Abstract
In eukaryotes, mitosis is tightly regulated to avoid the generation of numerical chromosome aberrations, or aneuploidies. The aneuploid phenotype is a consequence of chromosomal instability (CIN), i.e., an enhanced rate of chromosome segregation errors, which is frequently found in cancer cells and is associated with tumor aggressiveness and increased tumor cell survival potential. To avoid the generation of aneuploidies, cells rely on the spindle assembly checkpoint (SAC), a widely conserved mechanism that protects the genome against this type of error. This signaling pathway stops mitotic pro-gression before anaphase until all chromosomes are correctly attached to spindle microtubules. Howev-er, impairment of the SAC cannot account for the establishment of CIN because cells bearing this phe-notype have a functional SAC. Hence, in cells with CIN, anaphase is not triggered until all chromo-somes are correctly attached to spindle microtubules and congressed at the metaphase plate. Thus, an in-teresting question arises: What mechanisms actually mediate CIN in cancer cells? Recent research has shown that some pathways involved in chromosome segregation are closely associated to centromere-encoded non-coding RNA (cencRNA) and that these RNAs are deregulated in abnormal conditions, such as cancer. These mechanisms may provide new explanations for chromosome segregation errors. The present review discusses some of these findings and proposes novel mechanisms for the establish-ment of CIN based on regulation by cencRNA.
Keywords: Chomosome segregation, Chromosome instability, Centromere, Non-coding RNA
1. INTRODUCTION
The establishment of chromosomal instability in eukaryotic cells remains poorly understood despite intense research in this field during the past several decades. The classical protein-exclusive view of the kinetochore has provided several mechanisms that can lead to aneuploidy [1-4]. However, this perspective has to date failed to provide a comprehensive explanation of the changes in the function and composition of the kinetochores that lead to chromosomal instability (CIN).
Nevertheless, the recent boom in studies of non-coding RNAs has enriched our understanding of the kinetochore and centromere function [5, 6]. Progress in this field has refuted the old conception that both the centromere and mitotic cells are transcriptionally silent [6]. It has been demonstrated that centromeres are transcribed and that this transcriptional activity can occur during mitosis [7-9], adding an extra level of complexity to our models of centromere and kinetochore regulation.
Numerous RNAs transcribed from centromeric and pericentromeric DNA have been discovered. Although such RNAs vary widely in sequence and size [5, 10], the inhibition of their transcription or the manipulation of their concentration in the cell has detrimental effects on the kinetochore in multiple eukaryotic species. The novel mechanisms for regulating chromosome segregation discovered in these studies involve both the molecular interaction of the RNA molecules with proteins of the centromere and kinetochore and their transcription per se [11]. Interestingly, the expression of the centromeric and pericentromeric region is altered in cellular conditions associated with CIN, such as cancer [12-14].
Much of the research on the centromeric non-coding RNA function in chromosome segregation has focused the perpetuation of the centromeric chromatin identity [11]. These molecules and their transcription have been demonstrated to be essential for the deposition of CENP-A [15, 16] and have causal and consequential relationships with the epigenetic marks that characterize centromeric chromatin [5].
The relationship between disease and changes in the expression, stability, localization, or molecular interactions of centromere-encoded non-coding RNAs (cencRNAs) is beginning to gain attention as an explanation for the development of aneuploidy.
In the present review, we discuss recent advances in the study of centromeric transcripts and their importance in the regulation of chromosome segregation. We propose mechanisms by which deregulation of the transcription and abundance of these RNAs may lead to aneuploidy through the promotion of chromosome mis-segregation mechanisms that have been shown to occur in chromosomally unstable cells. Furthermore, we highlight that these RNA molecules and their transcriptional regulation may constitute a “missing piece” in our understanding of the establishment of CIN and ultimately contribute to cancer evolution.
2. ANEUPLOIDY, CANCER EVOLUTION AND NEWCIN MECHANISMS
The importance of proper centromere and kinetochore function lies in the conservation of the karyotype through cellular generations. Aneuploidy, the state of having a chromosome number different from a multiple of the wild type chromosome set of the species in question, is the result of chromosome segregation errors. Because this condition implies the gain or loss of entire chromosomes bearing hundreds of genes, it can dramatically impact the phenotype of individual cells, tissues, or whole organisms [17]. Thus, aneuploidy is thought to participate in many adaptation processes ranging from stress conditions in a cell or tissue to speciation [17].
One such adaptation process is cancer evolution. Tumor cells are under constant selective pressure and are accordingly characterized by aneuploidy and by an accelerated rate of generation of chromosome abnormalities [18] termed Chromosomal INstability (CIN). This phenotype is advantageous to cancer cell populations because it provides the variability that allows for adaptation to the harsh environment to which they are exposed [19, 20]. Consistently, CIN is associated with increased tumor aggressiveness and drug resistance in cancer [20]. Of note, the term CIN refers not only to an increased rate of errors in chromosome segregation but also to structural aberrations of the chromosomes. However, in the present review, when using the term CIN, we exclusively refer to numerical abnormalities.
The spindle assembly checkpoint (SAC) is a surveillance mechanism that ensures faithful chromosome segregation. The function of this pathway is crucial to preventing chromosome mis-segregation in eukaryotic cells. Its effector, the mitotic checkpoint complex (MCC), is composed of MAD2, BUBR1 and BUB3 and sequesters CDC20, an essential activator of the anaphase-promoting complex (APC) during metaphase, until sister chromatids are correctly congressed at the metaphase plate [21]. When bound to CDC20, the APC ubiquitinates several key proteins in mitosis to target them for degradation by the 26S proteasome. Most of the APC ubiquitination targets present during mitosis are proteins whose degradation is required to promote the metaphase-to-anaphase transition, such as cyclin B and SECURIN [21].
The MCC is activated during every prometaphase-to-metaphase transition and is silenced with the completion of chromosome congression at the metaphase plate. The signal derived from a single unattached kinetochore is sufficient to activate the SAC [22]. It has also been proposed that the SAC is able to sense when a centromere is not subject to stretching tension as spindle microtubules pull it towards the centrosome [23]. AURORA B is known to inhibit the microtubule depolymerase MCAK through phosphorylation, thus destabilizing microtubule-kinetochore unions in such centromeres. The resulting unattached kinetochore will be sensed by the SAC, and the metaphase-to-anaphase transition will be inhibited [24].
This model also proposes that there is an AURORA B phosphorylation gradient that peaks at the inner centromere (where AURORA B is localized in metaphase) and decreases towards the spindle poles. Accordingly, when the kinetochore is stretched towards a spindle pole, AURORA B targets are no longer reached by this phosphorylation gradient and remain active. In the case of MCAK, this will result in stabilization of the kinetochore-microtubule association [24]. However, there is some controversy as to whether AURORA B detects tension-free centromeres or some conformational change elicited by correct kinetochore-microtubule attachments [25].
The experimental ablation of the SAC genes has consistently been associated with aneuploidy in several eukaryotic models. The idea that mutations of the SAC genes constitute a major cause of CIN in cancer may have seemed straightforward. However, such alterations are rather rare in human cancer [1, 26], making them an unlikely etiology for the establishment of CIN. The contribution of changes in the expression of SAC genes in tumor cells is a matter of debate [1, 26]. Another known mechanism that can lead to CIN is the impairment of sister chromatid cohesion; however, as with the SAC genes, cohesin genes are rarely mutated in cancer [1]. Furthermore, it has been clearly established that CIN cells have a functional SAC. Experiments challenging such cells with spindle poisons have demonstrated that they display a functional SAC and that they do not reach anaphase until all chromosomes are aligned at the metaphase plate [1]. Although the contribution of CIN to cancer phenotypes is widely recognized, current evidence excludes malfunction of the SAC and mutation of cohesin genes from CIN mechanisms [1]. Thus, an interesting questions arises: what mechanisms actually mediate CIN in cancer cells? As mentioned above, the SAC pathway is activated by the presence of kinetochores that are either unattached from spindle microtubules or lacking tension that stretches them towards spindle poles. However, there are erroneous kinetochore-microtubule attachments that can go undetected by the SAC and cause chromosome lagging and, consequently, aneuploidy [21]. Merotelic kinetochore attachments are those in which a single kinetochore is anchored to microtubules emanating from both spindle poles. This type of attachment “satisfies” the SAC and hence does not elicit a mitotic arrest response. Furthermore, merotely is the primary mechanism of CIN in cancer cells [4].
However, cellular mechanisms of merotelic attachment prevention and correction exist, which rely on the proper function of several proteins and pathways, including AURORA B, heterochromatin formation pathways, and the regulation of sister chromatid cohesion [4]. These pathways and proteins are worth further exploration in the search for CIN mechanisms. Interestingly, some of these proteins are closely associated with cencRNAs and their transcription, providing support to the importance of cencRNA in cancer CIN cells.
3. FROM CENTROMERES TO KINETOCHORES
In most eukaryotes, the DNA sequence of the centromere consists of repeated units that extend for up to several millions of bases and is organized in two distinct domains: the core centromere domain, where the kinetochore is assembled, and the pericentromeric region, which flanks the core centromere [27].
For most organisms studied to date, there is no clear consensus on the difference between the repetitive sequences found in the centromere core and those found in the pericentromere [28]. Centromeric regions are highly variable between species, between different chromosomes of a single individual, and even between the same chromosome in different individuals [29]. This variability represents a major challenge to current genome assembly methods that rely on contig assembly [28]. Nonetheless, consensus sequences have been derived for the fundamental units of the repeat-containing regions underlying the centromere of different species. For example in humans, alpha satellite, the main monomeric sequence whose head-to-tail repeat arrangement constitutes the centromere core, consists of a ~171 bp AT-rich sequence [29, 30]. This unit contains a 17 bp CENP-B box, which serves as a tethering site for the CENP-B protein, a factor essential for the establishment and maintenance of the centromere identity [27, 31].
The localization of the kinetochore appears to be an epigenetic event rather than being exclusively sequence-dependent [27]. The centromere core is characterized by the presence of nucleosomes containing the histone H3 variant CENP-A (also called CID in Drosophila, Cse4 in Saccharomyces cerevisiae, Cnp1 in Saccharomyces pombe and CENH3 in some plants) interspersed with nucleosomes containing the canonical histone H3. The incorporation of CENP-A is the founding step of centromere specification [27].
The deposition of CENP-A can occur in regions lacking centromeric sequences. Conversely, there are no functional centromeres lacking CENP-A [32]. These findings, along with the diversity of centromeric regions found across eukaryotes, has led to the “centromere paradox”, i.e., the observation that the kinetochore has a widely conserved function in eukaryotes despite the diversity of the underlying DNA sequences in different species [33].
The overall mechanism leading to kinetochore specification and the perpetuation of centromeric chromatin remains obscure [11, 34, 35]. Nonetheless, the transcription of centromeric sequences is a conserved feature of every eukaryote studied to date, including yeast, beetles, maize, Arabidopsis, flies, mice, marsupials and humans [36-44]. In fact, centromeric transcription was reported as early as 1968 [45] and 1973 [46]; however, this phenomenon was ignored until recently, with the blooming of non-coding RNAs findings [6]. Currently, experimental evidence supports the concept that correct chromosome segregation is hindered by the inhibition, degradation, or overexpression of centromeric sequences. Thus, transcription of these regions is thought to participate in the perpetuation of centromeric histone marks as well as in centromeric localization and function [11, 35].
4. CENTROMERE-ENCODED NON-CODING RNA
Numerous centromeric and pericentromeric transcripts have been identified in different organisms under distinct conditions. Despite their molecular dissimilarities, these RNAs have a conserved centromeric regulatory function [47]. The size of these transcripts is probably their most diverse feature. This is partly because the size of DNA centromeric repeat monomers is disparate in different organisms but also because these RNAs can bear different multimers or a non-integer number of the corresponding satellite repeat. The length range of the transcripts reported to date extends from RNA interference molecules (~21 nt) to over 10 kb-long non-coding RNAs [48]. Table 1 presents some of the RNAs found in different species, in various conditions. Although this size span includes RNAs originating from diverse organisms, from yeast to higher vertebrates, it also reflects the diversity of transcripts that can be found in a single organism and may be the result of detecting different stages of their processing [10]. In many cases, the electrophoretic analysis of centromeric or pericentromeric transcripts from a single cell type under a single condition yields a smear that spans several kb [49, 50]. Nonetheless, it has been clearly established that the centromeric and pericentromeric transcriptional pattern can be shifted under different circumstances, such as stress [41, 51, 52], differentiation [41, 53], phases of the cell cycle [9, 15, 54, 55], senescence [56, 57] and cancer [12-14]. Changes in the expression patterns and sizes, or the processing of the pericentromeric and centromeric RNAs produced, have important repercussions in centromere function.
Table 1.
Some studies that have assessed the approximate size of cencRNAs in different organisms under distinct conditions.
| Organism | Originated From | Size | Observations | Technical Approach | References |
|---|---|---|---|---|---|
| Maize | CentC centromeric satellite repeats | 40 nt and 75 nt predominantly ∼75 nt |
Untreated cells | Northern blot | [68] |
| Maize | Centromeric retrotransposons (CRMs) and satellite repeats (CentC) | ∼ 40 – 250 nt | Enriched in CenH3-associated chromatin | CenH3 immunoprecipi-tation followed by blot hybridization | [69] |
| Chicken-human hybrid DT40 cell | Alphoid (centromeric) and Satellite III (pericentromeric) | - Smear spanning from ∼ 20nt to > 10 kb | Dicer depletion | Northern blot | [48] |
| Tammar wallaby | sat23, a centromere-specific repetitive satellite containing the CENP-B-binding domain | ∼ 40 nt | Untreated cells | Northern blot | [43] |
| Mouse | Minor satellite (centromere) | ∼ 2-4 kb ∼ 120 nt trancript accumulation |
- Untreated cells - Stress conditions, differentiation |
Northern blot | [70] |
| Human (HeLa cells) | α-satellite (centromeric repeats) | Discrete bands corresponding to multiples of the 171 nt monomer. | Actynomicin-treated cells (unchanged abundance as compared to untreated control) | PCR | [71] |
| Human (HeLa cells) | α-satellite (centromeric repeats) | ∼1.3 kb | Untreated cells | Northern blot | [15] |
| Human (HeLa cells) | α-satellite (centromeric repeats) | ∼171 bp | Enriched in chromosome-associated RNA fraction | Purification of chromosome-associated RNA fraction followed by Northern blot | [72] |
5. Mechanisms of regulating centromere function by centromeric non-coding RNAS
Centromere-encoded non-coding RNAs (cencRNAs) have been shown to physically associate with several centromeric proteins, including CENP-A [15, 37, 44, 54] and its chaperone HJURP [15], CENP-B [43], and CENP-C [40], as well as the chromosomal passengers AURORA B, INCENP and SURVIVIN [54, 58]. Interestingly, each one of these proteins is tightly associated with the DNA or inner centromere complexes and has an essential role in chromosome segregation. Moreover, the inhibition of centromeric transcription or destruction of satellite cencRNAs causes mistargeting of CENP-A [15, 37, 44], CENP-B [43] and CENP-C [9, 40]. Thus, it has been proposed that such RNAs can function as a scaffold for the assembly of the kinetochore [11].
CENP-A recruitment defects derived from changes in cencRNA transcription and abundance may contribute to the establishment of CIN. Cancer cells display overexpression and mistargeting of CENP-A as well as expanded alpha satellite centromeric arrays [59, 60]. The impairment of CENP-A incorporation caused by changes in cencRNA transcription rates and/or overall abundance may have permanent repercussions in establishing CIN. Because the same amount of CENP-A is renewed in every late mitosis/G1 stage [32], errors in its recruitment in a single incorporation event could permanently expand or reduce the amount of such protein in centromeres. Changes in cencRNA abundance have been shown to occur due to various types of stress [51, 52].
Hence, stress conditions affecting cencRNA expression may function as both an initiator and a promoter of CIN by perpetuating an altered CENP-A domain. Although this mechanistic model is merely speculative, it would be interesting to challenge these concepts. Fig. (1) shows a schematic representation of this theoretical model.
Fig. (1).
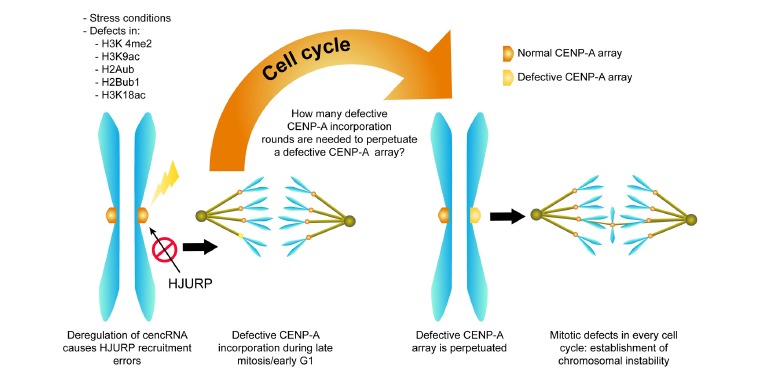
Model for the establishment of chromosomal instability through the deregulation of cencRNA and the deregulation of CENP-A deposition: several different factors can affect the regulation of cencRNAs, including errors in histone marks that control their expression or stress conditions (such as heat shock). The deregulation of such RNAs causes a defective CENP-A replenishment at the end of mitosis/early G1 as a consequence of inadequate CENP-A chaperone recruitment. Because the same amount of CENP-A is replenished in every cell cycle, it is arguable that a single cencRNA deregulation event may promote the perpetuation of an aberrant CENP-A array and consequently, induce chromosomal instability. However, this hypothesis has not been tested to date.
Furthermore, cencRNA is closely associated with epigenetic marks that regulate its transcription and participate in centromere organization. Centrochromatin is a dual region that contains post-translational histone modifications associated with both repression and active transcription [11, 35]. Studies in human artificial chromosomes (HACs) have shown that the centromere contains an H3 modification pattern characteristic of actively transcribed DNA regions: mono-, di- and trimethylated H3K4 and dimethylated H3K36 [61]. Nevertheless, H3K9me3, which is often associated with pericentromeric heterochromatin and transcriptional repression, is also present in this region [61]. This seemingly contradictory observation led to the concept of the so-called centrochromatin (the centromeric chromatin) as a distinctive type of chromatin that can bear epigenetic marks of both heterochromatin and active transcription [62].
Two elegant studies that specifically targeted histone-modifying enzymes to the centromere of an HAC showed that perturbations of centrochromatin that either downregulate or cause the overexpression of cencRNAs hinder centromeric function [61, 63]. In one of these studies, the CENP-B box of one in every two alpha satellite repeats of the HAC was replaced by elements that recruit histone H3 lysine 9 acetylases [63], thus promoting a strong overexpression of alpha satellite RNA. In the other paper, the authors specifically depleted histone H3 lysine 4 dymethylation (H3K4me2) with a similar strategy to repress alpha satellite cencRNAs [63]. In both cases, the consequential impairment of proper centromeric function was explained by a loss of CENP-A from the centromere. Notably, kinetochores depleted of H3K4me2 remained functional in the short term but were defective in the incorporation of CENP-A and were gradually inactivated [61]. This was a consequence of errors in the correct recruitment of HJURP, a chaperone that promotes CENP-A localization to the centromere [61]. Conversely, rather than merely inhibiting its correct incorporation, exacerbated centromeric transcription causes an active, rapid loss of CENP-A [63]. In this CENP-A deposition pathway, it has been demonstrated that both HJURP and CENP-A interact with RNAs transcribed from centromeric repeats in human cells to promote correct CENP-A deposition in early G1 [15]. In Drosophila, CAL1, a CENP-A chaperone also required centromeric transcription to promote CENP-A centromeric establishment [16].
In addition to classical histone epigenetic marks, other modifications have been implicated in the transcription of centrochromatin. Such is the case of H2B and H2A ubiquitination. Histone H2B ubiquitination is required for transcription of cencRNAs, whereas H2A ubiquitination by BRCA1 is required for their repression; both marks are deregulated in cancer [14, 64]. Impeding H2B ubiquitination with the use of interference RNA targeting RNF20, the ubiquitin ligase responsible for this epigenetic mark, causes an increase in H3 stability as well as unequal chromosome segregation in yeast and human cells [64]. Noteworthy, the exit of H3 from centrochromatin is essential for new CENP-A entry to the centromere [32, 65].
On the other hand, loss of H2A ubiquitination and the consequential de-repression of cencRNAs are associated with centrosome amplification, cell cycle checkpoint defects, DNA damage, and chromosome mis-segregation [14]. Moreover, BRCA1 deficiency is accompanied by a derepression of centromeric DNA and accumulation of centromeric transcripts. Accordingly, centrosome amplification, cell-cycle checkpoint defects, DNA damage, and genomic instability are observed [14]. Notably, these changes are associated with BRCA1 mutation in cancer [14].
Another recent study demonstrated that SIRT6, a member of the sirtuin family of deacetylases, prevents mitotic errors by silencing of pericentric transcripts through acetylation of histone H3 at lysine 18. Furthermore, downregulation of SIRT6 by siRNA caused an aberrant accumulation of pericentric transcripts and, consequentially, multipolar spindles, cellular senescence, and aneuploidy [66].
Moreover, alterations in the abundance of cencRNAs have important consequences in H3K9 methylation, a modification whose regulation is essential for their own transcription [37, 41, 43, 67], demonstrating an interesting feedback loop that is worthy of further study.
Besides, the interaction of cencRNAs with AURORA B has been associated with modifications of the kinase activity of AURORA B and is known to be necessary for the assembly of the CPC [54, 58]. On the other hand, the overexpression of cencRNAs has been shown to cause mislocalization of AURORA B, misaligned chromosomes at the metaphase plate, and abnormal numbers of chromosomes in murine erythroleukemia cells [41]. These findings directly link cencRNA expression to the regulation of chromosome segregation because Aurora B function is related to the detection of tension-free kinetochores at the metaphase plate [24]. Likewise, mistargeting of AURORA B could cause an abnormal tolerance to syntelic chromosomal attachments because, in this type of attachment, both sister chromatids are tethered to spindle microtubules emanating from the same pole, although tension is not generated because the kinetochores of both sister chromatids are pulled towards the same pole [24]. Importantly, cencRNA overexpression has been related to cancer and lagging chromosomes [12-14]. Furthermore, AURORA B has been shown to phosphorylate CENP-A at serine 7 [73]. Notably, these two proteins coincide at the centromere at the metaphase plate and later move to the contractile ring in cytokinesis. Phosphorylation of CENP-A on serine 7 and the consequent localization of CENP-A in the contractile ring have an as-yet unexplained function in correct cytokinesis. Moreover, cytokinesis failure induces CIN: its resulting polyploidization is accompanied by centrosome amplification and thus by merotelic kinetochore attachments, which are a common cause of aneuploidy [1]. A schematic representation of this model is depicted in Fig. (2). Whether a physical association of the CPC proteins with CENP-A is necessary for the localization of CENP-A at the contractile ring is currently unknown. However, CENP-A and the CPC proteins display cencRNA-binding activity. It is therefore interesting to hypothesize that the cencRNA could function as a scaffold to promote CENP-A localization at the contractile ring. These considerations suggest that changes in the expression of cencRNAs could provoke chromosome segregation defects by affecting AURORA B localization, kinase activity, and association with other chromosomal passengers during metaphase.
Fig. (2).
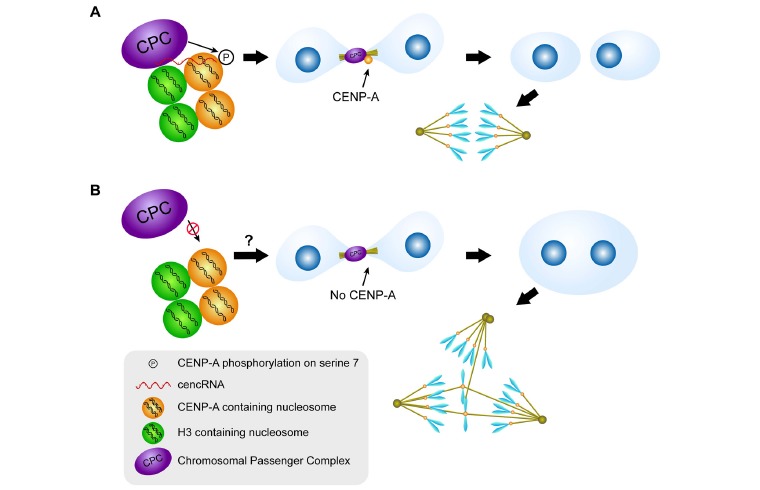
Model for the establishment of chromosomal instability through the deregulation of Aurora B activity and cytokinesis failure: both the localization and the kinase activity of Aurora B are promoted by its interaction with cencRNAs. Among other targets, Aurora B phosphorylates CENP-A on its serine 7 residue, and this phosphorylation is necessary for an as-yet unexplained function of CENP-A in cytokinesis. The pharmacological inhibition of Aurora B activity prevents such CENP-A phosphorylation, thereby promoting cytokinesis failure. This defect leads to polyploidy, which in turn promotes the development of multipolar spindles and/or merotelic kinetochore attachments and thus, chromosome mis-segregation. It would be interesting to test whether Aurora B inhibition through depletion of cencRNA has the same effect as its pharmacological inhibition.
Work on transcriptional regulation of cencRNAs has raised the question of whether cencRNAs or centromeric transcription per se is responsible for kinetochore regulation. Current evidence favors a model in which both are required: the RNA serves as a scaffold for centromeric proteins, such as CENP-A, HJURP, CENP-B, CENP-C and the chromosomal passengers [9, 15, 37, 40, 43, 44, 54], although the role of transcription itself remains a matter of debate [11] (discussed below). In any case, studies in this field have clearly established that the transcriptional activity of the centromere is tightly regulated and that both its over-activation and its repression are detrimental to centromere function.
6. IS CENCRNA SEQUENCE IMPORTANT FOR THEIR FUNCTION?
No sequence has been defined on cencRNAs that mediates their physical association with specific proteins. However, the fact that interactions have been detected between such transcripts and orthologous proteins of different organisms despite sequence divergence is noteworthy. For instance, Aurora B interacts with the cencRNAs of mouse [54], human [58], and Xenopus laevis [8], whereas CENP-A interacts with those of maize [44], mouse [54], fruit fly [37], and human [15]. In addition, Aurora B was shown to interact with RNAs (including mRNAs) other than cencRNAs in immunoprecipitation experiments followed by RNA sequencing. The RNA pool detected in this analysis showed an overrepresentation of adenines and the enrichment of transcripts previously known to localize to the mitotic spindle in Xenopus laevis [74]. However, the interaction of one of the RNAs detected with Aurora B was unaffected by a decrease of its adenine content in EMSA analysis [74]. Aurora B is a rather promiscuous RNA-interacting protein with modest sequence specificity. This was further supported by EMSA experiments in which X. laevis Aurora B showed a modestly higher binding affinity to alpha satellite (human cencRNA) and fcr1 (X. laevis cencRNA) over a control RNA transcribed from the multiple cloning sequence of a plasmid [8]. Importantly, animal centromeric repeats tend to be AT-rich [75], and neither human centromeric alpha satellite repeats nor X. laevis fcr1 centromeric repeats constitute an exception to this observation [30, 76]. Aurora B is an interesting example of a protein whose localization depends on cencRNA binding because RNA stimulates its kinase activity and there is a positive feedback loop between such kinase activity and its metaphase localization [77]. However, it is not the only protein whose centromeric localization depends on cencRNA but displays somewhat nonspecific RNA-binding; the same is true for CENP-C [39], and HP1α [78, 79], which share an RNA-interacting hinge domain. A common feature shared by these three proteins is that they have at least two chromatin-recruitment mechanisms, one of which is RNA-independent: Aurora B localization depends upon phosphorylation of histone H3 at threonine 3 and histone H2A at threonine 120 [80]. CENP-C is a promiscuous DNA binding protein; and HP1α binds to methylated histone H3 tails. Thus, cencRNA transcription/abundance at the centromere may serve as a secondary mechanism to hold RNA-binding centromeric proteins in their correct localization on chromatin without the need for sequence specificity. This mechanism could be crucial during mitosis, when the centromere displays transcriptional activity while the rest of the chromosome does not. According to this model, the recruitment of RNA-interacting proteins to mitotic centromeres would be determined in a cooperative fashion by, on one hand, the transcriptional regulation of the centromere and, on the other hand, an independent, mechanism specific for each protein. As a consequence, cencRNA deregulation would partially affect the recruitment of multiple proteins involved in chromosome segregation. This mechanism could explain the diversity of mitotic defects observed upon centromeric transcription impairment, upregulation, or exogenous cencRNA expression. Furthermore, this concept would explain why both the act of centromeric transcription and cencRNAs are crucial for the correct progression of chromosome segregation: cencRNAs contribute by functioning as a scaffold, and the act of transcription contributes by dictating the time and space in which cencRNAs exert their functions.
CONCLUSION
Several novel cencRNAs of different classes have recently been described. Both their transcription and molecular interactions have been shown to have functional implications in correct chromosome segregation. Although these RNAs have heterogeneous features, their association with certain proteins, including CENP-A, appears to be widely conserved.
Moreover, the deregulation of cencRNAs in cancer is beginning to gain attention as the mechanisms by which this deregulation causes aneuploidy are being elucidated. Intensive research is needed to determine whether current models by which cencRNAs are thought to regulate chromosome segregation are accurate and whether the deregulation of their expression constitutes a common cause of CIN or can promote aneuploidy only as an isolated phenomenon.
ACKNOWLEDGEMENTS
We thank Dr. Sara Frías Vázquez for critical comments on the manuscript. Rodrigo Cáceres-Gutiérrez is a doctoral fellow of the Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM) and of the Consejo Nacional de Ciencia y Tecnología (CONACYT, grant 290129).
LIST OF ABBREVIATIONS
- APC
Anaphase Promoting Complex
- cencRNA
Centromere- encoded non-coding RNA
- CIN
Chromosomal Instability
- HAC
Human Artificial Chromosome
- H3K4me2
Histone H3 dimethylated on lysine 4
- H3K9me3
Histone H3 trimethylated on lysine 9
- MCC
Mitotic Checkpoint Complex
- SAC
Spindle Assembly Complex
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Thompson S.L., Bakhoum S.F., Compton D.A. Mechanisms of chromosomal instability. Curr. Biol. 2010;20(6):285–295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon D.J., Resio B., Pellman D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 2012;13(3):189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 3.Weaver B.A., Silk A.D., Montagna C., Verdier-Pinard P., Cleveland D.W. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11(1):25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Gregan J., Polakova S., Zhang L., Tolic-Norrelykke I.M., Cimini D. Merotelic kinetochore attachment: Causes and effects. Trends Cell Biol. 2011;21(6):374–381. doi: 10.1016/j.tcb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biscotti M.A., Canapa A., Forconi M., Olmo E., Barucca M. Transcription of tandemly repetitive DNA: functional roles. Chromosome Res. 2015;23(3):463–477. doi: 10.1007/s10577-015-9494-4. [DOI] [PubMed] [Google Scholar]
- 6.Ferreira D., Meles S., Escudeiro A., Mendes-da-Silva A., Adega F., Chaves R. Satellite non-coding RNAs: The emerging players in cells, cellular pathways and cancer. Chromosome Res. 2015;23(3):479–493. doi: 10.1007/s10577-015-9482-8. [DOI] [PubMed] [Google Scholar]
- 7.Liu H., Qu Q., Warrington R., Rice A., Cheng N., Yu H. Mitotic Transcription Installs Sgo1 at Centromeres to Coordinate Chromosome Segregation. Mol. Cell. 2015;59(3):426–436. doi: 10.1016/j.molcel.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 8.Blower M.D. Centromeric transcription regulates Aurora-B localization and activation. Cell Reports. 2016;15(8):1624–1633. doi: 10.1016/j.celrep.2016.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan F.L., Marshall O.J., Saffery R., Kim B.W., Earle E., Choo K.H., Wong L.H. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl. Acad. Sci. USA. 2012;109(6):1979–1984. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enukashvily N.I., Ponomartsev N.V. Mammalian satellite DNA: A speaking dumb. Adv. Protein Chem. Struct. Biol. 2013;90:31–65. doi: 10.1016/B978-0-12-410523-2.00002-X. [DOI] [PubMed] [Google Scholar]
- 11.Chan F.L., Wong L.H. Transcription in the maintenance of centromere chromatin identity. Nucleic Acids Res. 2012;40(22):11178–11188. doi: 10.1093/nar/gks921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ting D.T., Lipson D., Paul S., Brannigan B.W., Akhavanfard S., Coffman E.J., Contino G., Deshpande V., Iafrate A.J., Letovsky S., Rivera M.N., Bardeesy N., Maheswaran S., Haber D.A. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331(6017):593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eymery A., Horard B., El Atifi-Borel M., Fourel G., Berger F., Vitte A.L., Van den Broeck A., Brambilla E., Fournier A., Callanan M., Gazzeri S., Khochbin S., Rousseaux S., Gilson E., Vourc'h C. A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells. Nucleic Acids Res. 2009;37(19):6340–6354. doi: 10.1093/nar/gkp639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Q., Pao G.M., Huynh A.M., Suh H., Tonnu N., Nederlof P.M., Gage F.H., Verma I.M. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477(7363):179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quenet D., Dalal Y. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. eLife. 2014;3:e03254. doi: 10.7554/eLife.03254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C.C., Bowers S., Lipinszki Z., Palladino J., Trusiak S., Bettini E., Rosin L., Przewloka M.R., Glover D.M., O'Neill R.J., Mellone B.G. Establishment of centromeric chromatin by the CENP-A assembly factor CAL1 requires FACT-mediated transcription. Dev. Cell. 2015;34(1):73–84. doi: 10.1016/j.devcel.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen G., Rubinstein B., Li R. Whole chromosome aneuploidy: big mutations drive adaptation by phenotypic leap. BioEssays. 2012;34(10):893–900. doi: 10.1002/bies.201200069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nowak M.A., Komarova N.L., Sengupta A., Jallepalli P.V., Shih Ie M., Vogelstein B., Lengauer C. The role of chromosomal instability in tumor initiation. Proc. Natl. Acad. Sci. USA. 2002;99(25):16226–16231. doi: 10.1073/pnas.202617399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heng H.H., Bremer S.W., Stevens J.B., Horne S.D., Liu G., Abdallah B.Y., Ye K.J., Ye C.J. Chromosomal instability (CIN): What it is and why it is crucial to cancer evolution. Cancer Metastasis Rev. 2013;32(3-4):325–340. doi: 10.1007/s10555-013-9427-7. [DOI] [PubMed] [Google Scholar]
- 20.Giam M., Rancati G. Aneuploidy and chromosomal instability in cancer: A jackpot to chaos. Cell Div. 2015;10(1):3. doi: 10.1186/s13008-015-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musacchio A., Salmon E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 22.Rieder C.L., Cole R.W., Khodjakov A., Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 1995;130(4):941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicklas R.B., Ward S.C., Gorbsky G.J. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J. Cell Biol. 1995;130(4):929–939. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lampson M.A., Cheeseman I.M. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21(3):133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Horst A., Lens S.M. Cell division: Control of the chromosomal passenger complex in time and space. Chromosoma. 2014;123(1-2):25–42. doi: 10.1007/s00412-013-0437-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holland A.J., Cleveland D.W. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 2012;13(6):501–514. doi: 10.1038/embor.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panchenko T., Black B.E. The epigenetic basis for centromere identity. Prog. Mol. Subcell. Biol. 2009;48:1–32. doi: 10.1007/978-3-642-00182-6_1. [DOI] [PubMed] [Google Scholar]
- 28.Hayden K.E. Human centromere genomics: Now it's personal. Chromosome Res. 2012;20(5):621–633. doi: 10.1007/s10577-012-9295-y. [DOI] [PubMed] [Google Scholar]
- 29.Waye J.S., Willard H.F. Nucleotide sequence heterogeneity of alpha satellite repetitive DNA: A survey of alphoid sequences from different human chromosomes. Nucleic Acids Res. 1987;15(18):7549–7569. doi: 10.1093/nar/15.18.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choo K.H., Vissel B., Nagy A., Earle E., Kalitsis P. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 1991;19(6):1179–1182. doi: 10.1093/nar/19.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohzeki J., Nakano M., Okada T., Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 2002;159(5):765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stellfox M.E., Bailey A.O., Foltz D.R. Putting CENP-A in its place. Cell. Mol. Life Sci. 2013;70(3):387–406. doi: 10.1007/s00018-012-1048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henikoff S., Ahmad K., Malik H.S. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293(5532):1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 34.Cao J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online. 2014;16(1):11. doi: 10.1186/1480-9222-16-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott K.C. Transcription and ncRNAs: At the cent(rome)re of kinetochore assembly and maintenance. Chromosome Res. 2013;21(6-7):643–651. doi: 10.1007/s10577-013-9387-3. [DOI] [PubMed] [Google Scholar]
- 36.Volpe T., Schramke V., Hamilton G.L., White S.A., Teng G., Martienssen R.A., Allshire R.C. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11(2):137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- 37.Rosic S., Kohler F., Erhardt S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol. 2014;207(5):673. doi: 10.1083/jcb.201404097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Neill R.J., Carone D.M. The role of ncRNA in centromeres: a lesson from marsupials. Prog. Mol. Subcell. Biol. 2009;48:77–101. doi: 10.1007/978-3-642-00182-6_4. [DOI] [PubMed] [Google Scholar]
- 39.Wong L.H., Brettingham-Moore K.H., Chan L., Quach J.M., Anderson M.A., Northrop E.L., Hannan R., Saffery R., Shaw M.L., Williams E., Choo K.H. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17(8):1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du Y., Topp C.N., Dawe R.K. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010;6(2):e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouzinba-Segard H., Guais A., Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl. Acad. Sci. USA. 2006;103(23):8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pezer Z., Ugarkovic D. RNA Pol II promotes transcription of centromeric satellite DNA in beetles. 2008. [DOI] [PMC free article] [PubMed]
- 43.Carone D.M., Longo M.S., Ferreri G.C., Hall L., Harris M., Shook N., Bulazel K.V., Carone B.R., Obergfell C., O'Neill M.J., O'Neill R.J. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma. 2009;118(1):113–125. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- 44.Topp C.N., Zhong C.X., Dawe R.K. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. USA. 2004;101(45):15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harel J., Hanania N., Tapiero H., Harel L. RNA replication by nuclear satellite DNA in different mouse cells. Biochem. Biophys. Res. Commun. 1968;33(4):696–701. doi: 10.1016/0006-291x(68)90352-5. [DOI] [PubMed] [Google Scholar]
- 46.Cohen A.K., Huh T.Y., Helleiner C.W. Transcription of satellite DNA in mouse L-cells. Can. J. Biochem. 1973;51(5):529–532. doi: 10.1139/o73-065. [DOI] [PubMed] [Google Scholar]
- 47.Ugarkovic D. Functional elements residing within satellite DNAs. EMBO Rep. 2005;6(11):1035–1039. doi: 10.1038/sj.embor.7400558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6(8):784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 49.Vourc'h C., Biamonti G. Transcription of satellite DNAs in mammals. Prog. Mol. Subcell. Biol. 2011;51:95–118. doi: 10.1007/978-3-642-16502-3_5. [DOI] [PubMed] [Google Scholar]
- 50.Pezer Z., Brajkovic J., Feliciello I., Ugarkovic D. Transcription of satellite DNAs in insects. Prog. Mol. Subcell. Biol. 2011;51:161–178. doi: 10.1007/978-3-642-16502-3_8. [DOI] [PubMed] [Google Scholar]
- 51.Valgardsdottir R., Chiodi I., Giordano M., Rossi A., Bazzini S., Ghigna C., Riva S., Biamonti G. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2008;36(2):423–434. doi: 10.1093/nar/gkm1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jolly C., Metz A., Govin J., Vigneron M., Turner B.M., Khochbin S., Vourc'h C. Stress-induced transcription of satellite III repeats. J. Cell Biol. 2004;164(1):25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terranova R., Sauer S., Merkenschlager M., Fisher A.G. The reorganisation of constitutive heterochromatin in differentiating muscle requires HDAC activity. Exp. Cell Res. 2005;310(2):344–356. doi: 10.1016/j.yexcr.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 54.Ferri F., Bouzinba-Segard H., Velasco G., Hube F., Francastel C. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 2009;37(15):5071–5080. doi: 10.1093/nar/gkp529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu J., Gilbert D.M. Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J. Cell Biol. 2007;179(3):411–421. doi: 10.1083/jcb.200706176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaubatz J.W., Cutler R.G. Mouse satellite DNA is transcribed in senescent cardiac muscle. J. Biol. Chem. 1990;265(29):17753–17758. [PubMed] [Google Scholar]
- 57.Shumaker D.K., Dechat T., Kohlmaier A., Adam S.A., Bozovsky M.R., Erdos M.R., Eriksson M., Goldman A.E., Khuon S., Collins F.S., Jenuwein T., Goldman R.D. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA. 2006;103(23):8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ideue T., Cho Y., Nishimura K., Tani T. Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes Cells. 2014;19(6):528–538. doi: 10.1111/gtc.12149. [DOI] [PubMed] [Google Scholar]
- 59.Tomonaga T., Matsushita K., Yamaguchi S., Oohashi T., Shimada H., Ochiai T., Yoda K., Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63(13):3511–3516. [PubMed] [Google Scholar]
- 60.Sullivan L.L., Boivin C.D., Mravinac B., Song I.Y., Sullivan B.A. Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res. 2011;19(4):457–470. doi: 10.1007/s10577-011-9208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bergmann J.H., Rodriguez M.G., Martins N.M., Kimura H., Kelly D.A., Masumoto H., Larionov V., Jansen L.E., Earnshaw W.C. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30(2):328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sullivan B.A., Karpen G.H. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004;11(11):1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bergmann J.H., Jakubsche J.N., Martins N.M., Kagansky A., Nakano M., Kimura H., Kelly D.A., Turner B.M., Masumoto H., Larionov V., Earnshaw W.C. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J. Cell Sci. 2012;125(Pt 2):411–421. doi: 10.1242/jcs.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sadeghi L., Siggens L., Svensson J.P., Ekwall K. Centromeric histone H2B monoubiquitination promotes noncoding transcription and chromatin integrity. Nat. Struct. Mol. Biol. 2014;21(3):236–243. doi: 10.1038/nsmb.2776. [DOI] [PubMed] [Google Scholar]
- 65.Jansen L.E., Black B.E., Foltz D.R., Cleveland D.W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176(6):795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tasselli L., Xi Y., Zheng W., Tennen R.I., Odrowaz Z., Simeoni F., Li W., Chua K.F. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 2016;23(5):434–440. doi: 10.1038/nsmb.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salvany L., Aldaz S., Corsetti E., Azpiazu N. A new role for hth in the early pre-blastodermic divisions in drosophila. Cell Cycle. 2009;8(17):2748–2755. doi: 10.4161/cc.8.17.9388. [DOI] [PubMed] [Google Scholar]
- 68.Du Y., Topp C.N., Dawe R.K. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010;6(2):e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Topp C.N., Zhong C.X., Dawe R.K. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. USA. 2004;101(45):15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bouzinba-Segard H., Guais A., Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl. Acad. Sci. USA. 2006;103(23):8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong L.H., Brettingham-Moore K.H., Chan L., Quach J.M., Anderson M.A., Northrop E.L., Hannan R., Saffery R., Shaw M.L., Williams E., Choo K.H. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17(8):1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang C., Wang X., Liu X., Cao S., Shan G. RNAi pathway participates in chromosome segregation in mammalian cells. Cell Discov. 2015;1(15029) doi: 10.1038/celldisc.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeitlin S.G., Shelby R.D., Sullivan K.F. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 2001;155(7):1147–1157. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jambhekar A., Emerman A.B., Schweidenback C.T., Blower M.D. RNA stimulates Aurora B kinase activity during mitosis. PLoS One. 2014;9(6):e100748. doi: 10.1371/journal.pone.0100748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Melters D.P., Bradnam K.R., Young H.A., Telis N., May M.R., Ruby J.G., Sebra R., Peluso P., Eid J., Rank D., Garcia J.F., DeRisi J.L., Smith T., Tobias C., Ross-Ibarra J., Korf I., Chan S.W. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013;14(1):R10. doi: 10.1186/gb-2013-14-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edwards N.S., Murray A.W. Identification of xenopus CENP-A and an associated centromeric DNA repeat. Mol. Biol. Cell. 2005;16(4):1800–1810. doi: 10.1091/mbc.E04-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang F., Ulyanova N.P., van der Waal M.S., Patnaik D., Lens S.M., Higgins J.M. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr. Biol. 2011;21(12):1061–1069. doi: 10.1016/j.cub.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maison C., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30(3):329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 79.Muchardt C., Guilleme M., Seeler J.S., Trouche D., Dejean A., Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002;3(10):975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamagishi Y., Honda T., Tanno Y., Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330(6001):239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]


