistent with the presence of HCN channels, both Sst-Cre and Pvalb-Cre positive EP neurons displayed sag potentials in response to hyperpolarizing current injections (Figure S5B–C). These data demonstrate that Sst+ EP neurons are more excitable, have different action potential shape, and fire action potentials more regularly than the more sporadically firing Pvalb+ EP neurons. Additionally, the described differences in AP shape could be used during in vivo single unit recordings to distinguish genetically distinct EP populations.
Figure 4. Differential electrophysiological properties of Sst+ and Pvalb+ expressing EP neurons.
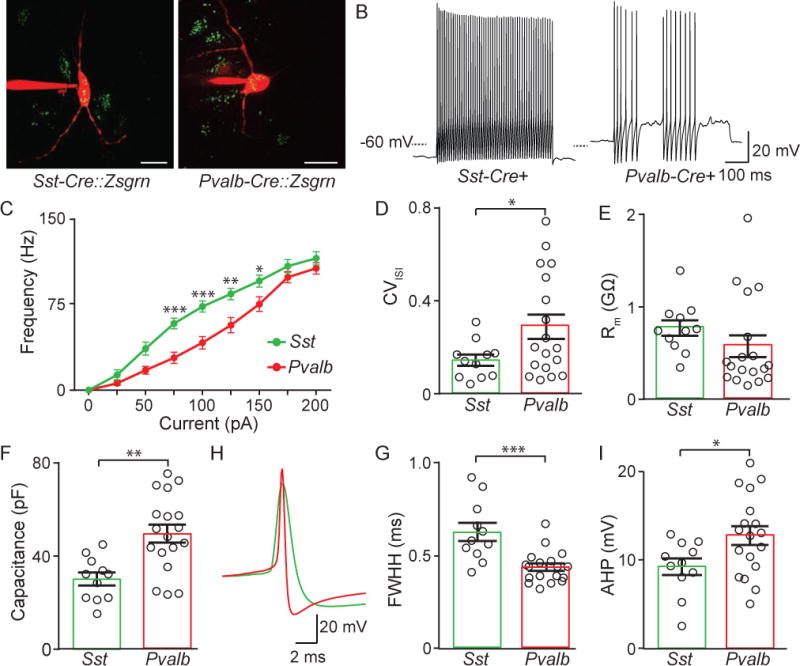
A. Images of Alexa Fluor-594 filled Zsgrn+ EP neurons in Sst-Cre and Pvalb-Cre mice crossed to Ai6 mouse.
B. Sample current-clamp recordings of action potential firing to +100 pA square wave current injection in Sst-Cre and Pvalb-Cre mice.
C. Action potential firing frequency vs current injection for neurons from Sst-Cre (green) (n=11 cells) and Pvalb-Cre (red) mice (n=18 cells).
D. The coefficient of variation of the interspike interval (CVISI) (averaged from ISIs across all current injections).
E, F. Membrane resistance (Rm) and capacitance across cell types.
G, H, I. Sample current-clamp recording of an action potential (G) and full width at half height (FWHH) (G) and after-hyperpolarization (AHP) (I) measurements from Sst-Cre (green) and Pvalb-Cre (red) neurons. All data are represented as mean ± SEM, *=p<0.05, **=p<0.01, ***=p<0.001. See also Figure S4 and S5.
Sst+ EP neurons exclusively innervate the LHb, but Pvalb+ neurons project to diverse downstream regions
As an output nucleus of the BG, the EP projects to many downstream structures such as the motor thalamus, central median/parafascicular thalamus (CM/PF), lateral habenula (LHb), and brain stem (Kha et al., 2000; Takada et al., 1994). To determine if EP projection patterns were cell-type specific, we injected Cre-dependent AAV encoding Synaptophysin-mCh. into the EP of Sst-Cre and Pvalb-Cre mice (Figure 5A). Injections into Sst-Cre and Pvalb-Cre mice both showed axonal labeling in the LHb albeit in different patterns (Figure 5B). Sst-Cre+ axons densely innervated the lateral portion of the LHb consistent with previous reports, whereas Pvalb-Cre+ axons were much less numerous and instead specifically targeted the oval nucleus of the LHb (Figure 5B) (Geisler et al., 2003). Pvalb-Cre+ axons were also found in the ventro-anterior lateral thalamus (VAL), ventro-medial thalamus (VM), anterior-dorsal thalamus (AD), PF, and brainstem consistent with previous reports (Figure 5C) (Rajakumar et al., 1994). Interestingly, Sst-Cre axons targeted both ipsilateral and contralateral LHb, whereas Pvalb-Cre axons only targeted ipsilateral LHb, and thalamus (Figure S6A–B).
Figure 5. Sst+ EP neurons target LHb, and Pvalb+ neurons target LHb and motor thalamus.
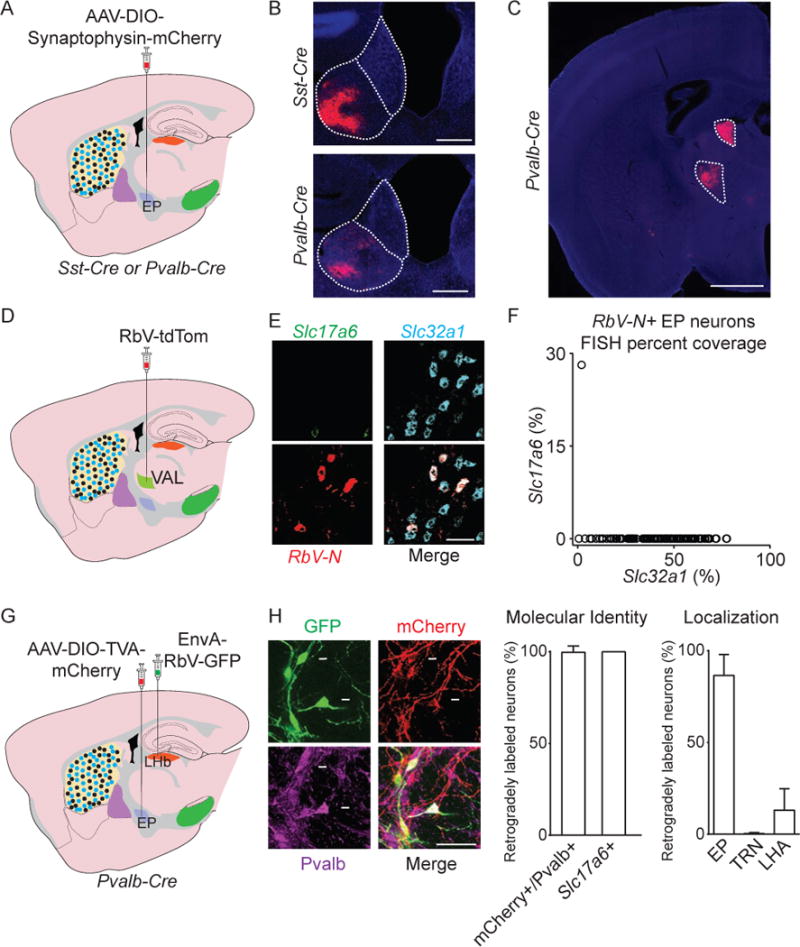
A. Illustration of a sagittal slice depicting AAV-DIO-Syn.-mCh. viral injection in EP in Sst-Cre or Pvalb-Cre mice.
B, C. Sample coronal image of axonal labeling (red) in the LHb (B) or VAL and AD thalamus (C) following viral injection into EP in Sst-Cre or Pvalb-Cre mice (DAPI in blue).
D. Illustration of a sagittal slice depicting RbV-tdTom viral injection in VAL.
E. Sample image of a coronal section of EP probed for RbV-N (red), Slc17a6 (green), and Slc32a1 (cyan).
F. Quantification of fluorescence coverage of Slc17a6 and Slc32a1 in RbV-N+ EP neurons (122 cells, n=3 animals).
G. Illustration of a sagittal slice depicting AAV-DIO-TVA-mCh. viral injection in EP, and EnvA-RbV-GFP injection in LHb in a Pvalb-Cre mouse.
H. (left) A sample image of a coronal section of EP showing RabV-GFP (green), mCh. (red), and Pvalb (magenta). (middle) Percentage of retrogradely labeled (GFP+) neurons that were also labeled for mCh. and Pvalb (169 cells, n=3 animals) or in separate FISH experiments Slc17a6 (25 cells, n=3 animals). (right) Quantification of soma location of GFP+ neurons following EnvA-RbV-GFP injection into LHb (186 cells, n=2 animals) (VAL=Ventral anterior lateral thalamus, AD=Anterior dorsal thalamus, LHA=Lateral hypothalamus, PF=Parafascicular nucleus of the thalamus, TRN=Thalamic reticular nucleus, MHb=medial habenula, V3=3rd ventricle). All data are represented as mean ± SEM. See also Figure S6.
Previous studies suggested that EP neurons that project to the LHb also send collaterals to the thalamus and other structures, and we also observed axonal labeling in PF following viral injections into EP of Sst-Cre mice (Figure S6F–H) (Kha et al., 2000; Takada et al., 1994). To test if axonal labeling observed in the PF was due to cells labeled in EP or due to viral leak into a nearby region, we injected a Cre-dependent AAV encoding TVA-mCh. into the EP. When expressed, the TVA receptor permitted axonal infection by glycoprotein (G) deleted rabies virus encoding eGFP and pseudotyped to express the EnvA viral coat (EnvA-RbV-GFP)(Wickersham et al., 2007). Following TVA-mCh. expression in Sst-Cre+ neurons in EP (~3 weeks), EnvA-RbV-GFP was injected into either LHb or PF (Figure S6F). The anatomical location of TVA-mCh. expressing neurons that were infected with EnvA-RbV-GFP was examined and quantified (Figure S6G and H). Following EnvA-RbV-GFP injection into LHb, infected (GFP+) neuronal somata were almost exclusively found in the EP and GFP+ axons were not observed in the PF or in any other structures (Figure S6G). Conversely, following EnvA-RbV-GFP injection into PF, infected (GFP+) neuronal somata were only found in the thalamic reticular nucleus (TRN), with no GFP+ axon collaterals in LHb (Figure S6H). Therefore, axonal labeling observed in PF in Sst-Cre mice likely arises from viral leak into the adjacent ventral TRN, which also expresses Cre and projects to PF (Kolmac and Mitrofanis, 1997). Additionally, these data suggest that Sst-Cre+ EP neurons that project to LHb do not send collaterals to other brain regions.
Unlike Sst-Cre+ neurons, Pvalb-Cre+ EP neurons innervate both limbic (LHb) and motor associated (VM/VAL thalamus) regions (Figure 5B and 5C). To determine if subclasses of Pvalb+ EP neurons specifically target limbic or motor associated regions we injected the retrograde tracer, G-deleted non-pseudotyped rabies virus encoding tdTom (RbV-tdTom), into VAL thalamus and 7 days later performed FISH in EP for Slc17a6, Slc32a1, and Rabies-Nucleoprotein (RbV-N) (Figure 5D–F). Nearly all RbV-N+ EP neurons were also Slc32a1+/Slc17a6− indicating they were GABAergic and likely correspond to the purely GABAergic Pvalb+ population described above (Figure 3B and S2B–F). To label the Pvalb+ EP neurons that projected to LHb we injected a virus encoding TVA-mCh. into EP of Pvalb-Cre mice and followed with injection of EnvA-RbV-GFP into the LHb (Figure 5G). Immunohistochemistry for Pvalb subsequently revealed, as expected, that all GFP+ EP neurons were Pvalb+ (Figure 5H). In parallel, FISH for RbV-N and Slc17a6 revealed that all RbV-N+ neurons were also positive for Slc17a6 (Figure 5H and S6E), indicating that the purely glutamatergic EP population described above projects exclusively to LHb. Therefore, purely GABAergic EP (Pvalb+/Slc32a1+/Slc17a6−) neurons project to VAL thalamus but not LHb, likely comprising the classic motor EP output, whereas the previously undescribed purely glutamatergic (Pvalb+/Slc17a6+) neurons project to the LHb but not to VAL thalamus. These data, and FISH experiments described above (Figure 3B), indicate that electrophysiological recordings from Pvalb-Cre+ neurons (Figure 4) are likely almost all from neurons that project to thalamus.
Functional connectivity and pharmacology of EP projections to distinct target regions
The anatomical and gene transcription data suggest that the EP has three functionally distinct outputs. These are a purely GABAergic output to motor regions, a purely glutamatergic output to LHb, and a GABA/glutamate coreleasing output to LHb. To test these predictions, we injected the EP of either Sst-Cre or Pvalb-Cre mice with Cre-dependent AAV encoding Channelrhodopsin2-mCh. (AAV-DIO-ChR2-mCh.; Boyden et al., 2005) and performed whole-cell voltage-clamp recordings in the downstream nuclei targeted by each cell class (Figure 6). Optogenetic stimulation of Sst-Cre+ EP axons while holding LHb neurons at the chloride reversal potential (−75 mV) evoked excitatory postsynaptic currents (EPSC) (Figure 6A–C). We confirmed that these currents were monosynaptic, with sequential application of TTX and TTX/4-AP, and glutamatergic, by application of the AMPA/NMDA receptor antagonists NBQX/CPP (Figure 6B and 6C) (Petreanu et al., 2009). Subsequent depolarization to 0 mV to increase the driving force for chloride, revealed an inhibitory postsynaptic current (IPSC) which was maintained in the continued presence of TTX/4-AP/NBQX/CPP but was blocked by the GABAA receptor antagonist SR95531 (gabazine) (Figure 6B and C). These data confirm previous results of glutamate and GABA corelease from EP axons in LHb (Shabel et al., 2014) and identify these previously genetically uncharacterized axons as arising from Sst-expressing EP neurons.
Figure 6. Sst+ and Pvalb+ EP neurons that target the LHb release GABA/glutamate or glutamate, respectively, and a distinct Pvalb+ population targets VM/VAL and releases GABA.
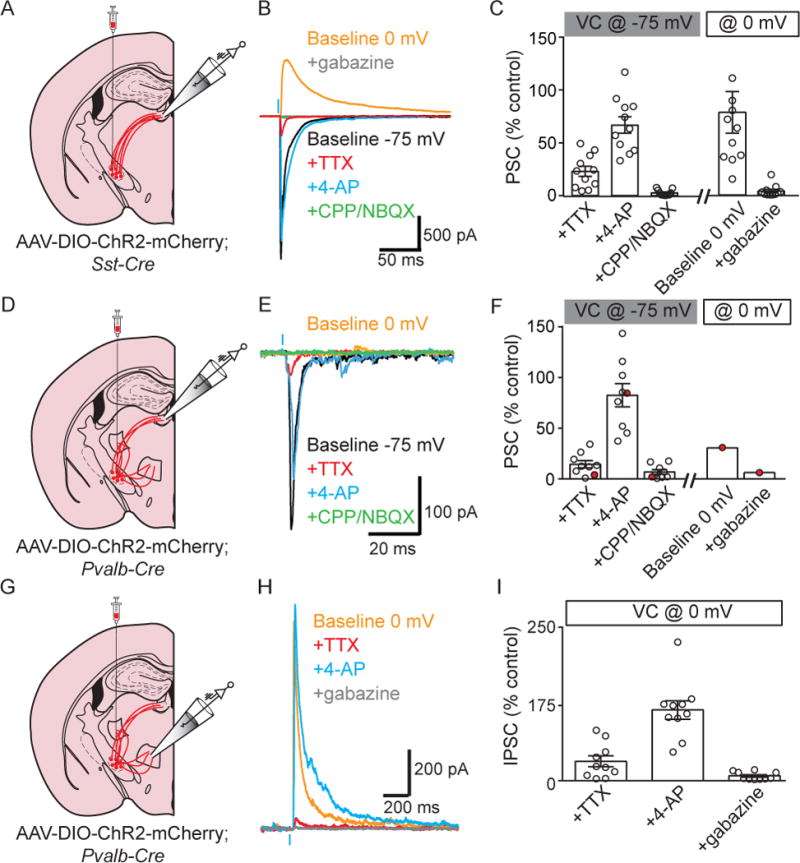
A. Illustration of a coronal slice depicting AAV-DIO-ChR2-mCh. viral injection into EP and recording location in LHb of a Sst-Cre mouse.
B. Sample voltage-clamp recordings in LHb during optogenetic activation of Sst-Cre+ EP axons. The cell was clamped at −75mV to record glutamatergic currents (black) and 0 mV to record GABAergic currents (orange).
C. Quantification of optogenetically-evoked PSC amplitude. TTX, 4-AP, CPP/NBQX and 0 mV baseline measurements are all normalized to the −75 mV pre-drug EPSC amplitude but the amplitude in “gabazine” is normalized to the 0 mV baseline IPSC (n=11).
D. Illustration of a coronal slice depicting AAV-DIO-ChR2-mCh. viral injection into EP and recording location in LHb of a Pvalb-Cre mouse.
E. Sample voltage-clamp recordings in LHb during optogenetic activation of Pvalb-Cre+ EP axons. The cell was clamped at −75mV to record glutamatergic currents (black) and 0 mV to record GABAergic currents (orange).
F. Quantification of optogenetically-evoked PSC amplitude analyzed as in panel C (n=9, red point represents a neuron that displayed glutamatergic and GABAergic currents).
G. Illustration of a coronal slice depicting AAV-DIO-ChR2-mCh. viral injection into EP and recording location in VM thalamus from a Pvalb-Cre mouse.
H. Sample voltage-clamp recordings in VM thalamus during optogenetic activation of Pvalb- Cre+ EP axons. The cell was clamped at 0mV to record GABAergic currents (orange).
I. Quantification of optogenetically-evoked IPSC amplitude. All measurements are all normalized to the 0 mV baseline IPSC amplitude (n=10). All data are represented as mean ± SEM. See also Figure S7.
Similar analysis of currents evoked in LHb neurons in Pvalb-Cre mice by optogenetic stimulation of Pvalb-Cre+ EP axons revealed exclusively glutamatergic EPSCs in all but one case (Figure 6D–F and S7A–C). This finding suggests that the small number of Pvalb+ neurons expressing only Slc17a6 identified by multi-color FISH (Figure 3C and 3D) mediate the sole Pvalb+ projection from EP to LHb. Finally, recordings in the VAL/VM thalamus in Pvalb-Cre mice while optogenetically stimulating Pvalb-Cre+ EP axons evoked only GABAergic IPSC in all cells examined (Figure 6G–I and S7D). These anatomical and electrophysiological findings confirm that the EP contains at least three distinct projection neuron subtypes, one that is Sst+ and coreleases GABA and glutamate in the LHb, a second that is Pvalb+ and releases glutamate in the LHb, and a third that is also Pvalb+ but releases GABA in the VAL/VM thalamus.
Viral genetic tracing of inputs to three EP neuron subclasses
To explore the possibility that different EP neuronal subtypes may be part of distinct microcircuits within the BG we performed monosynaptic retrograde tracing with EnvA-RbV-GFP. To restrict rabies virus infection to genetically defined subpopulations of EP neurons we again used Cre-dependent TVA-mCh., but coinjected a Cre-dependent AAV encoding the rabies glycoprotein (G) to allow for retrograde monosynaptic transfer of G-deleted EnvA-RbV-GFP. As only neurons in the EP will express G, GFP labeled neurons in other regions will be presynaptic to infected EP neurons (controls for specificity and spread of EnvA-RbV-GFP, see Figure S6C–D) (Wickersham et al., 2007). To label three distinct populations of EP neurons we injected AAVs encoding TVA-mCh. and G into the EP of Sst-Cre, Pvalb-Cre, and Rbp4-Cre mice. Rbp4-Cre was used because it labeled a subpopulation of VM/VAL projecting Pvalb+ EP neurons but not surrounding TRN neurons (Figure S8A and B) (Gong et al., 2007). After three weeks of AAV expression we injected EnvA-RabV-GFP into either the LHb (for Sst-Cre and Pvalb-Cre mice) or VAL thalamus (for Rbp4-Cre mice) (Figure 7A). EnvA-RabV-GFP injected into projection targets (i.e. LHb or VAL thalamus) only infects the TVA+ axonal fields of EP neurons projecting to that region, thus labeling the presynaptic partners of sub-populations EP neurons defined by their projection targets (Figure 7A). In all cases the vast majority of “starter cells” (cells that expressed mCh. and GFP) were located in the EP and robust retrograde labeling was observed in many BG nuclei including the striatum, GPe, STN, and SN (Figure 7A and Figure S8G–I).
Figure 7. LHb projecting EP neurons have patch biased striatal input.
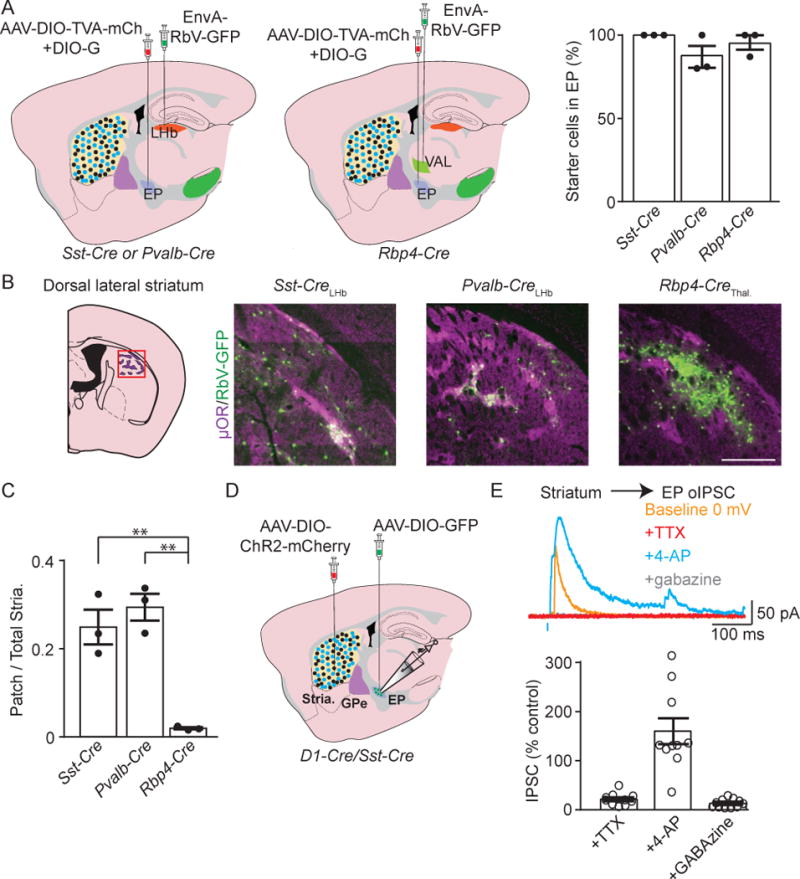
A. Illustration of sagittal slices depicting AAV-DIO-TVA-mCh. and AAV-DIO-G viral coinjection into EP. EnvA-RbV-GFP was injected into LHb of Sst-Cre or Pvalb-Cre mice (left), or into VAL thalamus in Rbp4-Cre mice (middle). Quantification of starter cell (mCh.+/GFP+) location (right) (Sst-Cre n=3, Pvalb-Cre n=3, Rbp4-Cre n=3 mice).
B. Illustration of a coronal section of striatum. The red box indicates region shown in sample images to right. In sample images, GFP+ cells are presynaptic to the indicated EP subpopulation and μOR immunostain (magenta) marks the patch (striosome) compartment.
C. Quantification of the proportion of retrogradely labeled striatal neurons that were within patches (Sst-Cre n=3, Pvalb-Cre n=3, Rbp4-Cre n=3 mice).
D. Illustration of a sagittal slice depicting viral injection targets and location of whole-cell recording. An AAV-DIO-ChR2-mCh. (Cre-ON) viral injection in striatum and an AAV-DIO-GFP injection in EP was made in a D1-Cre/Sst-Cre mouse.
E. top, Sample voltage-clamp recordings in an EP neuron during optogenetic activation of Drd1a-Cre+ striatal axons. The cell was clamped at 0 mV to record GABAergic IPSCs. bottom, Quantification of optogenetically-evoked IPSC amplitude. All measurements are normalized to the 0 mV baseline IPSC amplitude (n=10 cells). All data are represented as mean ± SEM *=p<0.05, **=p<0.01. See also Figure S6 and S8.
The striatum is the major input nucleus of the BG and is subdivided into the patch (striosome) and matrix compartments. The patch compartment can be distinguished by its high expression of the mu-opioid receptor (μOR) relative to surrounding matrix (Pert et al., 1976). Patches occupy approximately 10–20% of the striatum and receive input predominantly from limbic/associative cortical areas whereas cortical input to the matrix is largely sensorimotor (Gerfen, 1984; Ragsdale and Graybiel, 1981). We observed retrograde labeling in patches for starter neurons that projected to the LHb. Both LHb-projecting Sst-Cre+ and Pvalb-Cre+ neurons received approximately 25% of their striatal input from μOR labeled patches (Figure 7B and 7C). Note that the identification of patches by μOR staining likely results in an underestimate of the patch neurons because only large patches can be unambiguously identified. Using a Sst-Cre mouse crossed to a mouse expressing tdTom in dSPNs (Drd1a-tdTom, Ade et al., 2011) we repeated the monosynaptic retrograde tracing experiments with the advantage of being able to distinguish striatal neuron subclasses. We observed that 99% (530/539 of GFP+ neurons were tdTom+, n=2 mice) of retrogradely labeled striatal neurons also expressed tdTom indicating that dSPNs are the only striatal neuron class that projects to the Sst-Cre+ EP population (Figure S8E–F). Finally, in contrast to LHb projecting EP, Rbp4-Cre+ neurons (i.e. Pvalb+ neurons, Figure S8A–B) projecting to VAL thalamus received almost all of their striatal input from matrix (Figure 7B and 7C). We electrophysiologically confirmed input to the EP from striatum by injecting AAV-DIO-ChR2-mCh. into striatum of Drd1a-Cre mice and optogenetically stimulating dSPN axons while performing whole-cell voltage-clamp recordings from EP neurons (Figure 7D and 7E). Inputs to EP from dSPNs were pharmacologically confirmed to be monosynaptic and GABAergic (Figure 7E).
The EP also receives major input from the GPe (Rajakumar et al., 1993), which has recently been recognized as a cellularly heterogeneous nucleus (Abdi et al., 2015; Mastro et al., 2014; Saunders et al., 2015). The GPe contains two major neuronal subpopulations: the arkypallidal striatal projecting group marked by FoxP2, and the prototypic group marked by Pvalb (Abdi et al., 2015). This latter group is thought to mediate the canonical downstream projections of GPe to the EP, subthalamic nucleus (STN), and substantia nigra (SN) (note that a small population of Pvalb+ neurons that project to striatum also exists (Saunders et al., 2016)). Surprisingly, EP neurons that project to the LHb (from Sst-Cre and Pvalb-Cre mice), receive only 35% of their GPe input from Pvalb+ neurons (Figure 8A and 8B).
Figure 8. Pvalb negative GPe neurons innervate the LHb projecting EP.
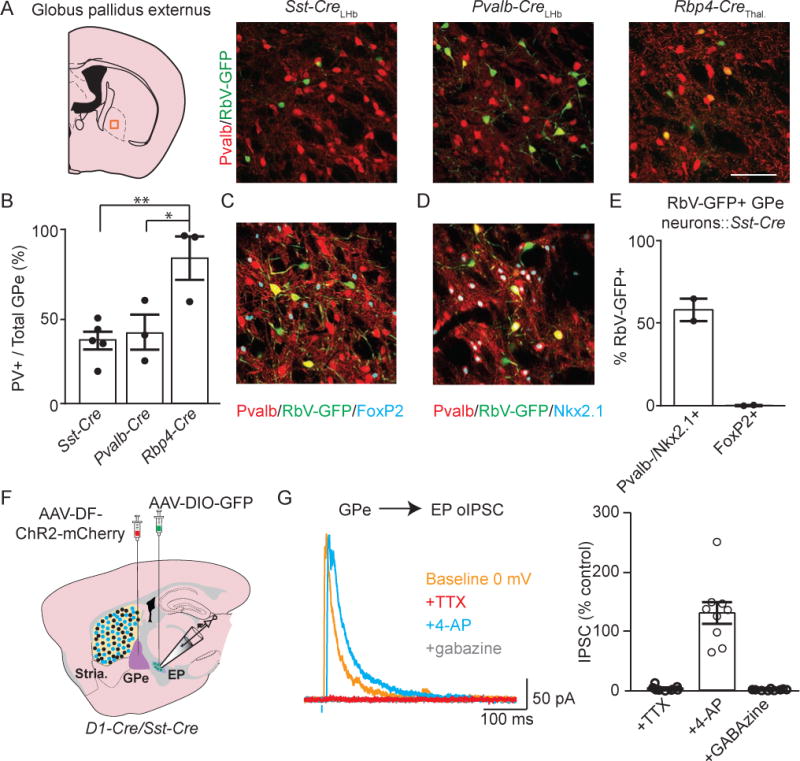
A. Illustration of a coronal section of GPe. The orange box indicates region shown in sample images to right. In sample images, GFP+ cells are presynaptic to different subpopulations of EP neurons and the Pvalb (red) marks the “prototypic” GPe neurons.
B. Quantification of the proportion of retrogradely labeled GPe neurons that were Pvalb+ (Sst-Cre n=5, Pvalb-Cre n=3, Rbp4-Cre n=3 animals).
C. Sample image of immunostaining in GPe for Pvalb (red) and FoxP2 (cyan), markers for prototypic and arkypallidal GPe neurons, respectively. GFP+ (green) are presynaptic to Sst+ EP neurons.
D. Sample image of immunostaining in GPe for Pvalb (red) and Nkx2.1 (cyan). GFP+ (green) are presynaptic to Sst+ EP neurons.
E. Proportions of retrogradely labeled GPe neurons that immunostained for each of the three markers shown in C and D (FoxP2+ (1/253 cells, n=2 animals), Pvalb-/Nkx2.1+ (117/218 cells, n=2 animals)).
F. Illustration of a sagittal slice depicting viral injection targets and location of whole-cell recording. An AAV-DF-ChR2-mCh. (Cre-OFF) viral injection into GPe, and AAV-DIO-GFP injection in EP was made in a D1-Cre/Sst-Cre mouse.
G. left, Sample voltage-clamp recordings in an EP neuron during optogenetic activation of GPe axons. The cell was clamped at 0 mV to record GABAergic currents. right, Quantification of optogenetically-evoked IPSC amplitude. All measurements are normalized to the 0 mV baseline IPSC amplitude (n=9 cells). All data are represented as mean ± SEM *=p<0.05, **=p<0.01. See also Figure S6 and S8.
We tested if the Pvalb− GPe neurons that project to Sst-Cre+ EP neurons expressed other markers of GPe neuron subclasses. We found that GPe neurons that project to Sst-Cre+ EP neurons were negative for FoxP2 (1/209 GFP+ neurons were FoxP2+, n=2 mice, Figure 8C and 8E). However, GFP+/Pvalb− GPe neurons were found to express Nkx2.1 (117/218 GFP+ neurons were Pvalb-/Nkx2.1+, n=2 mice, Figure 8D and 8E). Conversely, EP neurons that project to VAL thalamus received the majority of their input from prototypic GPe neurons that are Pvalb+ (Figure 8A and 8B). These data suggest that Pvalb-/FoxP2-/Nkx2.1+ are a third subtype of GPe neuron that may form a limbic channel within GPe, whereas Pvalb+ neurons may contribute towards a sensorimotor channel.
We functionally confirmed input to the EP from GPe by injecting AAV-DF-ChR2-mCh. into GPe of Drd1a-Cre mice. This virus turns off expression of ChR2-mCh. in the presence of Cre and minimizes contamination of striatal dSPN inputs to EP following injections into the GPe (Saunders et al., 2012) (Figure 8F). We optogenetically stimulated GPe axons while recording from EP neurons and pharmacologically confirmed GPe input to be monosynaptic and GABAergic (Figure 8G). These data provide a map of the input-output relationships of three classes of EP neurons, demonstrating the existence of distinct microcircuits that exist within the general BG framework.
DISCUSSION
Here we describe a comprehensive cellular and circuit analysis of the EP, a major output nucleus of the BG. We propose that the specific neuron-classes we have identified in EP channel separate inputs from striatum and GPe to downstream regions involved in reinforcement learning and motor control. We classify EP neurons into two major neuron types marked by either Sst or Pvalb using large-scale single cell transcriptional profiling. The Sst and Pvalb populations were confirmed to be non-overlapping by FISH. Anatomical and functional mapping of synaptic connectivity between each neuron-type and its limbic or motor related target region revealed additional heterogeneity in the neurotransmitter type(s) used by each class of neuron. Additionally, monosynaptic retrograde tracing placed the three neuron-types in one of two BG macrocircuits. Sst+ and Pvalb+ neurons that project to LHb receive input biased towards limbic associated regions of striatum (patches), whereas Pvalb+ neurons that project to motor thalamus received input exclusively from sensorimotor regions of striatum (matrix).
Glutamatergic and GABAergic output from the EP
Here we show that Sst+ EP neurons corelease GABA and glutamate in the LHb, whereas Pvalb+/Slc17a6+ neurons release only glutamate. It is unlikely that these two neuronal populations target regions outside the LHb as our anatomical and electrophysiological studies indicate minimal axonal collaterals and synaptic connections from these neurons in other regions. Therefore, glutamatergic output from EP is likely confined to the LHb. Within the LHb, the Pvalb+/Slc17a6+ input is concentrated in the oval nucleus subregion, and Sst+ GABA/glutamate input is distributed throughout various LHb subregions (Geisler et al., 2003).
Neurons of the lateral subnuclei of the LHb (including the oval nucleus) are positioned to receive EP input and project primarily to the rostral medial tegmental area (rMTg) (Quina et al., 2014). The rMTg is a primarily GABAergic nucleus that innervates the ventral tegmental area (VTA), is activated by the LHb, and is involved in aversive conditioning (Ji and Shepard, 2007; Stamatakis and Stuber, 2012). Therefore, it is likely that EP inputs to the LHb modulate this circuit and possibly provide sensory, motor, or other environmental related information (Hong and Hikosaka, 2008; Stephenson-Jones et al., 2016). Interestingly, a projection from the ventral tegmental area (VTA) to the LHb also coreleases GABA and glutamate suggesting that the LHb may be a hub of GABA/glutamate corelease in the brain, essential for normal LHb function (Root et al., 2014).
The firing rate of LHb-projecting EP neurons is reduced by rewarding outcomes and increased by aversive outcomes in monkeys (Hong and Hikosaka, 2008) and in mice (Stephenson-Jones et al., 2016). The stimulation of EP input to the LHb is aversive, and may serve to bias future choices based on previous outcomes (Shabel et al., 2012; Stephenson-Jones et al., 2016). However, behavioral changes resulting from phasic modulation of EP to LHb inputs rely on the combined action of Sst+ and Pvalb+/Slc17a6+ EP neurons as both populations are targeted in most previous behavioral experiments (Shabel et al., 2012; Stephenson-Jones et al., 2016). Therefore, we predict that the purely glutamatergic EP→LHb input would be intrinsically aversive, whereas the dual GABA/glutamate input could be aversive or rewarding depending on the ratio of the two transmitters released.
Synaptic plasticity in LHb circuits
The LHb receives input from many regions of the brain and may act as a gateway between forebrain structures and dopaminergic and serotonergic neuromodulatory centers (Hikosaka, 2010). Neurons of the LHb are highly plastic and both pre- and post-synaptic changes have been observed following prolonged exposure to aversive stimuli (Li et al., 2013; Meye et al., 2016; Shabel et al., 2014). Importantly, EP inputs titrate the amount of GABA or glutamate released to modulate firing of postsynaptic LHb neurons (Shabel et al., 2014). Therefore, this synapse can act as either inhibitory or excitatory depending on the ratio of GABA and glutamate it releases or, alternatively, depending on the number of GABA and glutamate receptors in the postsynaptic terminal. The effective GABA/glutamate ratio may be modified by input from a neuromodulatory teaching signal such as serotonin (Shabel et al., 2012, 2014). Whether other neuromodulators, such as dopamine, norepinephrine, or acetylcholine, have similar effects on EP inputs to LHb remains to be investigated.
Functions of BG output nuclei
LHb projecting EP does not fit the classical model of BG function, which stipulates inhibitory effects on downstream structures via the release of GABA (Alexander and Crutcher, 1990; DeLong, 1990). In contrast, Pvalb+/Gad+ neurons of the EP do fit this model as they inhibit VAL and VM thalamus, which project primarily to motor cortical regions. Therefore, the spontaneous activity of Pvalb+/Gad+ EP neurons likely restrains movement tonically and permits movements when transiently inhibited by upstream BG nuclei.
Classic BG models of motor control often describe EP and SN as performing the same function. Our results suggest that these two BG output nuclei are likely to perform different functions with differential effects on both limbic and sensorimotor subcircuits. Neurons of the EP that receive input from striatal patches project to the LHb, whereas patch-recipient zones of the SN (SNc dopamine neurons) are thought to project back to BG nuclei (Gerfen, 1984).
Curiously, LHb projecting EP neurons corelease GABA and glutamate, whereas SNc dopamine neurons are also capable of coreleasing GABA and glutamate along with their canonical transmitter dopamine (Chuhma et al., 2004; Tritsch et al., 2012). However, the mechanisms of GABA handling and release by EP and SNc neurons are fundamentally different as the latter lack classical GABA synthetic enzymes and vesicular transporters (Kim et al., 2015; Tritsch et al., 2012, 2014).
Sensorimotor regions of the EP and SN appear to be more similar in their functions as BG output nuclei. Both target motor thalamic regions and primarily consist of GABergic projection neurons. However, specific target regions of EP and SNr may differ. Pvalb+/Gad+ neurons in EP project to lateral portions of VAL thalamus (Figure 5C), and SNr neurons seem to project mostly to medial VAL, VM thalamus and superior colliculus (Oh et al., 2014). This anatomical heterogeneity may indicate that these two sensorimotor BG output regions sub serve different functions in BG-thalamo-cortical loops.
Sensorimotor and limbic segregation in the BG
The sensorimotor and limbic segregation of the striatum and substantia nigra has been well documented (Gerfen, 1984). dSPNs in the patch and matrix largely segregate their outputs onto dopaminergic or GABAergic SN neurons, respectively (Fujiyama et al., 2011; Watabe-Uchida et al., 2012). Limbic and sensorimotor segregation of the EP and GPe is more enigmatic. In agreement with recent reports, we show that LHb-projecting “limbic” EP receives striatal input from patches, but also receives input from matrix (Stephenson-Jones et al., 2016). In contrast, the sensorimotor EP receives its striatal input almost exclusively from matrix. This may indicate that LHb projecting EP integrates both sensorimotor and limbic input and that the motor thalamic projecting EP is specialized for control of specific sensorimotor tasks. This is supported by electrophysiological studies of LHb-projecting GPi neurons in primates that respond to reward related cues, but also to sensory cues related to the location/direction of a target (Hong and Hikosaka, 2008).
Input from GPe to EP also shows segregation that hints at a sensorimotor and limbic dichotomy. Our studies clearly show LHb projecting EP receives the majority of its external pallidal input from Pvalb-/Nkx2.1+ neurons. It is likely that these neurons are also positive for Lhx6, a distinct population of GPe neurons shown to target the SNc (Mastro et al., 2014). We hypothesize that these Pvalb-/Nkx2.1+ GPe neurons form a “limbic” neuron-type in GPe that may receive input from iSPNs in patches and target “limbic” EP and SNc. Conversely, sensorimotor EP receives GPe input from Pvalb+ prototypic neurons which also project to SNr (Mastro et al., 2014).
Importantly, in the monosynaptic retrograde tracing experiments (Figure 7 and 8), input-output specificity could be compromised because both Pvalb+ neurons that project to LHb and Pvalb+ neurons that project to thalamus are infected with TVA and G and therefore are competent to initiate retrograde spread of RbV-GFP. However, it is unlikely that there is significant cross contamination between these cell-types as retrograde tracing labels significantly different cell populations in striatum and GPe and different anatomical (Figure S8I).
Together our anatomical and physiological studies suggest a model in which phasic activation of patch dSPNs simultaneously inhibits SNc dopaminergic neurons and LHb projecting EP neurons. Decreased activity in EP neurons would decrease activity in the LHb, which sends an excitatory projection to rMTg GABAergic neurons. rMTg neurons would then disinhibit VTA dopaminergic neurons. Therefore, the net result of activity in patch dSPNs may result in a “stop and explore” signal through its combined direct inhibitory actions on SNc and polysynaptic excitatory actions on VTA dopaminergic neurons.
STAR METHODS TEXT
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to, and will be fulfilled by, the Lead Contact Bernardo L. Sabatini (bsabatini@hms.harvard.edu).
Experimental Model and Subject Details
Mice
Sst-Cre (JAX #013044), Pvalb-Cre (JAX #008069), Rbp4-Cre (GENSAT founder line KL100), Adora2a-Cre (A2A-Cre) (GENSAT founder line KG139), Drd1a-Cre (GENSAT founder line EY262), D1-tdTom (JAX #016204), mice bearing a Cre-dependent tdTomato transgene (Ai14; JAX #007908), and mice bearing a Cre-dependent ZsGreen transgene (Ai6; JAX #007906) were maintained on a C57BL/6 background and mice of both sexes (postnatal day 45–150) were used in all experiments. Mice were kept on a 12:12 light/dark cycle under standard housing conditions. All experimental manipulations were performed in accordance with protocols approved by the Harvard Standing Committee on Animal Care following guidelines described in the US NIH Guide for the Care and Use of Laboratory Animals.
Method Details
Virus preparation
Conditional expression of the light-gated non-selective cation channel channelrhodopsin-2 (ChR2, H134R variant) was achieved using a recombinant adeno-associated virus (AAV) encoding a doublefloxed inverted open reading frame (DIO, Cre-ON), a doublefloxed orientation (DF, Cre-OFF), or a doublefloxed orientation with FAS sites (FAS, Cre-OFF) of the ChR2-mCherry/YFP fusion protein under transcriptional control of the EF1α promoter (AAV-DIO-ChR2; http://www.stanford.edu/group/dlab/optogenetics/sequence_info.html#dio) (see Saunders et al., 2012 for full describtion of Cre-OFF constructs). A similar construct was used for Cre-dependent GFP and Synaptophysin-mCherry expression (Garfield et al., 2014). In viral tracing experiments, conditional expression of either the wildtype TVA receptor (TCB) (Miyamichi et al., 2013) or the rabies G protein was achieved using AAV encoding a flipexcision switch (FLEX) (Atasoy et al., 2008) under the control of the CAG promoter. These viral vectors were subsequently packaged (AAV8 or AAV9) by a commercial vector core facility (University of North Carolina, Boston Children’s Hospital Viral Core, or MIT Viral Core). All AAVs were stored in aliquots at a working concentration ~1012–13 genomic copies per ml at −80°C until intracranial injection. EnvA-pseudotyp ed, glycoprotein-deleted rabies virus carrying EGFP transgene (EnvA-RbV-GFP) and non-pseudotyped, glycoprotein-deleted rabies virus carrying tdTomato transgene (RbV-tdTom) were generated in house, using starting materials from Byungkook Lim (UCSD) (Lim et al., 2012). The recombinant rabies viruses were generated using BHK-B19G and BHK-EnvA cells using protocols similar to those previously described (Wickersham et al., 2010), and were used at a titer of approximately 1.0 × 109 infectious units/ml.
Stereotaxic Intracranial Injection
Adult mice were anesthetized with isoflurane and placed in a small animal stereotaxic frame (David Kopf Instruments). After exposing the skull under aseptic conditions, viruses were injected through a pulled glass pipette at a rate of 50 nl/min using a UMP3 microsyringe pump (World Precision Instruments). Injection coordinates from Bregma for EP were −1.1mm A/P, 2.1mm M/L, and 4.2mm D/V, for GPe −0.5mm A/P, 2.1mm M/L, and 4.0mm D/V, for LHb −1.65mm A/P, 0.5mm M/L, and 2.85mm D/V, for striatum 0.6mm A/P, 1.5mm M/L, and 3.0mm D/V, for VAL thalamus −0.55mm A/P, 1.5mm M/L, and 3.6–3.7mm D/V, and for PF −2.2mm A/P, 0.75mm M/L, and 3.25mm D/V. Injection volumes for specific anatomical regions and virus types were as follows EP: 50–100 nL AAV, GPe: 250nL AAV, LHb: 200nL RbV, striatum: 500 nL AAV, VAL thalamus: 250 nL RbV, PF: 200 nL RbV. After surgical procedures, mice were returned to their home cage for >21 days to allow for maximal gene expression. For EnvA-RbV-GFP tracing experiments, EnvA-RbV-GFP was injected 21 days after AAV helper virus injection and the animal was then perfused 7 days after EnvA-RbV-GFP injection in a biosafety level 2 animal facility.
Immunohistochemistry
Mice were deeply anesthetized with isoflurane and perfused transcardially with 4% paraformaldehyde in 0.1 M sodium phosphate buffer. Brains were post-fixed overnight, sunk in 30% (wt/vol) sucrose in phosphate buffered saline (PBS) and sectioned (50 m) coronally (Freezing Microtome, Leica). Free-floating sections were permeabilized/blocked with 5% normal goat/horse serum in PBS with 0.2% Triton X-100 (PBST) for 1 h at room temperature and incubated with primary antibodies at 4°C overni ght and with secondary antibodies for 1 h at room temperature in PBST supplemented with 5% normal goat/horse serum. Brain sections were mounted on superfrost slides, dried and coverslipped with ProLong antifade reagent containing DAPI (Molecular Probes). Primary antibodies used include: rabbit anti-OPRM1 (1:750; AB5511, Millipore), rabbit/goat anti-Pvalb (1:2000, Swant Pv 27/PVG 213), rat anti-SST (1:500, Millipore MAB354), rabbit anti-Nkx2.1 (1:500, Santa Cruz sc-13040), and rabbit anti-FoxP2 (1:1000, Sigma HPA000382). Alexa Fluor 594-, 488- and 647-conjugated secondary antibodies to rabbit, goat, and rat (Invitrogen) were diluted 1:500. tdTomato, mCherry, and GFP fluorescence were not immuno-enhanced. Whole sections were imaged with an Olympus VS120 slide scanning microscope. For colocalization analysis, high resolution images of regions of interest were acquired with an Olympus FV1200 confocal microscope using a 20X 0.75 NA air, or 60X 1.42 NA oil immersion objective (Neural Imaging Center, HMS). Individual imaging planes were overlaid and quantified for colocalization in ImageJ (NIH). Occasionally, images were linearly adjusted for brightness and contrast using ImageJ software. All images to be quantitatively compared underwent identical manipulations.
Fluorescence In-Situ Hybridization (FISH)
Mice were deeply anesthetized with isoflurane, decapitated, and their brains were quickly removed and frozen in tissue freezing medium on dry ice. Brains were cut on a cryostat (Leica CM 1950) into 30 μm sections, adhered to SuperFrost Plus slides (VWR), and immediately refrozen. Samples were fixed 4% paraformaldehyde and processed according to ACD RNAscope Fluorescent Multiplex Assay manual. Sections were incubated at room temperature for 30 seconds with DAPI, excess liquid was removed, and immediately coverslipped with ProLong antifade reagent (Molecular Probes). Antisense probes for RbV-N, Lypd1, Tbr1, Sst, Pvalb, Slc32a1, Slc17a6, Gad1, and Gad2 were purchased from Advanced Cell Diagonstics (ACD, http://acdbio.com/). Sections were imaged at 1024 × 1024 pixels on an Olympus FV1200 confocal microscope using a 20×, 0.75 NA air or a 60×, 1.42 NA oil immersion objective (Neural Imaging Center, HMS). Individual imaging planes were overlaid and quantified for colocalization in ImageJ (NIH) and Matlab (Mathworks).
Human Tissue
Coronal sections of fresh frozen, 14 m thick, postmortem (20 yr-old), human brain tissue was acquired from The Stanley Medical Research Institute Brain Collection (Torrey et al., 2000). Tissue sections included GPi, GPe, and striatum and were processed identically to mouse tissue except the tissue was incubated in protease for 45 mins instead of 30 mins prior to probe hybridization. Antisense probes for human SLC32A1, SLC17A6, SST, and PVALB were purchased from Advanced Cell Diagonstics (ACD, http://acdbio.com/).
Image Analysis
Quantification of colocalization was performed in ImageJ (NIH) and analysis was performed unblinded to genotype. Occasionally, images were linearly adjusted for brightness and contrast using ImageJ software. All images to be quantitatively compared underwent identical manipulations. In EnvA-RbV-GFP retrograde labeling studies (Figure 7) the patch compartment was defined by intense immunofluorescent labeling for MOR. ROI’s where drawn around areas of dorsal striatum with intense OR labeling and defined as patches. GFP+ cells were then counted automatically and assigned to either the patch or matrix compartment based upon previously drawn ROIs with a custom made macro in ImageJ.
FISH images were analyzed for “fluorescence coverage (%),” meaning the proportion of fluorescent pixels to total pixels in a cellular ROI, using a custom macro in Image J and custom scripts in Matlab (Figure 3, 5 and S3). 5–10 images from at least 3 mice were analyzed for each condition. Cell ROIs were automatically determined based on fluorescence signals in all three channels, and manually adjusted prior to analysis to ensure that all cell ROIs reflected individual cells and not clusters. After background subtraction (the signal outside of cell ROIs) and application of a fluorescence threshold (Renyi Entropy), the amount of fluorescent pixels in each optical channel were counted within the cellular ROI. All images compared underwent identical thresholding and no other manipulations were made. These data were used to generate X-Y plots displaying the percent coverage for each channel per cell.
Drop-seq methods/statistical analysis
Cell suspensions were prepared from acute brain slabs with microdissection of the EP, STN and surrounding regions followed by Papain (Worthington)/Proteinase XXIII(Sigma) digestion, physical titration, pelleting and debris-filtering using male C57BL/6 mice (P60–70, Charles River). In each of three replicates, slices from n=3–4 mice were processed in quick succession. Cell concentrations used to estimate STAMP numbers were ascertained with a haemocytometer (Propper). The cell suspension was allowed to warm to room temperature and processed for Drop-seq without further dilution ((Macosko et al., 2015); http://mccarrolllab.com/dropseq/). STAMP reverse transcription, library amplification, and Nextera tagmentation were performed as described previously (Macosko et al., 2015). The resulting n=3 libraries were sequenced using 75 cycle kits on the NextSeq500 (Illumina). Libraries from replicates 2 and 3 were consolidated into a single sequencing run. Each sequencing run contained between 7 – 10K cells and resulted in 199.6 – 302.4M high-quality (Q>30) mapped reads (mouse genome draft GRCm38.81). To generate the digital gene expression (DGE) matrix of transcript counts for each cell, reads were computationally grouped by cell and unique molecular identifier using the bead-based barcodes (Macosko et al., 2015). For clustering, only cells with >400 genes were retained, resulting in a DGE with a total of 16,484 cells and 19,945 genes. The DGE was then scaled to 10,000 transcripts per cell, log-transformed, and clustered following the Seurat work-flow (Satija et al., 2015) as described previously (Macosko et al., 2015), except that two rounds of clustering were performed. In the first round, neurons were isolated from other cell types by performing principal components analysis on 827 variable genes (selected by mean-variance analysis), and using the first five principal components (PCs) as input into tSNE. Density-based clustering of this graph, using a reachability distance parameter (eps) of 2, yielded a single neuronal cluster (as judged by expression of the canonical marker Rbfox3), composed of 3,230 cells. The 1,615 neuronal profiles from this cluster with the largest number of transcripts were subjected to a second round of principal components analysis using 1,167 variable genes, and the first 20 PCs used to generate the tSNE (Figure 1D). The graph was clustered using an eps of 2.2 and clusters with fewer than 20 cells removed, yielding a total of 10 populations. To determine which clusters were intrinsic to the EP (rather than derived from neighboring co-dissected nuclei), we screened marker genes from the 10 populations using the Allen Institute In Situ Hybridization Atlas (http://mouse.brain-map.org/). For genes included in Figure 2, we set a conservative threshold of 2.7-fold expression change; the ~60 genes with significantly different (p<5.0 × 10−5) gene expression between the two EP-intrinsic clusters (numbered 5 and 6) are included in Table S1. Expression levels for all genes and all clusters can be found in Table S1. Normalized data and analysis code can be found at Harvard Dataverse (http://dx.doi.org/10.7910/DVN/YPBBA1), raw data can be found at NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) accession number: GSE95133.
Acute brain slice preparation
Brain slices were obtained from 50–150 day old mice (both male and female) using standard techniques. Mice were anesthetized by isoflurane inhalation and perfused transcardially with ice-cold artificial cerebrospinal fluid (ACSF) containing (in mM) 125 NaCl, 2.5 KCl, 25 NaHCO3, 2 CaCl2, 1 MgCl2, 1.25 NaH2PO4 and 25 glucose (295 mOsm/kg). Cerebral hemispheres were removed, blocked and transferred into a slicing chamber containing ice-cold ACSF. Coronal slices of LHb or thalamus (250 μm thick) or sagittal slices of EP (200 μm thick) were cut with a Leica VT1000s vibratome in ice-cold ACSF, transferred for 10 min to a holding chamber containing choline-based solution (consisting of (in mM): 110 choline chloride, 25 NaHCO3, 2.5 KCl, 7 MgCl2, 0.5 CaCl2, 1.25 NaH2PO4, 25 glucose, 11.6 ascorbic acid, and 3.1 pyruvic acid) at 34°C then transferred to a secondary holding cha mber containing ACSF at 34°C for 10 mins and subsequently maintained at room temperature (20–22°C) until use. All recordings were obtained within 4 hours of slicing. Both choline solution and ACSF were constantly bubbled with 95% O2/5% CO2.
Electrophysiology
Individual slices were transferred to a recording chamber mounted on an upright customized 2-photon microscope and continuously superfused (4 ml/min) with room temperature ACSF. Cells were visualized through a 60X water immersion objective with infrared differential interference and epifluorescence to identify fluorescent neurons and regions displaying the highest density of ChR2+ axons. Epifluorescence was attenuated and used sparingly to minimize ChR2 activation prior to recording. Patch pipettes (2–4 MΩ) pulled from borosilicate glass (Sutter Instruments) were filled either with internal solution containing (in mM) 135 CsMeSO3, 10 HEPES, 1 EGTA, 3.3 QX-314 (Cl− salt), 4 Mg-ATP, 0.3 Na-GTP, 8 Na2-Phosphocreatine (pH 7.3 adjusted with CsOH; 295 mOsm/kg) for voltage-clamp recordings, or with an internal solution composed of (in mM) 135 KMeSO3, 3 KCl, 10 HEPES, 1 EGTA, 0.1 CaCl2, 4 Mg-ATP, 0.3 Na-GTP, 8 Na2-Phosphocreatine (pH 7.3 adjusted with KOH; 295 mOsm/kg) for current-clamp recordings. Membrane currents were amplified and low-pass filtered at 3 kHz using a Multiclamp 700B amplifier (Molecular Devices, Sunnyvale, CA), digitized at 10 kHz and acquired using National Instruments acquisition boards and a custom version of ScanImage (Pologruto et al., 2003) (available upon request or from https://github.com/bernardosabatinilab/SabalabSoftware_Nov2009) written in MATLAB (Mathworks, Natick, MA). Electrophysiology data were analyzed offline using Igor Pro (Wavemetrics, Portland, OR). Bath solutions for pharmacological isolation of excitatory and inhibitory currents in whole-cell voltage clamp recordings contained 2,3-dihydroxy-6-nitro-7-sulfamoyl-benzo(f)quinoxaline (NBQX; 10 μM), R,S-3-(2-carboxypiperazin-4-yl)propyl-1-phosphonic acid (CPP; 10 μM), SR95531 (gabazine) (10 μM), tetrodotoxin (TTX, 1 μM) and 4-Aminopyridine (4-AP, 0.5 mM). The approximate location of the recorded neuron was confirmed after termination of the recording using a 4× objective to visualize the pipette tip, while referencing an anatomical atlas (Figure S7) (Allen Institute Reference Atlas). For pharmacological analyses in Figure 6, 7, and 8, the peak amplitude of PSCs measured 4–5 min following the onset of drug perfusion were averaged, normalized to baseline averages obtained immediately prior to drug application. To activate ChR2-expressing cells and axons, light from a 473 nm laser (Optoengine) was focused on the back aperture of the microscope objective to produce wide-field illumination of the recorded cell. For voltage clamp experiments, brief pulses of light (1 ms duration; 2–10 mW·mm−2 under the objective) were delivered at the recording site at 20 s intervals under control of the acquisition software.
For excitability experiments described in Figure 4, neurons were hyperpolarized in current-clamp to −60 mV to reduce spontaneous firing during subsequent square wave current injections. Sag potential was measured by injecting a −100 pA square wave current for 500 ms and taking the difference of the peak of the negative-going membrane potential and mean voltage of the last 50 ms of the −100 pA step. The full-width at half-height (FWHH) of an action potential (AP) was calculated by finding the midpoint between the peak of the AP, and the initiation point of the AP (defined by the first peak of the 2nd derivative), and measuring the width of the AP at that membrane potential. After-hyperpolarization (AHP) was calculated by taking the difference of the resting membrane potential and the negative going peak of the falling phase of the AP. Max rate of rise and fall of the AP were determined by the 1st derivative of the AP. We measured the membrane time constant (Tm) in voltage clamp by fitting a double-exponential to the decay phase (peak + 10 ms) of the membrane capacitive transients evoked by −5 mV step in the holding potential. Neuronal capacitance (Cm) was calculated using Cm=Tm/Rseries. All drugs were purchased from Tocris Bioscience (https://www.tocris.com/) and bath applied. For all experiments, errors due to the voltage drop across the series resistance (< 25MΩ, changed <10% over the duration of the recording) and the liquid junction potential were left uncompensated.
Quantification and Statistical Analysis
Graphs represent mean values ± SEM. We represent p-values in all figures as *p≤0.05, **p≤0.01, and ***p≤0.001. We used the D’Agostino & Pearson omnibus normality test, and unpaired student’s t-test for significance (Figure 4), or one-way or two-way ANOVA with Tukey’s posthoc to test for significance (Figure 4, 7 and 8). We performed all statistical analyses using GraphPad Prism 6 software (GraphPad Software Inc., La Jolla, CA).
Data and Software Availability
Normalized data and analysis code can be found at Harvard Dataverse (http://dx.doi.org/10.7910/DVN/YPBBA1). The raw data have been deposited in the NCBI Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/), under accession number : GSE95133.
Supplementary Material
Highlights.
Drop-Seq and FISH define 3 EP/GPi neuron subclasses in mice and humans.
EP neuron subtypes have distinct targets and release different neurotransmitters.
EP neuron subtypes segregate into limbic and sensorimotor subcircuits.
Acknowledgments
The authors thank Terence Lee, Andrea Zanello, and Karina Bistrong for excellent technical assistance; Sarah Melzer and Adam Granger for assistance with FISH analysis; Lai Ding for writing the Image J macro used for RabV analysis; James Levasseur for animal husbandry and genotyping and the members of the Sabatini Lab for their helpful discussions and advice. Starting materials for generating pseudotyped rabies virus is a generous gift from B.K. Lim (UCSD). Human tissue samples were a generous gift from The Stanley Medical Research Institute Brain Collection. Confocal images were acquired at the Harvard Neurobiology Imaging Facility and Harvard Neurodiscovery Imaging Core. This work was supported by the NIH, National Institute of Neurological Disease and Stroke (NS007484-15 to M.L.W and NS046579-13 to B.L.S.), the Stanley Center for Psychiatric Research at the Broad Institute (S.A.M.) and the Helen Hay Whitney Foundation (A.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
M.L.W. conceived the study, designed and performed experiments, analyzed data and wrote the paper, A.S., A.P., E.Z.M., and M.G. designed and performed experiments, analyzed data and wrote the paper, K.W.H provided reagents and wrote the paper, S.A.M. wrote the paper, and B.L.S. conceived the study, designed experiments, analyzed data and wrote the paper.
References
- Abdi A, Mallet N, Mohamed FY, Sharott A, Dodson PD, Nakamura KC, Suri S, Avery SV, Larvin JT, Garas FN, et al. Prototypic and arkypallidal neurons in the dopamine-intact external globus pallidus. J Neurosci. 2015;35:6667–6688. doi: 10.1523/JNEUROSCI.4662-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ade KK, Wan Y, Chen M, Gloss B, Calakos N. An Improved BAC Transgenic Fluorescent Reporter Line for Sensitive and Specific Identification of Striatonigral Medium Spiny Neurons. Front Syst Neurosci. 2011;5:32. doi: 10.3389/fnsys.2011.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander GE, Crutcher MD. Functional architecture of basal ganglia circuits: neural substrates of parallel processing. Trends Neurosci. 1990;13:266–271. doi: 10.1016/0166-2236(90)90107-l. [DOI] [PubMed] [Google Scholar]
- Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX Switch Targets Channelrhodopsin-2 to Multiple Cell Types for Imaging and Long-Range Circuit Mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-Chinea P, Rico AJ, Pérez-Manso M, Roda E, López IP, Luis-Ravelo D, Lanciego JL. Glutamatergic pallidothalamic projections and their implications in the pathophysiology of Parkinson’s disease. Neurobiol Dis. 2008;31:422–432. doi: 10.1016/j.nbd.2008.05.019. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Chuhma N, Zhang H, Masson J, Zhuang X, Sulzer D, Hen R, Rayport S. Dopamine neurons mediate a fast excitatory signal via their glutamatergic synapses. J Neurosci. 2004;24:972–981. doi: 10.1523/JNEUROSCI.4317-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong MR. Primate models of movement disorders of basal ganglia origin. Trends Neurosci. 1990;13:281–285. doi: 10.1016/0166-2236(90)90110-v. [DOI] [PubMed] [Google Scholar]
- Fujiyama F, Sohn J, Nakano T, Furuta T, Nakamura KC, Matsuda W, Kaneko T. Exclusive and common targets of neostriatofugal projections of rat striosome neurons: a single neuron-tracing study using a viral vector. Eur J Neurosci. 2011;33:668–677. doi: 10.1111/j.1460-9568.2010.07564.x. [DOI] [PubMed] [Google Scholar]
- Garfield AS, Shah BP, Madara JC, Burke LK, Patterson CM, Flak J, Neve RL, Evans ML, Lowell BB, Myers MG, et al. A Parabrachial-Hypothalamic Cholecystokinin Neurocircuit Controls Counterregulatory Responses to Hypoglycemia. Cell Metab. 2014;20:1030–1037. doi: 10.1016/j.cmet.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Andres KH, Veh RW. Morphologic and cytochemical criteria for the identification and delineation of individual subnuclei within the lateral habenular complex of the rat. J Comp Neurol. 2003;458:78–97. doi: 10.1002/cne.10566. [DOI] [PubMed] [Google Scholar]
- Gerfen CR. The neostriatal mosaic: compartmentalization of corticostriatal input and striatonigral output systems. Nature. 1984;311:461–464. doi: 10.1038/311461a0. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, Heintz N. GENSAT BAC Cre-Recombinase Driver Lines to Study the Functional Organization of Cerebral Cortical and Basal Ganglia Circuits. Neuron. 2013;80:1368–1383. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillner S, Robertson B. The Basal Ganglia Over 500 Million Years. Curr Biol. 2016;26:R1088–R1100. doi: 10.1016/j.cub.2016.06.041. [DOI] [PubMed] [Google Scholar]
- Hikosaka O. The habenula: from stress evasion to value-based decision-making. Nat Rev Neurosci. 2010;11:503–513. doi: 10.1038/nrn2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Hikosaka O. The globus pallidus sends reward-related signals to the lateral habenula. Neuron. 2008;60:720–729. doi: 10.1016/j.neuron.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- Ji H, Shepard PD. Lateral habenula stimulation inhibits rat midbrain dopamine neurons through a GABA(A) receptor-mediated mechanism. J Neurosci. 2007;27:6923–6930. doi: 10.1523/JNEUROSCI.0958-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kha HT, Finkelstein DI, Pow DV, Lawrence AJ, Horne MK. Study of projections from the entopeduncular nucleus to the thalamus of the rat. J Comp Neurol. 2000;426:366–377. [PubMed] [Google Scholar]
- Kim JI, Ganesan S, Luo SX, Wu YW, Park E, Huang EJ, Chen L, Ding JB. Aldehyde dehydrogenase 1a1 mediates a GABA synthesis pathway in midbrain dopaminergic neurons. Science. 2015;350:102–106. doi: 10.1126/science.aac4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmac CI, Mitrofanis J. Organisation of the reticular thalamic projection to the intralaminar and midline nuclei in rats. J Comp Neurol. 1997;377:165–178. doi: 10.1002/(sici)1096-9861(19970113)377:2<165::aid-cne2>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lim BK, Huang KW, Grueter BA, Rothwell PE, Malenka RC. Anhedonia requires MC4R-mediated synaptic adaptations in nucleus accumbens. Nature. 2012;487:183–189. doi: 10.1038/nature11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Zhou T, Liao L, Yang Z, Wong C, Henn F, Malinow R, Yates JR, Hu H. βCaMKII in lateral habenula mediates core symptoms of depression. Science. 2013;341:1016–1020. doi: 10.1126/science.1240729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Maaten L, Hinton G. Visualizing Data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastro KJ, Bouchard RS, Holt HAK, Gittis AH. Transgenic mouse lines subdivide external segment of the globus pallidus (GPe) neurons and reveal distinct GPe output pathways. J Neurosci. 2014;34:2087–2099. doi: 10.1523/JNEUROSCI.4646-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, Roederer M, Gottardo R. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics. 2013;29:461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meye FJ, Soiza-Reilly M, Smit T, Diana MA, Schwarz MK, Mameli M. Shifted pallidal co-release of GABA and glutamate in habenula drives cocaine withdrawal and relapse. Nat Neurosci. 2016;19:1019–1024. doi: 10.1038/nn.4334. [DOI] [PubMed] [Google Scholar]
- Miyamichi K, Shlomai-Fuchs Y, Shu M, Weissbourd BC, Luo L, Mizrahi A. Dissecting Local Circuits: Parvalbumin Interneurons Underlie Broad Feedback Control of Olfactory Bulb Output. Neuron. 2013;80:1232–1245. doi: 10.1016/j.neuron.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Fukuda T. Immunohistochemical study on the neuronal diversity and three-dimensional organization of the mouse entopeduncular nucleus. Neurosci Res. 2015 doi: 10.1016/j.neures.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Nelson AB, Kreitzer AC. Reassessing models of Basal Ganglia function and dysfunction. Annu Rev Neurosci. 2014;37:117–135. doi: 10.1146/annurev-neuro-071013-013916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent A, De Bellefeuille L. Organization of efferent projections from the internal segment of globus pallidus in primate as revealed by fluorescence retrograde labeling method. Brain Res. 1982;245:201–213. doi: 10.1016/0006-8993(82)90802-2. [DOI] [PubMed] [Google Scholar]
- Parent M, Lévesque M, Parent A. Two types of projection neurons in the internal pallidum of primates: single-axon tracing and three-dimensional reconstruction. J Comp Neurol. 2001;439:162–175. doi: 10.1002/cne.1340. [DOI] [PubMed] [Google Scholar]
- Penney JB, Young AB. GABA as the pallidothalamic neurotransmitter: implications for basal ganglia function. Brain Res. 1981;207:195–199. doi: 10.1016/0006-8993(81)90693-4. [DOI] [PubMed] [Google Scholar]
- Pert CB, Kuhar MJ, Snyder SH. Opiate receptor: autoradiographic localization in rat brain. Proc Natl Acad Sci. 1976;73:3729–3733. doi: 10.1073/pnas.73.10.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L, Mao T, Sternson SM, Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pologruto TA, Sabatini BL, Svoboda K. ScanImage: flexible software for operating laser scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quina La, Tempest L, Ng L, Harris J, Ferguson S, Jhou T, Turner EE. Efferent pathways of the mouse lateral habenula. J Comp Neurol. 2014;00 doi: 10.1002/cne.23662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale CW, Graybiel AM. The fronto-striatal projection in the cat and monkey and its relationship to inhomogeneities established by acetylcholinesterase histochemistry. Brain Res. 1981;208:259–266. doi: 10.1016/0006-8993(81)90556-4. [DOI] [PubMed] [Google Scholar]
- Rajakumar N, Elisevich K, Flumerfelt BA. Compartmental origin of the striatoento-peduncular projection in the rat. J Comp Neurol. 1993;331:286–296. doi: 10.1002/cne.903310210. [DOI] [PubMed] [Google Scholar]
- Rajakumar N, Elisevich K, Flumerfelt BA. Parvalbumin-containing GABAergic neurons in the basal ganglia output system of the rat. J Comp Neurol. 1994;350:324–336. doi: 10.1002/cne.903500214. [DOI] [PubMed] [Google Scholar]
- Robinson RB, Siegelbaum SA. Hyperpolarization-activated cation currents: from molecules to physiological function. Annu Rev Physiol. 2003;65:453–480. doi: 10.1146/annurev.physiol.65.092101.142734. [DOI] [PubMed] [Google Scholar]
- Root DH, Mejias-aponte Ca, Zhang S, Wang H, Hoffman AF, Lupica CR, Morales M. Single rodent mesohabenular axons release glutamate and GABA. Nat Neurosci. 2014;17 doi: 10.1038/nn.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Johnson CA, Sabatini BL. Novel recombinant adeno-associated viruses for Cre activated and inactivated transgene expression in neurons. Front Neural Circuits. 2012;6:47. doi: 10.3389/fncir.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Oldenburg IA, Berezovskii VK, Johnson CA, Kingery ND, Elliott HL, Xie T, Gerfen CR, Sabatini BL. A direct GABAergic output from the basal ganglia to frontal cortex. Nature. 2015 doi: 10.1038/nature14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Huang KW, Sabatini BL. Globus Pallidus Externus Neurons Expressing parvalbumin Interconnect the Subthalamic Nucleus and Striatal Interneurons. PLoS One. 2016;11 doi: 10.1371/journal.pone.0149798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabel SJ, Proulx CD, Trias A, Murphy RT, Malinow R. Input to the lateral habenula from the basal ganglia is excitatory, aversive, and suppressed by serotonin. Neuron. 2012;74:475–481. doi: 10.1016/j.neuron.2012.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabel SJ, Proulx CD, Piriz J, Malinow R. GABA/glutamate co-release controls habenula output and is modified by antidepressant treatment. Science (80-) 2014;345:1494–1498. doi: 10.1126/science.1250469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis AM, Stuber GD. Activation of lateral habenula inputs to the ventral midbrain promotes behavioral avoidance. Nat Neurosci. 2012;15:1105–1107. doi: 10.1038/nn.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson-Jones M, Yu K, Ahrens S, Tucciarone JM, van Huijstee AN, Mejia LA, Penzo MA, Tai LH, Wilbrecht L, Li B. A basal ganglia circuit for evaluating action outcomes. Nature. 2016;539:289–293. doi: 10.1038/nature19845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada M, Tokuno H, Ikai Y, Mizuno N. Direct projections from the entopeduncular nucleus to the lower brainstem in the rat. J Comp Neurol. 1994;342:409–429. doi: 10.1002/cne.903420308. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The Stanley Foundation brain collection and Neuropathology Consortium. Schizophr Res. 2000;44:151–155. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
- Tritsch NX, Ding JB, Sabatini BL. Dopaminergic neurons inhibit striatal output through non-canonical release of GABA. Nature. 2012;490:262–266. doi: 10.1038/nature11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritsch NX, Oh WJ, Gu C, Sabatini BL. Midbrain dopamine neurons sustain inhibitory transmission using plasma membrane uptake of GABA, not synthesis. Elife. 2014;3:e01936. doi: 10.7554/eLife.01936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SR, Brown JC. Somatostatin immunoreactivity in the entopeduncular projection to the lateral habenula in the rat. Neurosci Lett. 1986;68:160–164. doi: 10.1016/0304-3940(86)90134-5. [DOI] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzelmann KK, Young JAT, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Sullivan HA, Seung HS. Production of glycoprotein-deleted rabies viruses for monosynaptic tracing and high-level gene expression in neurons. Nat Protoc. 2010;5:595–606. doi: 10.1038/nprot.2009.248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


