Abstract
Subcellular localization and translation of messenger RNAs are essential for the regulation of neuronal development and synaptic function. As post-transcriptional regulators, microRNAs (miRNAs) have been emerging as central players in the development and maturation of the nervous system. Recent discoveries reveal the critical functions of miRNAs in the axon of neurons via multiple pathways of molecular regulation. Here, we introduce methods for isolating axonal miRNAs and review recent findings on the localization and function as well as regulatory mechanism of axonal miRNAs during axon development.
Keywords: axonal miRNA, compartmentalized culture, axon development, RNA-binding protein
Introduction
Neurons are highly polarized cells that possess dendrites with vast, complicated spines to accept information and axons extending very distally to transmit signals (Martin and Ephrussi, 2009; Jung et al., 2012). Distinct subpopulations of messenger RNAs (mRNAs) have been identified in axons by high-throughput technologies such as microarrays and RNA sequencing (Andreassi et al., 2010; Zivraj et al., 2010; Gumy et al., 2011; Briese et al., 2015). Moreover, the local translation of mRNAs in axons has been found to be essential for functions such as axon elongation, regeneration, and viability (Cox et al., 2008; Yoo et al., 2009; Jung et al., 2014; Batista and Hengst, 2016; Tasdemir-Yilmaz and Segal, 2016).
As a class of small, non-coding RNA molecules, microRNAs (miRNAs) have been revealed to be involved in multiple biological processes through post-transcriptional regulation (Giraldez et al., 2005; McNeill and Van Vactor, 2012). Recent studies have demonstrated that miRNAs are distributed in the different subcellular compartment of neurons. For instance, miR-124 displays a soma-restricted pattern and regulates axonal pathfinding by targeting transcription factor (Kye et al., 2007; Baudet et al., 2012), suggesting a global and large change of gene expression mediated by miRNA in the cell body. However, some miRNAs are found to be spatially localized in the distal compartment of neurons such as the axon (Natera-Naranjo et al., 2010), allowing for the rapid and precise control of local mRNA translation without conveying signals to the soma to regulate transcription and mRNA translation as well as protein transport to axons.
Compartmentalized culture system is an efficient platform to separate axons from neuronal cell bodies and obtain pure axonal samples, which is recently adopted to explore the distribution of miRNAs in the axon of neurons. Several studies detected miRNA machinery proteins and miRNAs in the axon of distinct neurons in both the central nervous system and the peripheral nervous system by using compartmentalized culture system such as Campenot chamber and microfluidic chamber (Hengst et al., 2006; Kaplan et al., 2013; Hancock et al., 2014; Sasaki et al., 2014; Phay et al., 2015). In this review, we first introduce methods for isolating axonal miRNAs (Figure 1) and discuss the advantage and disadvantage of these methods (Table 1). Then, we summarize recent studies for the localization and function as well as regulatory mechanism of axonal miRNAs during axon development.
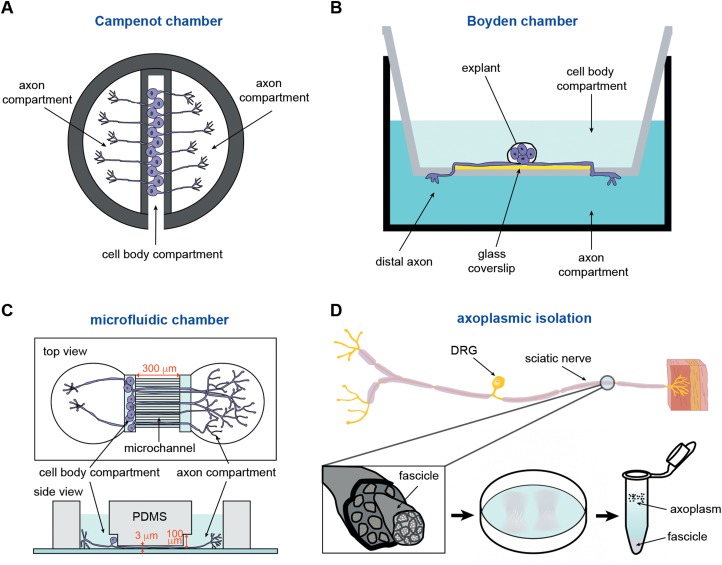
Figure 1.
Methods for isolating axonal miRNAs. (A) The Campenot chamber includes a scaffold made of Teflon, which is tightly adhered to a glass coverslip through silicone grease. The original three-chamber system consists of a central compartment and two side compartments. The dissociated neurons are plated in the central compartment. After several days in culture, only the long axons are able to pass through the silicone grease to both side compartments. (B) The Boyden chamber consists of a hollow plastic chamber sealed with a porous membrane containing pores of various sizes, allowing the motile cells to move to the other side. Explants or dissociated neurons are plated on a glass coverslip that is placed on the top of the microporous membrane. The growing axons cross through the membrane after several days. (C) The microfluidic chamber utilizes the replica-moulded transparent polydimethylsiloxane (PDMS) to establish a multi-compartment platform for cell culture. The chamber consists of separate compartments for the cell body and the axon, with microchannels (300 μm) linking the two compartments (top view). The cell body compartment is 100 μm high and is used for plating of neurons, whereas the microchannels are 3 μm in height and used for axon growth (side view). (D) The fascicles of axoplasm are mechanically separated from dissected sciatic nerve and incubated with a hypotonic solution (0.2× phosphate-buffered saline), either retaining intact axons or destroying Schwann cells. The ‘cloudy’ fascicles are incubated with the hypotonic solution for 2 h. After washing several times with the hypotonic solution, the axoplasm is eventually obtained in the supernatant of the solution (1× phosphate-buffered saline).
Table 1.
Comparison of different methods for isolating axonal miRNAs.
| Method | Advantage | Disadvantage | Optimal application |
|---|---|---|---|
| Campenot chamber | High purity, good for fluidic separation | Not easily manageable, low yield of axonal RNA | Cultured cells |
| Boyden chamber | Easily manageable, high yield of RNA | Not good for fluidic separation and live cell imaging | Cultured explants/cells |
| Microfluidic chamber | Easily manageable, high purity, very good for fluidic separation, good for live cell imaging | Low yield of axonal RNA | Cultured cells |
| Axoplasmic isolation | High yield of axonal RNA, good in vivo system | Not easily manageable | In vivo preparation of RNA granules |
Methods for isolating axonal miRNAs
Campenot chamber
The Campenot chamber is the first device applied in culture for the compartmentalization of the cell body and axon (Campenot, 1977). A scaffold made of Teflon (a material known for its use as a non-stick coating for cooking pans) is tightly coated on a glass coverslip through silicone grease (Kim and Jung, 2015). The original three-section chamber consists of a central compartment and two side compartments (Figure 1A). The dissociated neurons are plated in the central compartment. After several days in culture, only the long axons are able to pass through the silicone grease to both side compartments. The Campenot chamber is mostly used to study target-derived neurotrophin-initiated signalling, as well as retrograde signalling during axon development, neuronal survival, and synapse formation (Aschrafi et al., 2008; Tasdemir-Yilmaz and Segal, 2016). Using real-time quantitative reverse transciption polymerase chain reaction (qRT-PCR), a previous study has identified mRNA of cytochrome c oxidase IV (COXIV) and miR-338 in the axon of cultured sympathetic neurons (Aschrafi et al., 2008), supporting the utility of the Campenot chamber for detecting the local accumulation of mRNA and its corresponding regulators such as miRNAs. Importantly, this chamber is good for fluidic separation, which allows the central compartment and two side compartments selectively treated with drugs and siRNAs without interference on each other. However, there are some limitations for the Campenot chamber, such as low yield of the RNA and the difficulty of preparing the chamber (Table 1). A stable and complete seal formed by silicone grease between the Teflon barrier and the coverslip is a key step to limit the axons that can pass under the barrier, whereas an incomplete seal causes the entry of more axons and even leads to contamination with cell bodies.
Boyden chamber
The Boyden chamber is another device for compartmentalization of neurons, and it has previously been widely used in the cell migration assay to evaluate the motile activity of tumour cells (Kim and Jung, 2015). The classic Boyden chamber consists of a hollow plastic chamber sealed with a porous membrane containing pores of various sizes, allowing the motile cells to migrate to the other side (Figure 1B). For separation of axons from cell bodies, explants or dissociated neurons are plated on a glass coverslip that is placed on the top of the microporous membrane. The growing axons cross through the membrane after several days. Axons are harvested by scraping the underside of the membrane and can be used for quantitative analysis of miRNA by qRT-PCR (Kye et al., 2014). The advantages of the Boyden chamber are the simple culture of neuronal processes even from explants and the easy collection of axonal samples by scraping (Table 1). However, the Boyden chamber is not suitable for morphological experiments because the axon grows on the microporous membrane. The dendritic continuation of axonal sample is not able to be avoided due to the relatively short distance of membrane barrier between cell body compartment and axon compartment (Table 1).
Microfluidic chamber
The microfluidic chamber is currently a widely employed platform for the isolation of axonal fractions (Taylor et al., 2005). This method utilizes a replica-moulded transparent PDMS to establish a multi-compartment platform for cell culture. The cell body and axon of cultured neurons are separated by the physical compartments within the microchannels (Figure 1C). The dissociated neurons are plated in the cell body compartment of the microfluidic chamber, and the axons cross microchannels to reach the axon compartment after 2−4 days. The longer length of microchannels could efficiently avoid the pollution of dendritic processes into axon compartment (Taylor et al., 2005). Moreover, neuronal culture in a microfluidic chamber can be maintained for more than 3 weeks, allowing the investigation of synapse functions such as synapse formation and synaptic transmission (Jung et al., 2012). Commercial moulds are available with various sizes of microchannels for different types of neurons from the central or peripheral nervous system. More importantly, the transparency of microfluidic chamber is highly suitable for live cell imaging of axonal transport and for calcium imaging (Chen et al., 2012; Su et al., 2013) (Table 1). Similar to the Campenot chamber, the cell body compartment and axon compartments of this chamber could be separately treated. The major limitation of microfluidic chamber as well as Campenot chamber is the relatively low level of total RNA, which makes it difficult to perform genome-wide analysis (Table 1). More recently, an approach of highly sensitive sequencing for profiling small RNAs by optimizing the condition for amplification step (Yang et al., 2016) facilitates the investigation of the microRNome with small amount of RNA from the axon.
Axoplasmic isolation
In addition to in vitro systems of compartmentalized culture, the method for preparing axoplasm from the sciatic nerve has recently been improved. The fascicles of axoplasm are mechanically separated from dissected sciatic nerve and incubated in hypotonic solution, either retaining intact axons or destroying Schwann cells (Rishal et al., 2010) (Figure 1D). This modified procedure largely reduces non-neuronal contamination of the axoplasm and facilitates the investigation of proteome as well as the whole transcriptome. More interestingly, a recent study used RNA sequencing to reveal the dynamic alteration of axoplasmic miRNAs after sciatic nerve injury (Phay et al., 2015). Hence, this method for extraction of whole axoplasmic RNA will enable more efficient understanding regarding the axonal profile and dynamic expression level of local transcripts and their regulators such as miRNAs (Table 1). This approach has also been proposed for use in the characterization of axoplasmic miRNA profiles in various neurodegenerative diseases, such as Alzheimer's and Parkinson's diseases, which are related to defects in axonal transport, allowing further study of the regulatory roles of non-coding RNAs in these diseases (Eacker et al., 2009).
Localization and function of axonal miRNAs in different types of neurons
Over the last two decades, high-throughput technologies such as microarray and next-generation RNA sequencing have been developed to provide large-scale and quantitative analysis of genome and transcriptome (Chi et al., 2009; Phay et al., 2015). The data generated by technologies including microarrays and RNA sequencing have been utilized to systematically explore the profile of miRNAs and the quantitative relationship between miRNAs and transcripts. Due to different types of neurons and developmental stages, as well as various compartmentalized culture systems, profiles of axon-enriched miRNAs as reported in recent studies display distinct patterns (Natera-Naranjo et al., 2010; Hancock et al., 2014; Sasaki et al., 2014; Phay et al., 2015; Zhang et al., 2015b) (Table 2). Compared with the Boyden chamber, profiles of axonal miRNAs from the Campenot chamber and the microfluidic chamber are more comparable because of their similar architecture of compartmentalized culture (Figure 1). However, the precise localization of miRNA in the axon needs the confirmation with in situ hybridization and qRT-PCR.
Table 2.
High-throughput screening of axonal miRNAs.
| Species | Neuron type | Age | Number of axonal miRNAs | Highly axon-enriched miRNAs | Cultured method | Reference |
|---|---|---|---|---|---|---|
| Rat | Sympathetic neuron | P3 | 130 | miR-15b, miR-16, miR-204, miR-221 | Campenot chamber | Natera-Naranjo et al. (2010) |
| Mouse | Cortical neuron | E16 | 105 | miR-361, miR-532, miR-685, miR-720, miR-181a-1*, miR-709, miR-134 | Explant | Sasaki et al. (2014) |
| Mouse | Primary sensory neuron | E13.5 | 61 | miR-24, miR-191, miR-132, miR-138 | Microfluidic chamber | Hancock et al. (2014) |
E, embryonic day; P, postnatal day.
miRNAs in sympathetic axons
miR-338 is first found to be localized in the axon of sympathetic neuron and playing a key role in regulating mitochondrial function and axon growth (Aschrafi et al., 2008, 2012). A recent study shows the presence of approximately 130 miRNAs in the axon by using microarrays from the distal axons of cultured sympathetic neurons within the Campenot chamber (Natera-Naranjo et al., 2010) (Table 2). Quantitative detection by qRT-PCR further confirms that miR-16 and other 16 miRNAs are abundant in the axon of sympathetic neuron (Natera-Naranjo et al., 2010). The criteria by which a miRNA is defined as the axon-enriched miRNA include its abundance in the axon at a certain ratio higher than that in the cell body. Based on the average ratio of axonal enrichment by microarray analysis, most miRNAs display a similar abundance between the cell body and the axon. However, some miRNAs, such as miR-204, miR-221, miR-15b, and miR-16, are highly distributed in the axon (Table 2), whereas miR-297, miR-206, and miR-124a are mainly localized in the cell body (Natera-Naranjo et al., 2010). Although occasional inconsistencies in microarray and qRT-PCR analyses were existed for some miRNAs such as miR-329, miR-16 and miR-221 have been further validated as axon-enriched miRNAs by fluorescence in situ hybridization (FISH) and qRT-PCR (Natera-Naranjo et al., 2010). Subsequent investigation further demonstrates that axonal miR-16 contributes to axon growth by regulating the local activity of protein synthetic system, suggesting axonal miRNA-mediated effects via regulating translational machinery (Kar et al., 2013).
miRNAs in cortical axons
miR-9 has been shown to be localized in the axon of cortical neuron and to regulate axon growth and branching either in cultured microfluidic system or under the electroplated system of E14.5 brain in vivo, suggesting a crucial role of axonal miR-9 during axon development (Dajas-Bailador et al., 2012). Recently, six miRNAs (miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92) from the miR-17–92 cluster have been reported to be localized in the axon of cortical neuron and to contribute to axon outgrowth (Zhang et al., 2013).
Microarray technology is applied for screening axonal miRNAs in cultured cortical neurons. Embryonic explants are adopted to analyse the axonal distribution of miRNAs in the cortical neuron (Sasaki et al., 2014). By using neurosphere culture and TaqMan assays, seven miRNAs including miR-361, miR-532, miR-685, miR-720, miR-134, miR-181a-1*, and miR-709 have been detected to be enriched in the axon of cortical neuron (Sasaki et al., 2014) (Table 2). Although the dendritic localization of miR-134 and its role in synaptic plasticity has been established (Schratt et al., 2006), the function and mechanism of axonal miR-134 in the cortical neuron is elusive. Recently, miR-134 has been reported to be localized in the growth cone of Xenopus spinal neuron and involved in the chemotropic guidance of growth cone (Han et al., 2011), providing a possible role of axonal miR-134 in the axon guidance of cortical neuron. Similarly, despite unknown function of miR-181a-1* during axon development, miR-181d, another miR-181 family member, is enriched in the axon of dorsal root ganglion (DRG) neuron and involved in axon elongation by targeting MAP1B mRNA (Wang et al., 2015). Interestingly, MAP1B mRNA is also localized in the axon of cortical neuron (Dajas-Bailador et al., 2012), implying the potential role of axon-enriched miR-181a-1* during axon development in the cortical neuron.
Another recent microarray analysis reveals that axonal application of chondroitin sulphate proteoglycans (CSPGs), extracellular matrix molecules, exhibits an inhibitory effect on the axon growth of cortical neuron and alters the profile of axonal miRNAs (Zhang et al., 2015b). miR-29c, one of nine axonal miRNAs induced by CSPGs, inhibits the axon growth of cortical neurons in microfluidic culture (Zhang et al., 2015b). Future investigation is required for understanding the function of other miRNAs induced by axonal application of CSPGs in the cortical neuron.
miRNAs in the axon of primary sensory neuron
Appropriate projection of peripheral nerves is important for the establishment of functional connections between sensory neurons and skin or muscle (Kaplan et al., 2013; Chiu et al., 2014; Iyer et al., 2014). Although distinct molecular pathways are involved in accurate projection to peripheral tissues, direct evidence showing the miRNA-mediated post-transcriptional regulation of this process is rare. Conditional knockout of Dicer at embryonic day 9.5 (E9.5) of neural crest-derived cells causes aberrant axon trajectories in peripheral targets at E12.5 and E13.5, indicating that the Dicer-mediated maturation of miRNAs is partially necessary for the axon development in vivo (Hancock et al., 2014).
In the microfluidic culture of E13.5 DRG explants, the profile of axonal miRNAs assessed using the rodent TaqMan low density miRNA array shows that 61 of 375 miRNAs are distributed in the axon (Table 2) and that 26 miRNAs are axon-enriched by more than 1.5-fold (Hancock et al., 2014). Among these axon-enriched miRNAs, miR-132, miR-155, miR-365, and miR-410 were likely Dicer-regulated (Hancock et al., 2014), providing a possibility of these miRNAs exhibiting inhibitory effects on local transcripts. Furthermore, the axonal enrichment of miR-132 is the highest, with its level in axons at ~13 times higher than that in cell bodies, while the level of pre-miR-132 in axons is also enriched by 45-fold compared to that in cell bodies. The subsequent experiment further demonstrates that miR-132 contributes to axon extension as a positive regulator (Hancock et al., 2014). Axon-enriched and Dicer-regulated miR-181d has recently been reported to be involved in the axon elongation of primary sensory neuron (Zhao et al., 2010; Wang et al., 2015). Although the same type of neurons was used for the detection of axonal miRNAs, the microarray with 375 miRNAs missed some miRNAs, such as miR-181d.
Thus, the profile of the whole microRNome in the axon requires relatively complete miRNA array, more sensitive RNA-sequencing technology, and advanced bioinformatic tools to identify miRNAs.
Regulatory mechanisms of axonal miRNAs during axon development
Downstream targets of axonal miRNAs
Kaplan and colleagues reported that axonal miR-338 targets two mitochondrial mRNAs, COXIV (Aschrafi et al., 2008) and ATP synthase (ATP5G1) (Aschrafi et al., 2012) (Table 3), thereby locally regulating the metabolic rate and axon growth of sympathetic neuron (Kaplan et al., 2013). A recent study shows that miR-16 is one of the most abundant miRNAs in the axon of sympathetic neuron (Natera-Naranjo et al., 2010). Subsequent research has identified that miR-16 binds to the 3′ UTR of mRNAs, including those of the eukaryotic translation initiation factors eIF2B2 and eIF4G2 (Table 3); axonal application of pre-miR-16 decreases the axonal mRNA and protein levels of eIF2B2 and eIF4G2 and inhibits axon growth (Kar et al., 2013). In addition, inhibiting the local synthesis of eIF2B2 and eIF4G2 effectively impairs the system of local protein synthesis (Kar et al., 2013). These findings demonstrate that axon growth is regulated by local miRNAs through targeting key components of the biological processes.
Table 3.
Function and downstream targets of axonal miRNAs.
| Axonal miRNA | Species | Neuron type | Age | Function | Targeted mRNA | Cultured method | Reference |
|---|---|---|---|---|---|---|---|
| miR-338 | Rat | Sympathetic neuron | P3 | Mitochondrial oxidative phosphorylation | COXIV | Campenot chamber | Aschrafi et al. (2008) |
| miR-16 | Rat | Sympathetic neuron | P3 | Axon outgrowth | eIF2B2, eIF4G2 | Campenot chamber | Kar et al. (2013) |
| miR-181d | Rat | Primary sensory neuron | E13.5 | Axon elongation | MAP1B, calmodulin | Microfluidic chamber | Wang et al. (2015) |
| miR-132 | Mouse | Primary sensory neuron | E13.5 | Axon extension | Rasal | Microfluidic chamber | Hancock et al. (2014) |
| miR-9 | Mouse | Cortical neuron | E17 | Axon extension/branching | MAP1B | Microfluidic chamber | Dajas-Bailador et al. (2012) |
| miR-17-92 | Rat | Cortical neuron | E18 | Axon outgrowth | PTEN | Microfluidic chamber | Zhang et al. (2013) |
| miR-29c | Rat | Cortical neuron | E18 | Axon outgrowth | ITBG1 | Microfluidic chamber | Zhang et al. (2015b) |
COXIV, cytochrome c oxidase IV; eIF2B2, eukaryotic translation initiation factors 2B, subunit 2β; eIF4G2, eukaryotic translation initiation factors γ4, subunit 2; E, embryonic day; ITBG1, integrin, β1 subunit; MAP1B, microtubule-associated protein 1B; P, postnatal day; PTEN, phosphatase and tensin homology; Rasa1, RAS p21 protein activator 1.
miR-9 has been reported to be involved in the axon growth of cortical neuron by regulating axonal translation of MAP1B mRNA (Dajas-Bailador et al., 2012) (Table 3). Co-localization of miR-9 with the transcript of MAP1B by FISH further suggests that miR-9 locally targets MAP1B mRNA in the distal axon of cultured cortical neurons. Moreover, application of target protector (TP), which specifically blocks the interaction between miR-9 and the 3′ UTR of MAP1B mRNA, significantly increases the axon length and reduces the number of axon branches both in a compartmental culture of cortical neurons and in utero electroplated brain at E14.5 (Dajas-Bailador et al., 2012), revealing a local role of miRNA in microtubule stabilization. Interestingly, recent research has identified miR-181d as an axon-enriched miRNA that negatively regulates axon elongation by locally targeting two mRNAs, MAP1B and calmodulin, in the microfluidic culture of embryonic DRG neurons (Wang et al., 2015) (Table 3). These two studies highlight the importance of cytoskeletal mechanisms underlying local miRNA activity during axon development, further suggesting that one transcript could be co-targeted by multiple miRNAs in the axon of the central or peripheral nervous system.
As a typical example of a polycistronic miRNA cluster, the miR-17–92 cluster has been extensively investigated in tumours (Mi et al., 2010). Overexpression of the miR-17–92 cluster enhances axon outgrowth through downregulation of the PTEN protein level as well as attenuation of the mTOR signalling pathway in the axon (Zhang et al., 2013) (Table 3). Although substantial evidence suggests that PTEN is one of the downstream targets for the miR-17–92 cluster (Olive et al., 2009) and plays a critical role during axon regeneration (Liu et al., 2010), direct evidence supporting the existence of PTEN transcript in the axon is not provided via the high-throughput screening of axonal transcripts (Andreassi et al., 2010; Zivraj et al., 2010; Gumy et al., 2011; Briese et al., 2015). The localization and local translation of PTEN mRNA in the axon and the precise mechanism underlying the regulation of axon growth by miR-17–92 require further investigation.
As another axon-enriched miRNA in embryonic DRG neurons, miR-132 positively regulates axon outgrowth by locally targeting the mRNA of Rasal1, a Ras GTPase activator (Hancock et al., 2014) (Table 3). miR-132, as well as its precursor pre-miR-132, has been found to be localized in the axon of embryonic DRG neurons (Hancock et al., 2014). The amount of miR-132 in the axon is progressively increased during the embryonic development of the DRG in vivo, further suggesting that miR-132 is a positive regulator of the axon extension in DRG neurons (Hancock et al., 2014). Interestingly, miR-181d negatively regulates axon elongation by affecting the axonal translation of MAP1B and calmodulin mRNAs (Table 3) (Wang et al., 2015), showing that different miRNAs cooperatively regulate axon growth by targeting diverse downstream molecules.
Extrinsic signals involved in axonal miRNA-mediated axon development
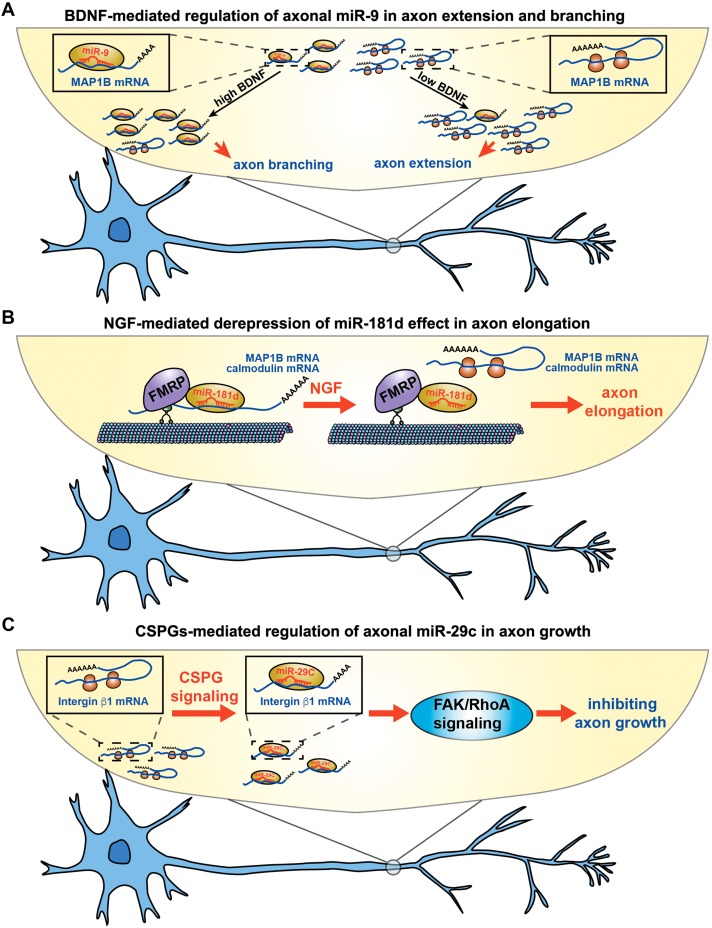
Extrinsic signals are essential for local mRNA translation during axon development in the central and peripheral nervous systems (Zivraj et al., 2010; Jung et al., 2014). It has been reported that miR-9 inhibits the development of cortical neuron, including axon extension and subsequent axon branching, by locally regulating the synthesis of MAP1B (Dajas-Bailador et al., 2012). Axonal application of a low concentration of brain-derived neurotrophic factor (BDNF) (10 ng/ml) for a short time (2 h) reduces the amount of miR-9 and increases axon growth by promoting local synthesis of MAP1B, whereas prolonged BDNF treatment (48 h) at a high concentration (100 ng/ml) elevates the amount of miR-9 in the axon and increases axon branching by inhibiting axonal synthesis of MAP1B (Dajas-Bailador et al., 2012) (Figure 2A). Therefore, miR-9 is a central component of BDNF downstream signalling during axon growth and branching, although evidence showing how BDNF changes the level of miR-9 in the axon is lacking. The distribution of pre-miRNAs in sensory axons (Kim et al., 2015) offers the possibility of RNA-binding protein (RBP)-mediated regulation for local biogenesis of miRNA (Huang et al., 2012), responding to signals for rapidly changing the level of miRNAs in axons. However, retrograde signalling mediated by target-derived factors may induce cell bodies to produce more pre-miRNAs or mature miRNAs and deliver to the axon. Future investigations into the BDNF-induced change of miR-9 and its precursor in the axon will improve the understanding of regulatory mechanisms for axonal miRNAs.
Figure 2.
Extrinsic signals are involved in the miRNA-mediated axon development. (A) BDNF-mediated regulation of axonal miR-9 in axon extension and branching by translational control of MAP1B mRNA. Axonal treatment with a low concentration of BDNF reduces the level of axonal miR-9 and increases axon extension by promoting local synthesis of MAP1B, whereas prolonged treatment with a high concentration of BDNF elevates the level of axonal miR-9 and increases axon branching by repressing axonal synthesis of MAP1B. (B) NGF-dependent derepression of axonal miR-181d affects MAP1B and calmodulin synthesis in axon elongation. NGF treatment in axons triggers the dissociation of MAP1B and calmodulin mRNAs from the miR-181d-repressing granules, thereby promoting axon elongation by increasing the axonal synthesis of MAP1B and calmodulin. (C) CSPGs-mediated regulation of axonal miR-29c in axon growth by translational control of integrin β1 mRNA. Axonal application of CSPGs elevates the level of axonal miR-29c and decreases local synthesis of integrin β1, thereby inhibiting the axon outgrowth in cortical neuron through downstream FAK/RhoA signalling.
Another axonal miRNA that responds to local signalling is miR-181d, which is involved in the nerve growth factor (NGF)-mediated axon elongation of embryonic DRG neurons (Wang et al., 2015). Interestingly, local synthesis of MAP1B and calmodulin in the axon of primary sensory neuron is regulated by miR-181d and NGF treatment. Unlike the BDNF-mediated regulation of miR-9 in the axon of cortical neuron, NGF treatment does not change the level of miR-181d in either the cell body or the axon, instead triggering the dissociation of MAP1B and calmodulin mRNAs from the miR-181d-repressing granules, thereby promoting axon elongation by controlling the local synthesis of MAP1B and calmodulin (Figure 2B). The precise mechanism by which NGF regulates the dissociation of targets from miR-181d-repressing granules requires further elucidation, although FMRP (fragile X mental retardation protein, encoded by Fmr1) has been suggested as a scaffold protein to connect transcripts with the miRNA-associated RNA-induced silencing complex (RISC) (Muddashetty et al., 2011). However, in retina, TGF-β induces the expression of miR-181a/b in the cell body and then inhibits axon growth through targeting ERK2 mRNA (Carrella et al., 2015). These studies indicate that extrinsic signals regulate the miRNA-mediated function by different ways even though influencing the member from same miRNA family.
Axonal miRNAs have also been found to be locally involved in extracellular matrix-induced axon growth (Zhang et al., 2015b). Axonal application of CSPGs elevates the level of miR-29c in the axon of cortical neuron and decreases axonal synthesis of integrin β1 mRNA, thereby inhibiting axon outgrowth through the FAK/RhoA signalling pathway (Zhang et al., 2015b) (Table 3 and Figure 2C). Moreover, increased synthesis of cGMP induced by sildenafil, an inhibitor of phosphodiesterase type 5, reverses the effects of CSPGs on the majority of altered miRNAs, including miR-29c, and promotes axon outgrowth (Zhang et al., 2015b). Furthermore, axonal application of CSPGs also induces the decreased level of axonal miRNAs such as miR-132, which is enriched in developing axon and promotes axon extension (Hancock et al., 2014). Therefore, rapid recovery of these miRNAs in the axon may facilitate growing axon to overcome the inhibitory action of extrinsic signals. The miRNAs originated from local biogenesis and remote supply of cell bodies are possibly required for the maintenance of these axonal miRNAs such as miR-132. Importantly, this study suggests that extrinsic signals other than neurotrophins also control the miRNA-mediated signalling pathways to regulate axon development.
RBP-mediated delivery of axonal miRNAs
RBPs have been suggested to play a pivotal role in the multiple aspects of RNA processing including RNA splicing and subcellular localization (Darnell, 2013). Several RBPs have been detected in the neuronal axon and implicated in the stabilization and localization of mRNAs (Darnell, 2013; Hornberg and Holt, 2013; Wang et al., 2015). RBPs are also reported to participate in almost every step of miRNA biogenesis and maturation in concert with the RISC complex (Muddashetty et al., 2011; Hornberg and Holt, 2013). Unlike the relatively small extent of the somatodendritic region, a long distance is existed between the cell body and the distal axon. Recently, FMRP has been revealed to contribute to axonal delivery of miR-181d in embryonic DRG neurons (Wang et al., 2015). In the microfluidic culture of DRG neurons, FMRP deficiency causes a decrease in the level of miR-181d in the axon but not in the cell body, indicating that FMRP is required for axonal localization but not the biogenesis of miR-181d. The association with the RISC complex and motor proteins provides the possibility that FMRP enables the long-distance delivery and targeting of the miRNA−RISC complex to the distal axon, although some evidence shows local assembly of the RISC complex in the dendrite of hippocampal neuron (Huang et al., 2012). Furthermore, ~40% of miR-181d co-localized with FMRP in the axon of DRG neuron, suggesting that miRNA granules are heterogeneous similar to mRNA granules. Thus, high-throughput technologies such as immunoprecipitation of RBPs followed by microarrays (RIP-chips) or high-throughput sequencing of RNAs by crosslinking immunoprecipitation (HITS-CLIP) specifically in the axon will facilitate our understanding of the intricate network of miRNAs and RBPs in the axon (Chi et al., 2009; Kim and Jung, 2015).
More recently, FMRP has been reported to block non-telomeric isoform of telomere repeat-binding factor 2 (TRF2-s, a novel RNA- and FMRP-binding protein)-mediated delivery of mRNAs in the axon of cortical neuron (Zhang et al., 2015a). However, TRF2-s is almost undetectable in embryonic (our unpublished data) and adult DRG neurons (Li et al., 2016), indicating that FMRP plays distinct roles in the central and peripheral nervous systems. A very recent study identifies a splicing factor, SFPQ, coordinates neurotrophin-dependent and anterograde transport of axonal transcripts including Bclw and LmnB2 mRNAs that are involved in axon survival (Cosker et al., 2016). Indeed, both Bclw and LmnB2 mRNAs are specifically associated with SFPQ rather than with FMRP in the axon, further supporting the existence of selective mechanisms for the axonal targeting of transcripts and related miRNAs, as well as the heterogeneity of mRNA granules.
Concluding remarks
Subcellular localization and translation of mRNAs in the axon are essential for axon elongation, branching, and survival. Recent studies have detected the vast diversity of miRNAs compartmentalized in the axon of different neurons and have identified miRNA-based functions in the translational control of local protein synthesis. However, the localization, function, and regulatory mechanism of numerous miRNAs in the axon remain unknown. Advanced technologies such as high-throughput RNA sequencing for axonal RNAs present at low levels may provide an integrated map for the miRNA-mediated regulation of mRNAs during axon development. Future investigations will be needed to explore the miRNA-mediated pathways regulated by extrinsic signals and to determine the roles of RBP-mediated interactions between miRNAs and mRNAs in the axon for functional coordination and axonal targeting of miRNAs. Increasing numbers of studies on miRNA-mediated regulation of mRNAs will greatly expand our understanding of the local control during axon development and even during axon regeneration in neurodegenerative diseases.
Acknowledgements
We thank Lei Diao for assistance with the design and accomplishment of schematics in this paper.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31271141 and 31330046), National Basic Research Program of China (2014CB942802), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000), China Postdoctoral Science Foundation (2012M510096 and 2013T60469), Postdoctoral Foundation from Shanghai Institutes for Biological Sciences (2012KIP505), and Shanghai Postdoctoral Science Foundation (12R21417300).
References
- Andreassi C., Zimmermann C., Mitter R., et al. (2010). An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat. Neurosci. 13, 291–301. [DOI] [PubMed] [Google Scholar]
- Aschrafi A., Kar A.N., Natera-Naranjo O., et al. (2012). MicroRNA-338 regulates the axonal expression of multiple nuclear-encoded mitochondrial mRNAs encoding subunits of the oxidative phosphorylation machinery. Cell. Mol. Life Sci. 69, 4017–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschrafi A., Schwechter A.D., Mameza M.G., et al. (2008). MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J. Neurosci. 28, 12581–12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista A.F., and Hengst U (2016). Intra-axonal protein synthesis in development and beyond. Int. J. Dev. Neurosci. 55, 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudet M.L., Zivraj K.H., Abreu-Goodger C., et al. (2012). miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nat. Neurosci. 15, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese M., Saal L., Appenzeller S., et al. (2015). Whole transcriptome profiling reveals the RNA content of motor axons. Nucleic Acids Res. 44, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campenot R.B. (1977). Local control of neurite development by nerve growth factor. Proc. Natl Acad. Sci. USA 74, 4516–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrella S., Barbato S., D'Agostino Y., et al. (2015). TGF-β controls miR-181/ERK regulatory network during retinal axon specification and growth. PLoS One 10, e0144129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.Q., Wang B., Wu C., et al. (2012). Endosome-mediated retrograde axonal transport of P2X3 receptor signals in primary sensory neurons. Cell Res. 22, 677–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi S.W., Zang J.B., Mele A., et al. (2009). Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H., Alqadah A., and Chang C (2014). The role of microRNAs in regulating neuronal connectivity. Front. Cell. Neurosci. 7, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker K.E., Fenstermacher S.J., Pazyra-Murphy M.F., et al. (2016). The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat. Neurosci. 19, 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox L.J., Hengst U., Gurskaya N.G., et al. (2008). Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat. Cell Biol. 10, 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajas-Bailador F., Bonev B., Garcez P., et al. (2012). microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat. Neurosci. 15, 697–699. [DOI] [PubMed] [Google Scholar]
- Darnell R.B. (2013). RNA protein interaction in neurons. Annu. Rev. Neurosci. 36, 243–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eacker S.M., Dawson T.M., and Dawson V.L (2009). Understanding microRNAs in neurodegeneration. Nat. Rev. Neurosci. 10, 837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., et al. (2005). MicroRNAs regulate brain morphogenesis in zebrafish. Science 308, 833–838. [DOI] [PubMed] [Google Scholar]
- Gumy L.F., Yeo G.S., Tung Y.C., et al. (2011). Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA 17, 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Wen Z., Lynn R.C., et al. (2011). Regulation of chemotropic guidance of nerve growth cones by microRNA. Mol. Brain 4, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock M.L., Preitner N., Quan J., et al. (2014). MicroRNA-132 is enriched in developing axons, locally regulates Rasa1 mRNA, and promotes axon extension. J. Neurosci. 34, 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst U., Cox L.J., Macosko E.Z., et al. (2006). Functional and selective RNA interference in developing axons and growth cones. J. Neurosci. 26, 5727–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberg H., and Holt C (2013). RNA-binding proteins and translational regulation in axons and growth cones. Front. Neurosci. 7, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Ruiz C.R., Eyler E.C., et al. (2012). Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis. Cell 148, 933–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A.N., Bellon A., and Baudet M.L (2014). microRNAs in axon guidance. Front. Cell. Neurosci. 8, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., Gkogkas C.G., Sonenberg N., et al. (2014). Remote control of gene function by local translation. Cell 157, 26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., Yoon B.C., and Holt C.E (2012). Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat. Rev. Neurosci. 13, 308–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan B.B., Kar A.N., Gioio A.E., et al. (2013). MicroRNAs in the axon and presynaptic nerve terminal. Front. Cell. Neurosci. 7, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar A.N., MacGibeny M.A., Gervasi N.M., et al. (2013). Intra-axonal synthesis of eukaryotic translation initiation factors regulates local protein synthesis and axon growth in rat sympathetic neurons. J. Neurosci. 33, 7165–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., and Jung H (2015). Local protein synthesis in neuronal axons: why and how we study. BMB Rep. 48, 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.H., Kim P., Phay M., et al. (2015). Identification of precursor microRNAs within distal axons of sensory neuron. J. Neurochem. 134, 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kye M.J., Liu T., Levy S.F., et al. (2007). Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA 13, 1224–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kye M.J., Niederst E.D., Wertz M.H., et al. (2014). SMN regulates axonal local translation via miR-183/mTOR pathway. Hum. Mol. Genet. 23, 6318–6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.L., Li K.C., Wu D., et al. (2016). Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res. 26, 83–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Lu Y., Lee J.K., et al. (2010). PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat. Neurosci. 13, 1075–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K.C., and Ephrussi A (2009). mRNA localization: gene expression in the spatial dimension. Cell 136, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill E., and Van Vactor D (2012). MicroRNAs shape the neuronal landscape. Neuron 75, 363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., Li Z., Chen P., et al. (2010). Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc. Natl Acad. Sci. USA 107, 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty R.S., Nalavadi V.C., Gross C., et al. (2011). Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell 42, 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O., Aschrafi A., Gioio A.E., et al. (2010). Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA 16, 1516–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive V., Bennett M.J., Walker J.C., et al. (2009). miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 23, 2839–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phay M., Kim H.H., and Yoo S (2015). Dynamic change and target prediction of axon-specific microRNAs in regenerating sciatic nerve. PLoS One 10, e0137461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishal I., Michaelevski I., Rozenbaum M., et al. (2010). Axoplasm isolation from peripheral nerve. Dev. Neurobiol. 70, 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y., Gross C., Xing L., et al. (2014). Identification of axon-enriched microRNAs localized to growth cones of cortical neurons. Dev. Neurobiol. 74, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G.M., Tuebing F., Nigh E.A., et al. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439, 283–289. [DOI] [PubMed] [Google Scholar]
- Su Y.Y., Ye M., Li L., et al. (2013). KIF5B promotes the forward transport and axonal function of the voltage-gated sodium channel Nav1.8. J. Neurosci. 33, 17884–17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir-Yilmaz O.E., and Segal R.A (2016). There and back again: coordinated transcription, translation and transport in axonal survival and regeneration. Curr. Opin. Neurobiol. 39, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.M., Blurton-Jones M., Rhee S.W., et al. (2005). A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat. Methods 2, 599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Pan L., Wei M., et al. (2015). FMRP-mediated axonal delivery of miR-181d Regulates axon elongation by locally targeting Map1b and Calm1. Cell Rep. 13, 2794–2807. [DOI] [PubMed] [Google Scholar]
- Yang Q., Lin J., Liu M., et al. (2016). Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Sci. Adv. 2, e1501482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S., van Niekerk E.A., Merianda T.T., et al. (2009). Dynamics of axonal mRNA transport and implications for peripheral nerve regeneration. Exp. Neurol. 223, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Abdelmohsen K., Liu Y., et al. (2015. a). Novel RNA- and FMRP-binding protein TRF2-S regulates axonal mRNA transport and presynaptic plasticity. Nat. Commun. 6, 8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chopp M., Liu X.S., et al. (2015. b). MicroRNAs in the axon locally mediate the effects of chondroitin sulfate proteoglycans and cGMP on axonal growth. Dev. Neurobiol. 75, 1402–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ueno Y., Liu X.S., et al. (2013). The microRNA-17-92 cluster enhances axonal outgrowth in embryonic cortical neurons. J. Neurosci. 33, 6885–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Lee M.C., Momin A., et al. (2010). Small RNAs control sodium channel expression, nociceptor excitability, and pain thresholds. J. Neurosci. 30, 10860–10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivraj K.H., Tung Y.C., Piper M., et al. (2010). Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J. Neurosci. 30, 15464–15478. [DOI] [PMC free article] [PubMed] [Google Scholar]