Abstract
Type I interferon (IFN) production by the proper activation of nucleic acid sensors is essential for hosts to eliminate invading viruses. Among these sensors, RIG-I-like receptors (RLRs) are well-known viral RNA sensors in the cytoplasm that recognize the nonself signatures of viral RNAs to trigger IFN responses. Recent accumulating evidence has clarified that some specific and atypical self-RNAs also cause activation of RLRs independently of virus infection. Importantly, when RLR-activation by these RNAs or a conformational change via missense mutations is sustained, the resulting continuous production of type I IFN will lead to autoimmune disorders. We, herein, focus on autoimmune diseases caused by chronic activation of RLRs and discuss possible mechanisms of their onset.
Keywords: : RIG-I-like receptors (RLRs), interferon (IFN), autoimmune diseases
Introduction
Nucleic acid sensors sense viral invasion by the recognition of nonself motifs and/or abnormal localization of viral nucleic acids and trigger the induction of antiviral interferon (IFN) for the elimination. Three kinds of sensors, Toll-like receptors (TLRs) (Takeuchi and Akira 2010), RIG-I-like receptors (RLRs) (Yoneyama and others 2004, 2005), and cytosolic DNA sensors, such as cGAS (Sun and others 2013), have been identified as essential for the induction of type I IFNs (IFN-α, IFN-β), and deficiency in these sensors drastically increases the susceptibility to virus infection.
Transmembrane protein TLRs recognize viral nucleic acids in the endosomal compartment of immune cells and induce interferon. Especially, professional IFN-producing cells designated as plasmacytoid dendritic cells utilize TLR7 and TLR9 for eliciting the high production of IFN in response to endosomal single-stranded (ss) RNA and unmethylated CpG DNA, respectively (Diebold and others 2004; Hemmi and others 2000). TLRs are viral sensing systems that trigger IFN as an alert for surrounding cells before the viral infection is established in virus-invading cells. On the contrary, the other cytoplasmic sensors, RLRs, which consist of 3 RNA helicases, designated RIG-I, MDA5, and LGP2, and DNA sensors, such as cGAS, respectively, detect replication of intermediate RNA and DNA after the establishment of virus infection and trigger IFN responses to suppress and eliminate invading viruses. Detailed signaling pathways are indicated in Fig. 1. Among RLRs, RIG-I specifically recognizes nonself motifs of viral RNAs, including double-strand, 5′triphosphates, and absence of N1-2'O-methylation (Hornung and others 2006; Pichlmair and others 2006; Schlee and others 2009; Schuberth-Wagner and others 2015). MDA5 requires a long length of dsRNA (over 2 kbp) for its activation, and the filament formation (head-to-tail MDA5 oligomers) along dsRNA has been reported in the activation process (Kato and others 2008; Peisley and others 2011). In the case of cGAS, it detects cytoplasmic localization of dsDNA and also long ssDNA in some virus cases for IFN induction (Gao and others 2013; Herzner and others 2015; Sun and others 2013).
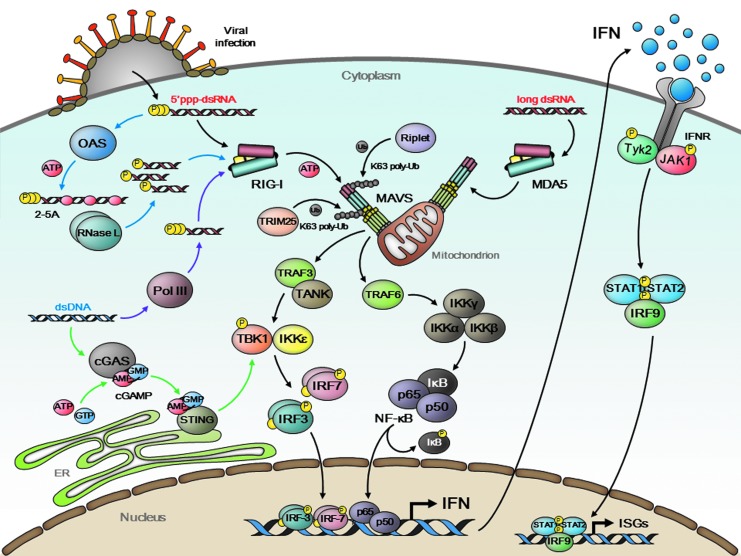
FIG. 1.
IFN production and signaling upon viral infection. Viral infection causes cytoplasmic exposure of viral nucleic acids. RIG-I and MDA5 are RNA helicases that directly bind to viral dsRNA species to trigger a downstream signal cascade (black arrows). Ubiquitin ligases, Riplet and TRIM25, mediate K63-linked polyubiquitination of RIG-I and facilitate its activation and self-oligomerization. MDA5 is activated by forming filamentous structure around dsRNA in an ubiquitin-independent manner. MAVS, a mitochondrial adaptor protein, associates with activated RIG-I/MDA5 with accompanying prion-like aggregation on the mitochondrial outer membrane, and further activates downstream molecules, including TRAF3/TANK, TRAF6, TBK1/IKKɛ, and IKKα/β/γ. IRF3/7 and NF-κB (p65 + p50) transcription factors are then translocated into the nucleus and undertaken IFN gene expression. Secreted IFNs bind to cognate IFN receptor (IFNR) expressed on neighboring cells and elicit JAK-STAT signal cascade to induce expression of hundreds of ISGs. OAS is an ISG-encoded antiviral enzyme that processes viral dsRNA into 2′,5′-linked oligoadenylate (2-5A) (blue arrows). This unique RNA is subsequently cleaved by RNase L into small product, which potentially activates RIG-I for secondary IFN induction. AT-rich viral dsDNA is a latent template of a DNA-dependent RNA polymerase, Pol III, to produce immunostimulatory 5′ppp-dsRNA (purple arrows). cGAS also binds to viral dsDNA and catalyzes cyclic GMP-AMP (cGAMP) synthesis (green arrows). This second messenger activates ER-associated STING for the IFN induction.
Secreted IFNs act both autocrinely and paracrinely by binding with receptors on the cell surface and promoting the expression of hundreds of antiviral IFN-stimulated genes (ISGs), including 2′-5′-oligoadenylate synthetases, RNase L, PKR, and IFITs, for the elimination of invading viruses (Fig. 1) (Diamond and Farzan 2013). This production of IFN is critical for the suppression of viruses, but tightly regulated to be transient by negative feedback mechanisms, since excess production is harmful to hosts.
Recently, several lines of evidence have shown that aberrant antiviral signaling from nucleic acid sensors or caused by a failure in the clearance of nucleic acids can result in the excess production of IFN, leading to disorders designated as interferonopathies. For instance, studies on Y-linked autoimmune accelerator (Yaa) mice, which harbor a duplication of the TLR7 gene, suggested its role in an amplification loop for the production of IFN and also the activation of B cells in autoimmune disorders such as systemic lupus erythematosus (SLE) (Pisitkun and others 2006; Deane and others 2007). Deficiency of DNases, including DNase I, DNase II, and TREX1 (DNase III), has been reported to cause abnormal accumulation of DNAs, which activates DNA sensors and leads to IFN signatures and autoimmune disorders (Ahn and others 2012, 2014; Jacob and others 2002; Kawane and others 2001, 2006). More recent studies demonstrated autoimmune diseases are also caused by abnormal IFN signaling by constitutive activation of cytoplasmic RNA sensors MDA5 and RIG-I in both ligand-dependent and ligand-independent manners. In this review, focusing on viral RNA sensors RLRs, we will provide an overview of recent studies on single-nucleotide polymorphisms (SNPs) in RLRs, mice with RLR mutations, and autoimmune disease patients with RLR mutations, as well as discuss how aberrant activation of RLRs possibly cause autoimmune symptoms.
SNPs of Viral RNA Sensors RLRs Are Involved in Susceptibility to Autoimmune Disorders
Since RLRs are essential viral RNA sensors, polymorphisms in RLR can be a risk factor for RNA viruses. For instance, a polymorphism in the IFIH1 gene encoding MDA5 was reported to be associated with the severity of enterovirus 71 infection (Cinek and others 2012). The involvement of RLR polymorphisms in the susceptibility to human diseases has also been reported, especially, in the case of autoimmune diseases. Among RLRs, several SNPs of the IFIH1 gene have been repeatedly identified in type I diabetes patients, suggesting a strong association of IFIH1 SNPs with the susceptibility to type I diabetes (de Azevedo Silva and others 2015; Liu and others 2009; Plagnol and others 2011; Smyth and others 2006; Zouk and others 2014). One SNP (rs1990760, A946T) shows the strongest correlation, and interestingly this SNP has also been identified in the case of other autoimmune diseases, including SLE, Graves' disease, multiple sclerosis (MS), rheumatoid arthritis, and psoriasis, and also in immunodeficiencies such as IgA deficiency (Bergamaschi and others 2010; Enevold and others 2014; Ferreira and others 2010; Molineros and others 2013; Robinson and others 2011; Sutherland and others 2007). Although many groups have also assessed the significance of RIG-I and LGP2 SNPs in the susceptibility to human diseases, such as SLE and MS, no significant difference has been reported so far. SNPs in an essential adaptor MAVS have also been examined and several groups have reported some correlation in the case of SLE and MS (Pothlichet and others 2011).
Discovery of Mice with an Ifih1 Missense Mutation (G821S)
Previous studies reported that human SNPs in IFIH1 encoding MDA5, such as rs1990760 (A946T), were correlated with an increase in the susceptibility to autoimmune disorders (Robinson and others 2011), such as SLE, suggesting that the atypical activation of RLR signaling may lead to SLE. Funabiki and others (2014) recently reported that mice with the Ifih1 missense mutation spontaneously developed lupus-like symptoms, including nephritis, systemic IFN signature, skin rash, and production of antinuclear and dsDNA antibody in the blood. This is the first study demonstrating that mutations in RLRs directly lead to an autoimmune disease.
The upregulation of type I IFNs, IFN-inducible genes, and inflammatory cytokines, including IL-6, IL-1β, and TNF-α, was detected in multiple organs of this MDA5 mutant mouse, reflecting the ubiquitous expression of MDA5. This missense mutation enhanced the basal activation level of IFN by MDA5, but abrogated responsiveness to viral infection as well as the ATPase activity induced by an MDA5 ligand dsRNA. More importantly, the autoimmune phenotype was not observed in the mouse background of Mavs−/−. These findings indicate that the mutant MDA5 may confer constitutive activity, rather than being hypersensitive to endogenous or viral RNA. Similar to Yaa mice with TLR7 gene duplication (Pisitkun and others 2006), a simple increase in the wild-type Ifih1 gene dosage was insufficient to cause spontaneous nephritis (Crampton and others 2012), suggesting that a dysregulation of MDA5 by a mutation may be essential for triggering autoimmunity.
Autoimmune Disease Patients with RLR Mutations
Discovery of MDA5 mutant mice in 2014 was followed by the discovery of mutations in the IFIH1 gene in patients with autoimmune diseases. The first evidence of human MDA5 mutations was reported in patients with Aicardi–Goutieres syndrome (AGS), an inflammatory disease that particularly affects the brain. After the report on AGS patients, mutations in the IFIH1 gene were also reported in 2 autoimmune diseases, SLE and Singleton–Merten syndrome (SMS). Details of each autoimmune disease related to RLR mutations are described below.
Aicardi–Goutieres Syndrome
Patients with the neuroinflammatory autoimmune disease AGS exhibit profound intellectual disabilities, dystonia and IFN signatures, and high lethality by the age of 5 years (approximately 20%). Detailed characterization of disease phenotypes in AGS patients revealed that approximately 30% of patients exhibited chilblains in addition to neurological phenotypes, and other symptoms, including nephritis, glaucoma, hypothyroidism, and cardiomyopathy, have also been observed in some cases, implicating phenotypic overlap with other autoimmune diseases such as SLE and SMS (Crow YJ 2013; Crow MK 2014).
AGS has been shown to develop from mutations in genes encoding nucleotide processing proteins, including TREX1, SAMHD1, ADAR1, RNASEH2A, RNASEH2B, and RNASEH2C (Crow and Rehwinkel 2009). Importantly deficiency in TREX1, RNaseH 2A, 2B, and 2C has been shown by gene targeting to accumulate abnormal DNAs and cause IFN signature through the activation of cytsolic DNA sensor cGAS (Ablasser and others 2014; Gray and others 2015; Mackenzie and others 2016; Pokatayev and others 2016) as described in Fig. 2. Recently, the IFIH1 gene encoding viral RNA sensor MDA5 was identified by 2 groups as the 7th responsible gene for AGS onset (Oda and others 2014; Rice and others 2014). Rice and others (2014) identified mutations in IFIH1 in AGS patients, which conferred the constitutive activity of MDA5, but occurred at different positions from the mouse Ifih1 mutation reported by Funabiki and others (2014). Human MDA5 mutants exhibited hyper-responsiveness to ligand stimulation, thereby suggesting the possible involvement of endogenous and/or viral RNA in the onset of AGS (Fig. 2).
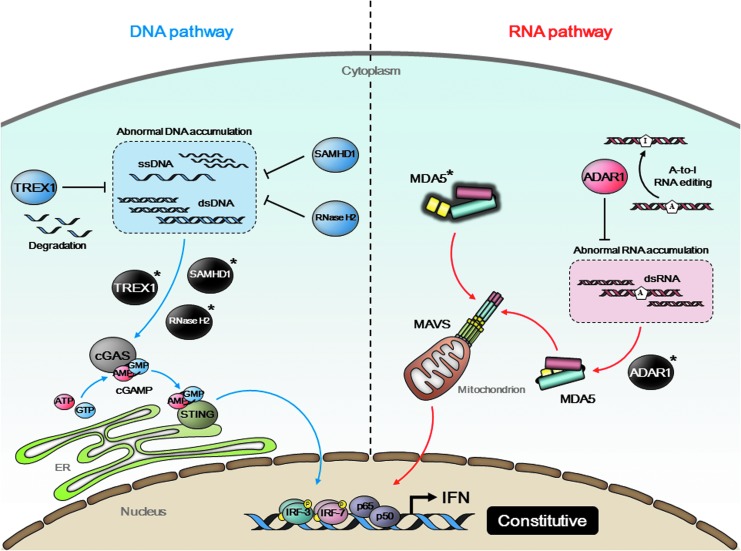
FIG. 2.
Type I interferonopathy caused by dysfunction of regulatory proteins. A cytoplasmic exonuclease TREX1 degrades extranuclear ss/dsDNA accumulated within intrinsic DNA metabolism. A dGTP-dependent triphosphohydrolase SAMHD1 and a heterotrimeric ribonuclease complex RNase H2 (RNase H2A, B, and C) also function as DNA metabolizing enzymes. Mutations in their respective genes (TREX1, SAMHD1, and RNASEH2A/B/C) abrogate their enzymatic activities and cause redundant accumulation of self-DNA in the cytoplasm. This abnormal DNA pool potentially stimulates cGAS-STING signaling axis (blue arrows). ADAR1, which is encoded by ADAR1, is an RNA-editing enzyme that binds to dsRNA and converts adenosine bases to inosines to prevent MDA5 from sensing endogenous dsRNA as nonself. Mutations in ADAR1 result in constant activation of MDA5 (red arrows). Gain-of-function mutation in IFIH1, which encodes MDA5, is also identified. All these mutations lead to constitutive activation of IFN and are strongly associated with Aicardi–Goutières syndrome (AGS), a genetic disorder referred to as type I interferonopathy.
The other group, Oda and others (2014), also identified IFIH1 heterozygous missense mutations in AGS patients, and there was 1 mutation (R779H) identical to the reported mutation by Rice and others (2014). The encoded MDA5 mutants exhibited constitutive activity, but failed to respond to a viral stimulus similar to the mouse MDA5 mutation. These findings indicated a link between constitutive MDA5 activity and AGS; however, responsiveness to viral infections remains controversial, particularly, the same mutation (G2336A:R779H) included in these studies. This discrepancy needs to be reexamined using a common assay.
Recently, several groups clearly demonstrated that RNA editing of endogenous RNAs by ADAR1 prevents the activation of MDA5. Adar1-deficient mice and mice with an editing-deficient knock-in mutation (E861A/E861A) have been shown to be embryonic lethal, but their lethality was rescued by concurrent depletion of MDA5 (Liddicoat and others 2015; Pestal and others 2015), indicating that aberrant activation of wild-type MDA5 by unedited endogenous RNAs is the cause in AGS patients with ADAR1 mutations (Fig. 2). It will be interesting to explore the responsible RNAs for MDA5 activation, which are usually edited by ADAR1, but not in AGS patients with ADAR1 mutations.
Systemic lupus erythematosus
SLE is an autoimmune disease that causes a chronic inflammation affecting many organs, including skin, joints, kidneys, lung, and nervous system. Nephritis, especially, has been reported in severe cases. IFN signature has been reported in SLE patients, and a central role of type I IFN in disease pathogenesis has been suggested. SNPs in several molecules involved in the production of IFN have been correlated with the onset of SLE. As described above, the strong association of IFIH1 SNPs with the susceptibility to autoimmune diseases, such as SLE, has been reported (Robinson and others 2011).
Importantly, de novo gain-of-function mutation in the IFIH1 gene (R779H) has recently been identified by whole-exome sequencing in a patient with severe early-onset SLE and selective IgA deficiency (Van Eyck and others 2015). Together with the SLE-like phenotypes observed in MDA5 G821S mutant mice (Funabiki and others 2014), these findings indicate a strong correlation between aberrant activation of MDA5 and SLE. Of note, IFIH1 SNPs have been shown to correlate with IgA deficiency as well, and a shared genetic predisposition between SLE and IgA deficiency through IFIH1 is suggested. According to the mouse study, macrophages and dendritic cells were reported to be responsible for IFN production, leading to overproduction of antibodies, including autoantibodies from B cells via T cell activation, deposition of antibodies in kidney, and finally, nephritis. Thus, it is important to clarify the pathogenesis and the mechanisms of disease onset in SLE patients with MDA5 mutation, referring to the investigation on MDA5 mutant mice. Furthermore, the R779H mutation was also identified in an AGS patient, as described above. Thus, it is intriguing to investigate how the same mutation causes different phenotypes of diseases, SLE and AGS.
Singleton–Merten Syndrome
Patients with SMS, an extremely rare and multisystem disorder, generally develop dental dysplasia, calcifications of the aorta, glaucoma, and osteoporosis. SMS patients also exhibit IFN signature.
Recently Rutsch and others (2015) reported a novel missense mutation (R822G) in IFIH1 in an SMS patient. MDA5 R822G exhibits constitutive activity (Rutsch and others 2015). Considering that some AGS patients exhibit glaucoma or premature tooth loss, further clinical examination in AGS patients, especially, those with an IFIH1 mutation may help us to understand the pathogenesis of SMS, since, only 1 SMS patient with an IFIH1 mutation has been reported so far. It is worth noting that this mutation is next to the G821S mutation in Ifih1. Therefore, it would be interesting to explore the SMS-like phenotypes in MDA5 G821S mutant mice, which were reported to exhibit lupus-like nephritis.
Jang and others (2015) recently described a family affected by these symptoms and identified a missense mutation (E373A) in DDX58, encoding RIG-I. This is the first report to show mutations of RIG-I in patients with an autoimmune disease. RIG-I E373A leads to the constitutive production of type I IFN. Moreover, a further analysis of DDX58 in 100 individuals with congenital glaucoma resulted in the identification of another mutation (C268F). Two amino acids, Glu373 and Cys268, of RIG-I reside in the ATP-binding motifs I and II in the Hel-1 domain, respectively. The constitutive activities and responsiveness to ligand RNA of these mutants have not yet been characterized.
Future Perspectives
Recent intensive studies have revealed a strong correlation of IFIH1 mutations with autoimmune diseases. However, there remain several important and specific questions to be solved.
(1) How do MDA5 mutants cause a wide variety of autoimmune disorders? Also how can identical IFIH1 mutation, such as R779H, cause different autoimmune defects?
(2) What would be an effective treatment for patients with RLR mutations?
(3) What kinds of RNA species activate MDA5 in AGS patients with ADAR1 mutations? Are endogenous RNAs involved in the chronic activation of mutant RLRs in patients?
(4) Although RIG-I shares a common adaptor molecule MAVS for signaling, why is RIG-I mutation less frequently found in autoimmune disease patients compared to MDA5?
All IFIH1 mutations found in humans and mice cause amino acid substitutions within the helicase domain of MDA5; however, no obvious hot spot has been identified to date. These mutations may commonly induce conformational changes, resulting in the unmasking of CARD for constitutive activity. Some mutations may also enhance the detection of ligand RNA derived from host cells, which might include chronic viral infections. There is much possibility that chronic IFN production level determines pathogenesis in organs, in other words, the suppression of local IFN signature at a certain level will help to delay the onsets or ameliorate autoimmune symptoms. Thus, it is important to investigate the involvement of endogenous ligand RNAs for RLR activation in each affected organ, and if any, to identify the endogenous ligand RNAs of RLRs, and understand how this is prevented by homeostatic mechanisms. Studies on MDA5 ligand RNAs in ADAR1-deficient mice may especially help us to characterize what kinds of endogenous RNAs have the potential for MDA5 activation. In addition, to reveal the mechanisms of pathogenesis in RLR-related interferonopathies, MDA5 G821S mice will be useful to investigate what cell types are the major and primary source of IFN, the mechanisms by which overproduced IFN leads to autoimmune disorders, and for screening drugs to ameliorate symptoms of affected organs. Also, the detailed molecular phenotypes of these mutations need to be categorized in relation to the different manifestations in patients.
Acknowledgments
This study was supported by research grants from Ministry of Education, Culture, Sports, and Science and Technology of Japan: Innovation Areas ‘Infection Competency’ #24115004 (TFujita), Japan Society for the Promotion of Science: Grants-in-Aid for Scientific Research ‘A’ #23249023; Japan Science and Technology Agency (PRESTO); and the Practical Research Project for Rare/Intractable Diseases from Japan Agency for Medical Research and Development, AMED.
Author Disclosure Statement
No competing financial interests exist.
References
- Ablasser A, Hemmerling I, Schmid-Burgk JL, Behrendt R, Roers A, Hornung V. 2014. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J Immunol 192(12):5993–5997 [DOI] [PubMed] [Google Scholar]
- Ahn J, Gutman D, Saijo S, Barber GN. 2012. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci U S A 109(47):19386–19391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Ruiz P, Barber GN. 2014. Intrinsic self-DNA triggers inflammatory disease dependent on STING. J Immunol 193(9):4634–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi L, Ban M, Leone MA, Sawcer SJ, D'Alfonso S. 2010. No evidence of association of the rare nsSNP rs35667974 in IFIH1 with multiple sclerosis. J Neuroimmunol 221(1–2):112–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinek O, Tapia G, Witso E, Kramna L, Holkova K, Rasmussen T, Stene LC, Ronningen KS. 2012. Enterovirus RNA in peripheral blood may be associated with the variants of rs1990760, a common type 1 diabetes associated polymorphism in IFIH1. PloS One 7(11):e48409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton SP, Deane JA, Feigenbaum L, Bolland S. 2012. Ifih1 gene dose effect reveals MDA5-mediated chronic type I IFN gene signature, viral resistance, and accelerated autoimmunity. J Immunol 188(3):1451–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow MK. 2014. Advances in understanding the role of type I interferons in systemic lupus erythematosus. Curr Opin Rheumatol 26(5):467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ. 2013. Aicardi-Goutieres syndrome. Handbook Clin Neurol 113:1629–1635 [DOI] [PubMed] [Google Scholar]
- Crow YJ, Rehwinkel J. 2009. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum Mol Genet 18(R2):R130–R136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo Silva J, Tavares NA, Santos MM, Moura R, Guimaraes RL, Araujo J, Crovella S, Brandao LA. 2015. Meta-analysis of STAT4 and IFIH1 polymorphisms in type 1 diabetes mellitus patients with autoimmune polyglandular syndrome type III. Genet Mol Res 14(4):17730–17738 [DOI] [PubMed] [Google Scholar]
- Deane JA, Pisitkun P, Barrett RS, Feigenbaum L, Town T, Ward JM, Flavell RA, Bolland S. 2007. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity 27(5):801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Farzan M. 2013. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 13(1):46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303(5663):1529–1531 [DOI] [PubMed] [Google Scholar]
- Enevold C, Kjær L, Nielsen CH, Voss A, Jacobsen RS, Hermansen ML, Redder L, Oturai AB, Jensen PE, Bendtzen K, Jacobsen S. 2014. Genetic polymorphisms of dsRNA ligating pattern recognition receptors TLR3, MDA5, and RIG-I. Association with systemic lupus erythematosus and clinical phenotypes. Rheumatol Int 34(10):1401–1408 [DOI] [PubMed] [Google Scholar]
- Ferreira RC, Pan-Hammarström Q, Graham RR, Gateva V, Fontán G, Lee AT, Ortmann W, Urcelay E, Fernández-Arquero M, Núñez C, Jorgensen G, Ludviksson BR, Koskinen S, Haimila K, Clark HF, Klareskog L, Gregersen PK, Behrens TW, Hammarström L. 2010. Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nat Genet 42(9):777–780 [DOI] [PubMed] [Google Scholar]
- Funabiki M, Kato H, Miyachi Y, Toki H, Motegi H, Inoue M, Minowa O, Yoshida A, Deguchi K, Sato H, Ito S, Shiroishi T, Takeyasu K, Noda T, Fujita T. 2014. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity 40(2):199–212 [DOI] [PubMed] [Google Scholar]
- Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341(6148):903–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Treuting PM, Woodward JJ, Stetson DB. 2015. Cutting edge: cGAS is required for lethal autoimmune disease in the Trex1-deficient mouse model of Aicardi-Goutieres Syndrome. J Immunol 195(5):1939–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408(6813):740–745 [DOI] [PubMed] [Google Scholar]
- Herzner AM, Hagmann CA, Goldeck M, Wolter S, Kubler K, Wittmann S, Gramberg T, Andreeva L, Hopfner KP, Mertens C, Zillinger T, Jin T, Xiao TS, Bartok E, Coch C, Ackermann D, Hornung V, Ludwig J, Barchet W, Hartmann G, Schlee M. 2015. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat Immunol 16(10):1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 2006. 5'-Triphosphate RNA is the ligand for RIG-I. Science 314(5801):994–997 [DOI] [PubMed] [Google Scholar]
- Jacob M, Napirei M, Ricken A, Dixkens C, Mannherz HG. 2002. Histopathology of lupus-like nephritis in Dnase1-deficient mice in comparison to NZB/W F1 mice. Lupus 11(8):514–527 [DOI] [PubMed] [Google Scholar]
- Jang MA, Kim EK, Now H, Nguyen NT, Kim WJ, Yoo JY, Lee J, Jeong YM, Kim CH, Kim OH, Sohn S, Nam SH, Hong Y, Lee YS, Chang SA, Jang SY, Kim JW, Lee MS, Lim SY, Sung KS, Park KT, Kim BJ, Lee JH, Kim DK, Kee C, Ki CS. 2015. Mutations in DDX58, which encodes RIG-I, cause atypical Singleton-Merten Syndrome. A J Hum Genet 96(2):266–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205(7):1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawane K, Fukuyama H, Kondoh G, Takeda J, Ohsawa Y, Uchiyama Y, Nagata S. 2001. Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science 292(5521):1546–1549 [DOI] [PubMed] [Google Scholar]
- Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, Yoshikawa H, Nagata S. 2006. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature 443(7114):998–1002 [DOI] [PubMed] [Google Scholar]
- Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR. 2015. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349(6252):1115–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang H, Jin Y, Podolsky R, Reddy MV, Pedersen J, Bode B, Reed J, Steed D, Anderson S, Yang P, Muir A, Steed L, Hopkins D, Huang Y, Purohit S, Wang CY, Steck AK, Montemari A, Eisenbarth G, Rewers M, She JX. 2009. IFIH1 polymorphisms are significantly associated with type 1 diabetes and IFIH1 gene expression in peripheral blood mononuclear cells. Hum Mol Genet 18(2):358–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Lettice L, Tarnauskaitė Ž, Reddy K, Dix F, Revuelta A, Abbondati E, Rigby RE, Rabe B, Kilanowski F, Grimes G, Fluteau A, Devenney PS, Hill RE, Reijns MA, Jackson AP. 2016. Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J 35(8):831–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineros JE, Maiti AK, Sun C, Looger LL, Han S, Kim-Howard X, Glenn S, Adler A, Kelly JA, Niewold TB, Gilkeson GS, Brown EE, Alarcón GS, Edberg JC, Petri M, Ramsey-Goldman R, Reveille JD, Vilá LM, Freedman BI, Tsao BP, Criswell LA, Jacob CO, Moore JH, Vyse TJ, Langefeld CL, Guthridge JM, Gaffney PM, Moser KL, Scofield RH, Alarcón-Riquelme ME, Williams SM, Merrill JT, James JA, Kaufman KM, Kimberly RP, Harley JB, Nath SK, Network B. 2013. Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PLoS Genet 9(2):e1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Nakagawa K, Abe J, Awaya T, Funabiki M, Hijikata A, Nishikomori R, Funatsuka M, Ohshima Y, Sugawara Y, Yasumi T, Kato H, Shirai T, Ohara O, Fujita T, Heike T. 2014. Aicardi-Goutieres Syndrome is caused by IFIH1 mutations. Am J Hum Genet 95(1):121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, Walz T, Hur S. 2011. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci U S A 108(52):21010–21015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. 2015. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity 43(5):933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314(5801):997–1001 [DOI] [PubMed] [Google Scholar]
- Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. 2006. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science 312(5780):1669–1672 [DOI] [PubMed] [Google Scholar]
- Plagnol V, Howson JM, Smyth DJ, Walker N, Hafler JP, Wallace C, Stevens H, Jackson L, Simmonds MJ, Bingley PJ, Gough SC, Todd JA, Consortium TDG. 2011. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet 7(8):e1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokatayev V, Hasin N, Chon H, Cerritelli SM, Sakhuja K, Ward JM, Morris HD, Yan N, Crouch RJ. 2016. RNase H2 catalytic core Aicardi-Goutiéres syndrome-related mutant invokes cGAS-STING innate immune-sensing pathway in mice. J Exp Med 213(3):329–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothlichet J, Niewold TB, Vitour D, Solhonne B, Crow MK, Si-Tahar M. 2011. A loss-of-function variant of the antiviral molecule MAVS is associated with a subset of systemic lupus patients. EMBO Mol Med 3(3):142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, del Toro Duany Y, Jenkinson EM, Forte GM, Anderson BH, Ariaudo G, Bader-Meunier B, Baildam EM, Battini R, Beresford MW, Casarano M, Chouchane M, Cimaz R, Collins AE, Cordeiro NJ, Dale RC, Davidson JE, De Waele L, Desguerre I, Faivre L, Fazzi E, Isidor B, Lagae L, Latchman AR, Lebon P, Li C, Livingston JH, Lourenço CM, Mancardi MM, Masurel-Paulet A, McInnes IB, Menezes MP, Mignot C, O'Sullivan J, Orcesi S, Picco PP, Riva E, Robinson RA, Rodriguez D, Salvatici E, Scott C, Szybowska M, Tolmie JL, Vanderver A, Vanhulle C, Vieira JP, Webb K, Whitney RN, Williams SG, Wolfe LA, Zuberi SM, Hur S, Crow YJ. 2014. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet 46:503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson T, Kariuki SN, Franek BS, Kumabe M, Kumar AA, Badaracco M, Mikolaitis RA, Guerrero G, Utset TO, Drevlow BE, Zaacks LS, Grober JS, Cohen LM, Kirou KA, Crow MK, Jolly M, Niewold TB. 2011. Autoimmune disease risk variant of IFIH1 is associated with increased sensitivity to IFN-alpha and serologic autoimmunity in lupus patients. J Immunol 187(3):1298–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutsch F, MacDougall M, Lu C, Buers I, Mamaeva O, Nitschke Y, Rice GI, Erlandsen H, Kehl HG, Thiele H, Nurnberg P, Hohne W, Crow YJ, Feigenbaum A, Hennekam RC. 2015. A specific IFIH1 gain-of-function mutation causes Singleton-Merten Syndrome. Am J Hum Genet 96(2):275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, Juranek S, Kato H, Kawai T, Poeck H, Fitzgerald KA, Takeuchi O, Akira S, Tuschl T, Latz E, Ludwig J, Hartmann G. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31(1):25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth-Wagner C, Ludwig J, Bruder AK, Herzner AM, Zillinger T, Goldeck M, Schmidt T, Schmid-Burgk JL, Kerber R, Wolter S, Stumpel JP, Roth A, Bartok E, Drosten C, Coch C, Hornung V, Barchet W, Kummerer BM, Hartmann G, Schlee M. 2015. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N-2′O-methylated self RNA. Immunity 43(1):41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, Guja C, Ionescu-Tirgoviste C, Widmer B, Dunger DB, Savage DA, Walker NM, Clayton DG, Todd JA. 2006. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet 38(6):617–619 [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339(6121):786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland A, Davies J, Owen CJ, Vaikkakara S, Walker C, Cheetham TD, James RA, Perros P, Donaldson PT, Cordell HJ, Quinton R, Pearce SH. 2007. Genomic polymorphism at the interferon-induced helicase (IFIH1) locus contributes to Graves' disease susceptibility. J Clin Endocrinol Metab 92(8):3338–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140(6):805–820 [DOI] [PubMed] [Google Scholar]
- Van Eyck L, De Somer L, Pombal D, Bornschein S, Frans G, Humblet-Baron S, Moens L, de Zegher F, Bossuyt X, Wouters C, Liston A. 2015. Brief report: IFIH1 mutation causes systemic lupus erythematosus with selective IgA deficiency. Arthritis Rheumatol 67(6):1592–1597 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr., Akira S, Yonehara S, Kato A, Fujita T. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 175(5):2851–2858 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5(7):730–737 [DOI] [PubMed] [Google Scholar]
- Zouk H, Marchand L, Li Q, Polychronakos C. 2014. Functional characterization of the Thr946Ala SNP at the type 1 diabetes IFIH1 locus. Autoimmunity 47(1):40–45 [DOI] [PubMed] [Google Scholar]