Abstract
One primary agenda of the developmental evolution field is to elucidate molecular mechanisms governing differences in animal form. While mounting evidence has established an important role for mutations in transcription controlling cis-regulatory elements (CREs), the underlying mechanisms that translate these alterations into differences in gene expression are poorly understood. Emerging studies focused on pigmentation differences among closely related Drosophila species have provided many examples of phenotypically relevant CRE changes, and have begun to illuminate how this process works at the level of regulatory sequence function and transcription factor binding. We review recent work in this field and highlight the conceptual and technical challenges that currently await experimental attention.
Introduction
Many insights into the molecular events that drive the appearance of complex organismal traits have come from integrating developmental genetics into the study of evolution [1]. Multicellular animals and their distinguishing features are built during the course of development, and by examining developmental processes, one can determine how such mechanisms change. A key driver of development is differential gene activity: the specification of which subset of the genome is activated, or expressed at each step of the developmental sequence. Work in the developmental evolution field over the last several years has established an important role for transcriptional regulatory DNA mutations in altering gene expression patterns and thereby phenotypic traits [2–8]. As the molecular details of cis-regulatory mutations emerge, we can compose a deeper mechanistic understanding of complex evolutionary problems such as trait origination, coordination, pleiotropy, and phenotypic plasticity. Here we review how recent work on fruit fly pigmentation has contributed to the current view of developmental evolution, and highlight the challenges and rewards for addressing these systems in more sophisticated mechanistic detail.
The Drosophila pigmentation model
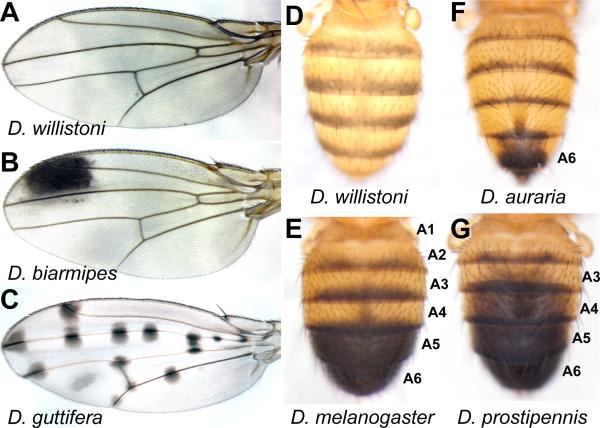
The pigmentation patterns decorating the bodies of fruit flies are remarkably diverse [9–12]. These patterns have been associated with various aspects of organismal fitness, such as resistance to desiccation [13,14], ultra violet radiation protection [15,16], and courtship success [17,18]. The genetic composition for three pigmentation traits in particular has received considerable experimental attention (Figure 1). In the abdomen of D. melanogaster, male-specific pigmentation on the entire A5 and A6 tergites evolved from an ancestor that displayed only thin stripes of pigment along each tergite. The male-specific spot on the antero-distal region of the D. biarmipes wing, and the spotted pattern adorning the wings of D. guttifera both evolved from ancestors that produced uniformly unpigmented wings. Several of the genes that must be specifically expressed to generate these pigmentation patterns are now known, and many details of their transcriptional regulation have been recently characterized.
Figure 1. Fruit fly pigmentation traits that serve as models for gene regulatory evolution.
(A) The wings of D. willistoni are unpigmented. (B) D. biarmipes displays an antero-distal black pigmentation spot. (C) The wings of D. guttifera flies are adorned with numerous spots including those positioned where multiple veins connect, and at the positions of campaniform sensilla. (D) Among Drosophila species, the dorsal surfaces of abdominal segments 1 through 6 are covered by cuticle plates known as tergites that are typically only partially pigmented, as shown for D. willistoni. (E) In D. melanogaster males, the A5 and A6 segment tergites are fully pigmented. Male-specific pigmentation has (F) contracted to the male A6 segment of D. auraria and (G) expanded to include the male A3-A6 segments of D. prostipennis.
Each pigmentation feature is associated with the patterned expression of enzymes that produce melanin pigments. The gene yellow is expressed in zones where dark melanic pigmentation occurs on the cuticle [19–21]. While several hypotheses exist about the molecular function of yellow [22], its expression appears to be a general requirement for black melanin formation. For the three traits considered here (Figure 1), there also exists a reciprocal exclusion of ebony gene expression [8,19,23]. ebony encodes an enzyme that converts Dopamine into N-ß-alanyl dopamine (NBAD), which generates yellow-colored cuticle [24]. While other pigmentation enzymes contribute to cuticle coloration, only ebony and yellow have thus far been associated with strong expression patterns that predict the wing phenotypes. For male-specific abdomen pigmentation, an additional gene, tan, is expressed in a pattern which closely matches that of yellow [21]. tan encodes an enzyme with NBAD hydrolase activity which converts NBAD into Dopamine, promoting the formation of darker cuticle pigments [25].
Many of the upstream regulators responsible for the patterns of yellow, tan and ebony expression have been identified. In the abdomen, the Hox gene Abd-B is expressed in the epidermis underlying the pigmented A5 and A6 tergites and is required for the activation of yellow and tan [21,26]. The expression of yellow and tan in the unpigmented female abdomen is prevented by the Bab1 and Bab2 transcriptional repressors [27]. In the wing of D. biarmipes, the transcription factor Distalless (Dll) has been identified as a major input mediating activation and mutual exclusion of yellow and ebony respectively [28]. The wing spots of D. guttifera were associated with the specific expression of the wingless (wg) morphogen gene. RNAi screens of the majority of transcription factors in the D. melanogaster genome have revealed dozens of additional factors required for pigmentation phenotype and/or gene activity in the wing and abdomen [28,29] whose direct connections to the regulatory regions of pigmentation genes await elucidation.
Regulatory control of the Drosophila pigmentation genes
The expression of each pigmentation gene during development is governed by at least one cis-regulatory element (CRE) sequence that controls its timing and spatial extent of transcription. CREs often extend over a hundred base pairs in length and include short 5-20 base pair motifs that function as binding sites for a specific set of transcription factors that form their regulatory logic. The main pigmentation gene CREs have been defined, and a portion of their regulatory logic has been elucidated. For the tan and yellow genes, single CREs known as the tan Male Specific Element or “t_MSE” [21,30] and the yellow Body Element or “yBE” [7,26] drive their male-limited abdominal patterns respectively. ebony on the other hand requires the activity of an activating enhancer and two repressive silencer elements to produce a reciprocal pattern of expression in the D. melanogaster abdomen [23]. For wing spots, yellow is the only gene for which CREs have been characterized. In D. biarmipes, an antero-distal spot of expression in the pupal wing is driven by a CRE known as the spot element [8]. In D. guttifera, the 16 spots of expression are driven by a CRE known as “vein spot” [20]. The identification of these CRE architectures has facilitated mechanistic studies of several complex evolutionary processes, such as the origination, coordination, and constraint of their developmental programs.
Elucidating the origins of pigmentation traits and the CREs that drive them
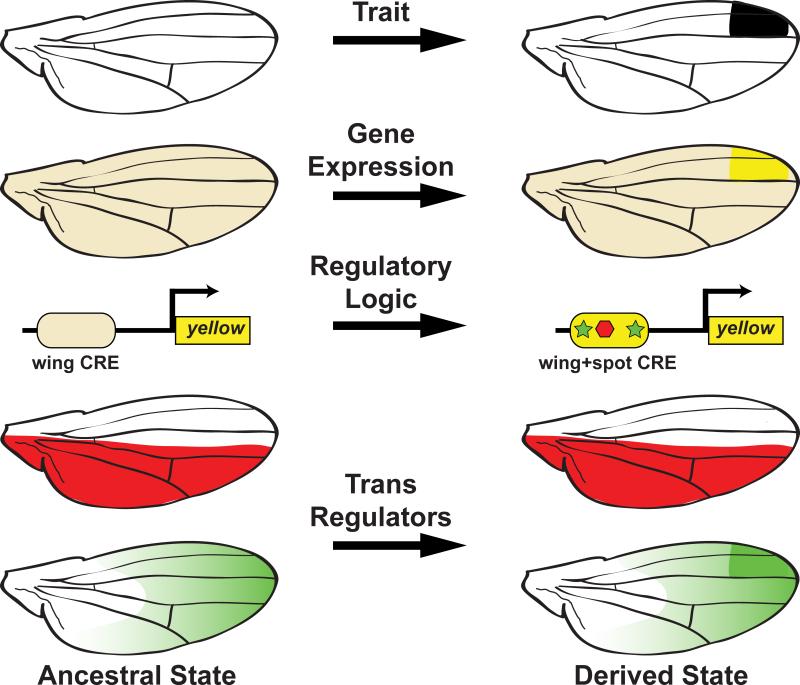
One way to study the origin of a trait is to examine the evolutionary history of the CREs that drive patterned expression of key trait-building genes. For example, in a seminal study of the wing spot of D. biarmipes, it was found that the yellow spot element evolved upon a CRE that ancestrally functioned to direct a low level of expression across the entire wing epidermis [8]. The origin of the spot element occurred through the co-option of Dll activation and En repression which required the evolution of binding sites for these two transcription factors in the spot CRE (Figure 2) [28]. Regarding the 16 pigmentation spots in D. guttifera, the origin of the vein spot CRE appears to have involved the evolution of connections to the Wingless signaling pathway through binding inputs that still await identification [20]. Recent dissection of CREs in the wg locus has revealed that its expression in four vein tips evolved in the locality of a conserved enhancer active in the crossveins of diverse species, including D. melanogaster [31]. In contrast, a CRE that drives five spot-associated foci of wg expression in cells of the campaniform sensilla evolved in the intron of the nearby Wnt10 gene, where no CRE activity has yet been characterized. These findings demonstrate how new CREs are associated with new expression patterns and traits, and may often originate through the co-option of pre-existing activities.
Figure 2. The origin of wing spots in D. biarmipes involved changes in cis and trans to the pigmentation-promoting gene yellow.
A novel pattern of yellow gene expression accompanied the evolution of the D. biarmipes wing spot. The evolutionary path to this expression change involved converting a pre-existing wing CRE into the wing+spot CRE by evolving sites for Dll (green) as an activator and En (red) as a repressor. Along this evolutionary path, the Dll activator evolved a strong spot-prefiguring pattern of expression.
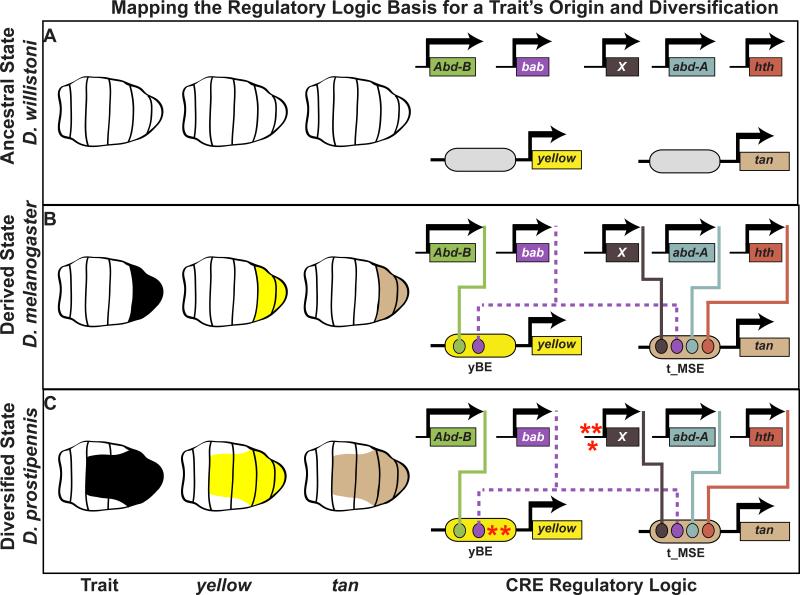
The full tergite pigmentation of the D. melanogaster male posterior abdomen (Figure 1E) evolved from an ancestor that lacked this trait, as is observed in current day D. willistoni (Figure 1D) [26]. Tests of the yBE and t_MSE from D. willistoni revealed an inability to drive expression in the abdomen [21]. This supports a model whereby the origin of this trait required the evolution of these CREs. Within the yBE and t_MSE, a regulatory logic evolved that at a minimum involved direct binding of Abd-B to the yBE, and connected Abd-A and Hth to the t_MSE. Furthermore, additional inputs for sex-specific expression must have formed at both CREs (Figure 3). At present, these studies cannot resolve whether these evolutionary transitions occurred through the de novo evolution of a functional CRE or by the repurposing of an ancestral CRE, as was the case for the spot and vein spot/tip CREs. In the case of yellow, this CRE evolved next to a CRE that produces a thin stripe of expression at the posterior edges of tergites, raising the possibility that this activity may have been co-opted [21], but this model warrants further examination. Collectively, each of these cases of trait origination was accompanied by the insertion of one or more transcription factors into the regulatory logic of a differentiation gene CRE. However, the coordinated evolution of co-expressed CRE activities can arise by evolving very different regulatory inputs.
Figure 3. Mapping the origins and coordinated evolution of the Drosophila abdominal pigmentation network.
(A) In the lineage of D. melanogaster, the ancestral state was inferred to have lacked male-specific tergite pigmentation in part because the yellow and tan genes were not expressed in the underlying male epidermis. The absence of yellow and tan expression corresponds with an absence of abdomen-specific CREs. (B) The derived male-specific tergite pigmentation originated through the gain of yellow and tan expression in the abdomen, driven by CREs containing binding sites for body plan-patterning transcription factors. (C) The expanded pigmentation observed on the abdomen of D. prostipennis males required coordinated expansion of yellow and tan gene expression. The maintenance of their co-expressed relationship during evolution involved modifications of the yBE, in the case of yellow, and alterations to an unknown trans-regulator(s), that altered the output of the t_MSE and thereby expression of tan. Red asterisks indicate the presence of function-modifying mutations.
The evolution of coordinately regulated expression programs
An impressive degree of orchestration underlies the hundreds of gene expression decisions that mark the progression of development. Pigmentation traits provide excellent models to address how the coordinated deployment of multiple genes evolves to alter a phenotype. In the case of D. biarmipes, several closely related species possess spots of different shapes and intensities, and the patterns of Yellow expression closely pre-configures the adult pattern of pigmentation [28]. In each species tested, the pattern of Dll expression closely matches the pattern of the adult wing pigmentation pattern (Figure 2). This suggests that upon the origin of the Dll-regulated pigmentation spot, diversity has been largely shaped by evolving patterns of Dll expression. However, it is currently not known whether the causative mutations reside within the Dll locus, or alternatively reside in an upstream regulator of Dll expression in the wing.
The sexually dimorphic abdominal pigmentation of D. melanogaster and its relatives provide a particularly convenient model to study the coordinated evolution of a gene expression program. Within the melanogaster species group, tergite pigmentation ranges from the A6 segment in species like D. auraria (Figure 1F), the A5 and A6 segments of species such as D. melanogaster (Figure 1E), to the expanded A3-A6 segment pigmentation found in D. prostipennis (Figure 1G). In D. prostipennis, yellow and tan expression has expanded beyond A5 and A6 to include the A3 and A4 segments, and likewise ebony expression has receded from these segments [32]. The CREs for these three genes were tested for their ability to regulate GFP reporter expression in transgenic D. melanogaster. For the yBE CRE, the D. prostipennis reporter recapitulated the male expression of yellow in the A3-A6 segments indicating cis-evolutionary change at this CRE. However, the t_MSE and ebony CREs drove patterns of GFP expression indistinguishable from those driven by the D. melanogaster CREs (Figure 3C) [32]. These outcomes are consistent with evolutionary changes altering a trans-regulator of tan activation and ebony repression in D. prostipennis. However, the evolved trans-regulatory factor or factors currently remain unknown.
In another study, tan and yellow CREs from D. auraria were tested for their regulatory capability in transgenic D. melanogaster. While yellow and tan expression is limited to the A6 segment of this species, the CREs drove GFP expression in the A5 and A6 segments in a manner indistinguishable from that of D. melanogaster [21]. This outcome indicates that the regulatory logic with these two species yBE and t_MSE has not evolved, and imply modifications to the trans-regulatory landscape of this species. Moreover, in two independent cases in which male specific pigmentation was lost, it was found that the t_MSE for one species and the yBE for the other can still function in transgenic D. melanogaster. Thus, the lack of yellow and tan expression in these species likely results from changes to a trans-regulator. The importance of trans-regulatory evolution is further highlighted by two recent studies on convergent evolution, in which the underlying changes mapped in one case to the pdm3 transcription factor gene [33], and in the other case to the Abd-B transcription factor gene and ebony [34]. Collectively, these studies highlight how coordinated gene expression programs can often evolve by modifying trans-regulators that can affect only a subset of the co-expressed genes.
The role of pleiotropy in shaping the logical evolution of CREs
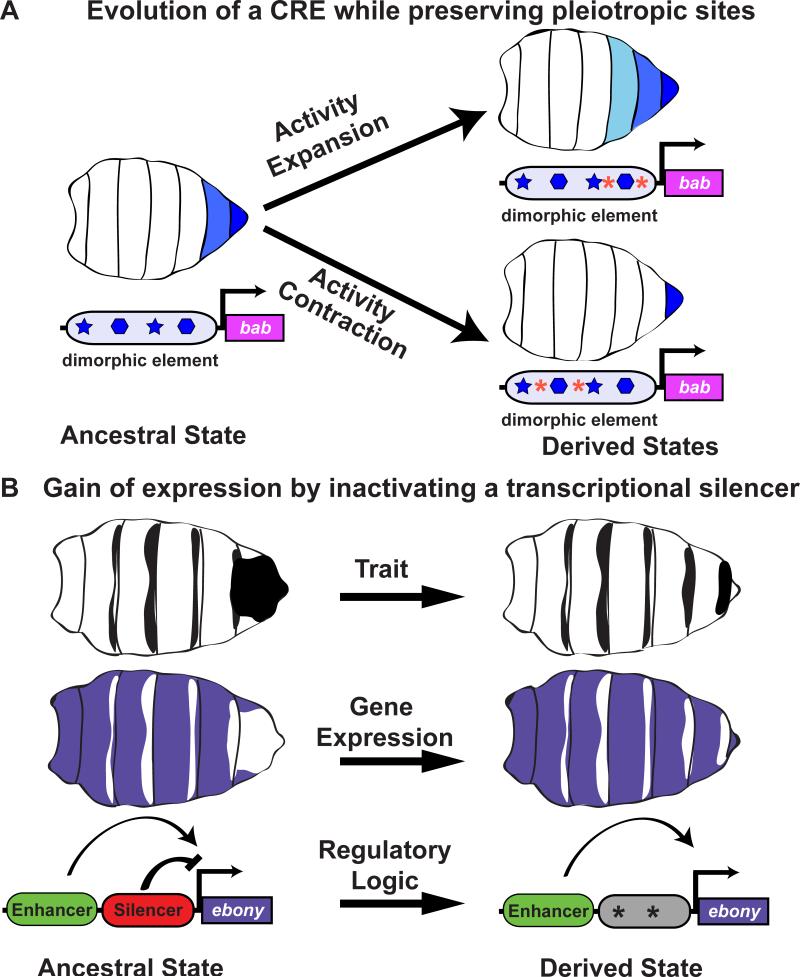
While much can be learned about trait evolution without knowing the specific transactions that mutations make with respect to the coming and going of transcription factor binding sites in CREs, this added depth of understanding allows one to probe increasingly complex phenomena, such as evolutionary constraint. In one recent study, it was found that D. melanogaster females with different extents of tergite pigmentation posses alleles of a CRE known as the “dimorphic element” that drives the pigment-repressing Bab transcription factor in the poster female abdomen [35]. These alleles differ in the number of segments and level of transcriptional activity, but share the ability to drive expression in the adjacently situated female genitalia (Figure 4A). The regulatory logic for this CRE possesses 14 binding sites for the Hox factor Abd-B, providing body-axis positional information, and two sites for Doublesex, the sex-specific regulator that restricts this CRE to the female abdomen and genitalia [36]. However, despite detailed knowledge of sixteen inputs to this CRE, the function-altering mutations of each allele occurred outside of these known binding sites, a trend which was also observed among other species at this CRE [35]. These outcomes indicate how the ancestral pleiotropic function of this CRE and its core regulatory logic could be maintained in the genitalia, while alterations occurred at other sites to modulate its more malleable abdominal function (Figure 4A). These observations demonstrate how knowledge of a CRE's logic can provide a mechanistic understanding of the constraints imposed on regulatory sequence evolution by pleiotropy.
Figure 4. Understanding CRE logic and architecture informs the nature of developmental constraints on evolution.
(A) The dimorphic element is a CRE that activates the pigment repressor Bab in the female abdomen. Within D. melanogaster populations, the dimorphic element drives activity in a varying number of segments, ranging from many (light blue) to few (dark blue). Reconstruction of an ancestral enhancer for this species revealed that high and low activities evolved from an intermediate ancestor without disrupting 16 conserved binding sites (star and hexagon shapes) for two transcription factors. Hence, significant changes in abdominal activity evolved by function-altering mutations (red asterisks) that had no effect on the binding sites that activate the enhancer in its ancestral genital setting (dark blue). (B) D. auraria males commonly have a dorsal pattern of pigmentation on the A6 tergite, which anti-correlates with the expression of the ebony (purple). Within Japan, some strains lack this pigmentation pattern, due to the gain of ebony expression throughout the A6 segment epidermis. This change in expression evolved through mutational changes (*) that abrogated the activity of a CRE that functions as a silencer element.
A great degree of variation in male-specific pigmentation is seen for the A6 tergite of D. auraria males in Japan. For this intraspecific difference, yellow and tan expression similarly occurs in both highly pigmented and non-pigmented males. However, in the non-pigmented males, A6 expression of ebony is notably upregulated and this up-regulation is due to mutations that occurred in a CRE that functions as a transcriptional silencer element [37]. Thus, for this gene, the gain of expression occurred through mutational changes that attenuated the function of a silencer. It is possible that silencer inactivation provides a streamlined path to evolved increases in ebony expression (Figure 4B). Consistent with this scenario, the independent loss of pigmentation in D. serrata was similarly shown to result from increased ebony expression and the mutational modification of this species’ silencer element [37]. Thus, the modification of a tissue-specific silencer, when present may be the easiest and least pleiotropic way to expand expression activities.
Conclusions
The results reviewed here show how obtaining a mechanistic view of CRE evolution and its relationship to phenotypes can reveal the molecular logic by which developmental systems change. We propose that the elucidation of how mutations affect a CRE's regulatory logic is a key to answering many of the looming questions in developmental evolution. Transcription factor binding events are the language by which the genome's instructions are read, and as we now know, they play an important part in how those instructions change. Deciphering this language will allow us to understand why some passages change, while others cannot be edited. With the growing recognition that phenomena such as developmental plasticity, constraint, and robustness can play important evolutionary roles [38,39], traits like fruit fly pigmentation [40,41], for which the molecular basis of their construction and evolution have been worked out to the finest resolution will lead the way in providing concrete details of how these processes work mechanistically. Indeed, pigmentation of the female D. melanogaster posterior abdomen is influenced by temperature, and both bab [41] and tan [42] have been implicated in the environmental response. An understanding of these genes’ CRE regulatory logic should allow insights to be made about the role of transcriptional regulatory sequences in the evolution of plasticity.
However, resolving regulatory logic presents a daunting challenge to the field. For example, the D. melanogaster genome has around 750 transcription factor genes [43] whose expression patterns are generally poorly characterized. Thus, connecting a single transcription factor to a functional binding event in a CRE remains laborious to establish [44]. In the face of these challenges though, the diversity of Drosophila pigmentation traits and their underlying CREs present an excellent model system to better understand transcriptional regulatory evolution, and are likely to lead the way into the mechanistic understanding regulatory logic evolution.
Highlights.
Modification of cis-regulatory elements (CREs) often underlie morphological evolution
Novel gene expression patterns can emerge via the modification of preexisting CREs
CREs with distinct transcription factor binding inputs can drive gene co-expression
Pleiotropic roles for transcription factor binding sites shape paths of CRE evolution
Mechanistic grasp of CRE evolution will inform constraints, plasticity, canalization
Acknowledgements
TMW and MR were supported by a grant from the National Science Foundation (IOS-1555906). MR's work on Drosophila pigmentation was additionally supported by a grant from the National Institutes of Health (5R01GM114093-02). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
- 1.Carroll SB, Grenier J, Weatherbee SD. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Blackwell Publishing; 2004. [Google Scholar]
- 2.Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–90. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- 4.Cretekos CJ, Wang Y, Green ED, Martin JF, Rasweiler JJ, Behringer RR. Regulatory divergence modifies limb length between mammals. Genes Dev. 2008;22:141–51. doi: 10.1101/gad.1620408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–5. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, Kingsley DM. cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–89. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wittkopp PJ, Vaccaro K, Carroll SB. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr. Biol. 2002;12:1547–56. doi: 10.1016/s0960-9822(02)01113-2. [DOI] [PubMed] [Google Scholar]
- 8.Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–7. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 9.Wittkopp PJ, Carroll SB, Kopp A. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends Genet. 2003;19:495–504. doi: 10.1016/S0168-9525(03)00194-X. [DOI] [PubMed] [Google Scholar]
- 10.Prud'homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc. Natl. Acad. Sci. U. S. A. 2007;104(Suppl):8605–12. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salomone JR, Rogers WA, Rebeiz M, Williams TM. The evolution of Bab paralog expression and abdominal pigmentation among Sophophora fruit fly species. Evol. Dev. 2013;15:442–57. doi: 10.1111/ede.12053. [DOI] [PubMed] [Google Scholar]
- 12.Gompel N, Carroll SB. Genetic mechanisms and constraints governing the evolution of correlated traits in drosophilid flies. Nature. 2003;424:931–935. doi: 10.1038/nature01787. [DOI] [PubMed] [Google Scholar]
- 13.Rajpurohit S, Parkash R, Ramniwas S. Body melanization and its adaptive role in thermoregulation and tolerance against desiccating conditions in drosophilids. Entomol. Res. 2008;38:49–60. [Google Scholar]
- 14.Parkash R, Kalra B, Sharma V. Impact of body melanisation on contrasting levels of desiccation resistance in a circumtropical and a generalist Drosophila species. Evol. Ecol. 2010;24:207–225. [Google Scholar]
- 15.Matute DR, Harris A. The influence of abdominal pigmentation on desiccation and ultraviolet resistance in two species of drosophila. Evolution (N. Y) 2013;67:2451–2460. doi: 10.1111/evo.12122. [DOI] [PubMed] [Google Scholar]
- 16.Bastide H, Yassin A, Johanning EJ, Pool JE. Pigmentation in Drosophila melanogaster reaches its maximum in Ethiopia and correlates most strongly with ultra-violet radiation in sub-Saharan Africa. BMC Evol. Biol. 2014;14:179. doi: 10.1186/s12862-014-0179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopp A, True JR. Evolution of male sexual characters in the oriental Drosophila melanogaster species group. Evol. Dev. 2002;4:278–91. doi: 10.1046/j.1525-142x.2002.02017.x. [DOI] [PubMed] [Google Scholar]
- 18.Yeh S-D, True JR. The genetic architecture of coordinately evolving male wing pigmentation and courtship behavior in Drosophila elegans and Drosophila gunungcola. G3 (Bethesda) 2014;4:2079–93. doi: 10.1534/g3.114.013037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wittkopp PJ, True JR, Carroll SB. Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. Development. 2002;129:1849–58. doi: 10.1242/dev.129.8.1849. [DOI] [PubMed] [Google Scholar]
- 20.Werner T, Koshikawa S, Williams TM, Carroll SB. Generation of a novel wing colour pattern by the Wingless morphogen. Nature. 2010;464:1143–8. doi: 10.1038/nature08896. [DOI] [PubMed] [Google Scholar]
- 21•.Camino EM, Butts JC, Ordway A, Vellky JE, Rebeiz M, Williams TM. The Evolutionary Origination and Diversification of a Dimorphic Gene Regulatory Network through Parallel Innovations in cis and trans. PLOS Genet. 2015;11:e1005136. doi: 10.1371/journal.pgen.1005136. [This publication presents evidence showing how the male-specific tergite pigmentation originated by pigmentation gene CREs co-opting regulation by unique combinations of body plan transcription factors.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferguson LC, Green J, Surridge A, Jiggins CD. Evolution of the insect yellow gene family. Mol. Biol. Evol. 2011;28:257–272. doi: 10.1093/molbev/msq192. [DOI] [PubMed] [Google Scholar]
- 23.Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science (80−. ) 2009;326:1663–1667. doi: 10.1126/science.1178357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hovemann BT, Ryseck RP, Walldorf U, Stortkuhl KF, Dietzel ID, Dessen E. The Drosophila ebony gene is closely related to microbial peptide synthetases and shows specific cuticle and nervous system expression. Gene. 1998;221:1–9. doi: 10.1016/s0378-1119(98)00440-5. [DOI] [PubMed] [Google Scholar]
- 25.True JR, Yeh S-D, Hovemann BT, Kemme T, Meinertzhagen IA, Edwards TN, Liou S-R, Han Q, Li J. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genet. 2005;1:e63. doi: 10.1371/journal.pgen.0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong S, Rokas A, Carroll SB. Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell. 2006;125:1387–99. doi: 10.1016/j.cell.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 27.Kopp A, Duncan I, Carroll SB. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature. 2000;408:553–9. doi: 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- 28•.Arnoult L, Su K, Manoel D, Minervino C, Magrina J, Gompel N, Prud'homme B. Emergence and Diversification of Fly Pigmentation Through Evolution of a Gene Regulatory Module. Science. 2013;339:1423–1426. doi: 10.1126/science.1233749. [The authors show how the D. biarmipes spot evolved by the co-option of Dll activation by the gain of CRE binding sites and how the diversification of the spot phenotype involved the evolution of Dll expression.] [DOI] [PubMed] [Google Scholar]
- 29.Rogers WA, Grover S, Stringer SJ, Parks J, Rebeiz M, Williams TM. A survey of the trans-regulatory landscape for Drosophila melanogaster abdominal pigmentation. Dev. Biol. 2014;385:417–432. doi: 10.1016/j.ydbio.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 30.Jeong S, Rebeiz M, Andolfatto P, Werner T, True J, Carroll SB. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 31•.Koshikawa S, Giorgianni MW, Vaccaro K, Kassner VA, Yoder JH, Werner T, Carroll SB. Gain of cis -regulatory activities underlies novel domains of wingless gene expression in Drosophila. Proc. Natl. Acad. Sci. 2015 doi: 10.1073/pnas.1509022112. doi:10.1073/pnas.1509022112. [This paper dissected the entire regulatory region of the wingless locus of D. guttifera, identifying two enhancers that combine to generate spot associated expression patterns in this species.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Ordway A, Hancuch KN, Johnson W, Wiliams TM, Rebeiz M. The expansion of body coloration involves coordinated evolution in cis and trans within the pigmentation regulatory network of drosophila prostipennis. Dev. Biol. 2014 doi: 10.1016/j.ydbio.2014.05.023. doi:10.1016/j.ydbio.2014.05.023. [The expansion of D. prostipennis tergite pigmentation was connected to changes in the CRE controlling yellow expression, whereas the expression of two other pigmentation enzyme genes evolved through changes in one or more upstream regulators of their CREs.] [DOI] [PubMed] [Google Scholar]
- 33•.Yassin A, Delaney EK, Reddiex AJ, Seher TD, Bastide H, Appleton NC, Lack JB, David JR, Chenoweth SF, Pool JE, et al. The pdm3 Locus Is a Hotspot for Recurrent Evolution of Female-Limited Color Dimorphism in Drosophila. Curr. Biol. 2016;0:998–1001. doi: 10.1016/j.cub.2016.07.016. [In this paper the authors show that female abdomen color dimorphism evolved in multiple species of the montium subgroup of fruit flies though evolutionary changes to the transcription factor-encoding gene pdm3.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Signor SA, Liu Y, Rebeiz M, Signor SA, Liu Y, Rebeiz M, Kopp A. Genetic Convergence in the Evolution of Male- Specific Color Patterns in Drosophila Article Genetic Convergence in the Evolution of Male-Specific Color Patterns in Drosophila. Curr. Biol. 2016 doi: 10.1016/j.cub.2016.07.034. doi:10.1016/j.cub.2016.07.034. [Multiple transitions in male-specific pigmentation in the Drosophila ananassae species subgroup were connected by mapping studies to cis-regulatory changes that altered ebony expression in each case.] [DOI] [PubMed] [Google Scholar]
- 35•.Rogers WA, Salomone JR, Tacy DJ, Camino EM, Davis KA, Rebeiz M, Williams TM. Recurrent Modification of a Conserved Cis-Regulatory Element Underlies Fruit Fly Pigmentation Diversity. PLoS Genet. 2013;9:e1003740. doi: 10.1371/journal.pgen.1003740. [This study presented evidence that variation in female tergite pigmentation has evolved through changes in the regulation of the bab genes. Of importance, the CRE controlling bab expression was shown to be pleiotropic and its function evolved in spite of the conservation of pleiotropic transcription factor binding sites.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, Carroll SB. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–23. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Johnson WC, Ordway AJ, Watada M, Pruitt JN, Williams TM, Rebeiz M. Genetic Changes to a Transcriptional Silencer Element Confers Phenotypic Diversity within and between Drosophila Species. PLOS Genet. 2015;11:e1005279. doi: 10.1371/journal.pgen.1005279. [The authors demonstrated how a transcriptional silencer element was attenuated in two lineages of fruit flies resulting in increased ebony expression and thereby eliminated male tergite pigmentation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moczek AP, Sultan S, Foster S, Ledòn-Rettig C, Dworkin I, Nijhout HF, Abouheif E, Pfennig DW, Ledón-Rettig C, Dworkin I, et al. The role of developmental plasticity in evolutionary innovation. Proc. R. Soc. London B Biol. Sci. 2011;278:2705–2713. doi: 10.1098/rspb.2011.0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laland KN, Uller T, Feldman MW, Sterelny K, Muller GB, Moczek A, Jablonka E, Odling-Smee J. The extended evolutionary synthesis: its structure, assumptions and predictions. Proc Biol Sci. 2015;282:20151019. doi: 10.1098/rspb.2015.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibert P, Moreteau B, Munjal a, David JR. Phenotypic plasticity of abdominal pigmentation in Drosophila kikkawai: multiple interactions between a major gene, sex, abdomen segment and growth temperature. Genetica. 1999;105:165–76. doi: 10.1023/a:1003704315194. [DOI] [PubMed] [Google Scholar]
- 41.Gibert JM, Peronnet F, Schlötterer C. Phenotypic plasticity in Drosophila pigmentation caused by temperature sensitivity of a chromatin regulator network. PLoS Genet. 2007;3:0266–0280. doi: 10.1371/journal.pgen.0030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibert J-M, Mouchel-Vielh E, De Castro S, Peronnet F. Phenotypic Plasticity through Transcriptional Regulation of the Evolutionary Hotspot Gene tan in Drosophila melanogaster. PLOS Genet. 2016;12:e1006218. doi: 10.1371/journal.pgen.1006218. [This study shows how the temperature-induced plasticity of D. melanogaster female tergite pigmentation involves the plastic behavior of the CRE controlling tan expression.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adryan B, Teichmann SA. FlyTF: a systematic review of site-specific transcription factors in the fruit fly Drosophila melanogaster. Bioinformatics. 2006;22:1532–3. doi: 10.1093/bioinformatics/btl143. [DOI] [PubMed] [Google Scholar]
- 44.Kalay G, Lusk R, Dome M, Hens K, Deplancke B, Wittkopp PJ. Potential Direct Regulators of the Drosophila yellow Gene Identified by Yeast One-Hybrid and RNAi Screens. G3. 2016 doi: 10.1534/g3.116.032607. doi:10.1534/g3.116.032607. [A high throughput yeast one-hybrid assay was used to identify novel candidate transcription factors that interact with cis-regulatory regions of the yellow gene.] [DOI] [PMC free article] [PubMed] [Google Scholar]