Abstract
Faithful chromosome segregation relies on kinetochores, the large protein complexes that connect chromatin to spindle microtubules. Although human and yeast kinetochores are largely homologous, they track microtubules with the unrelated protein complexes Ska (Ska-C, human) and Dam1 (Dam1-C, yeast). We here uncovered that Ska-C and Dam1-C are both widespread among eukaryotes, but in an exceptionally inverse manner, supporting their functional analogy. Within the complexes, all Ska-C and various Dam1-C subunits are ancient paralogs, showing that gene duplication shaped these complexes. We examined various evolutionary scenarios to explain the nearly mutually exclusive patterns of Ska-C and Dam1-C in present-day species. We propose that Ska-C was present in the last eukaryotic common ancestor, that subsequently Dam1-C displaced Ska-C in an early fungus and was horizontally transferred to diverse non-fungal lineages, displacing Ska-C in these lineages too.
Keywords: kinetochore, analogs, gene displacement, horizontal gene transfer, gene duplication, protein complex evolution
Distributions of Ska-C and Dam1-C Are Wide, Phylogenetically Coherent and Inversely Correlated
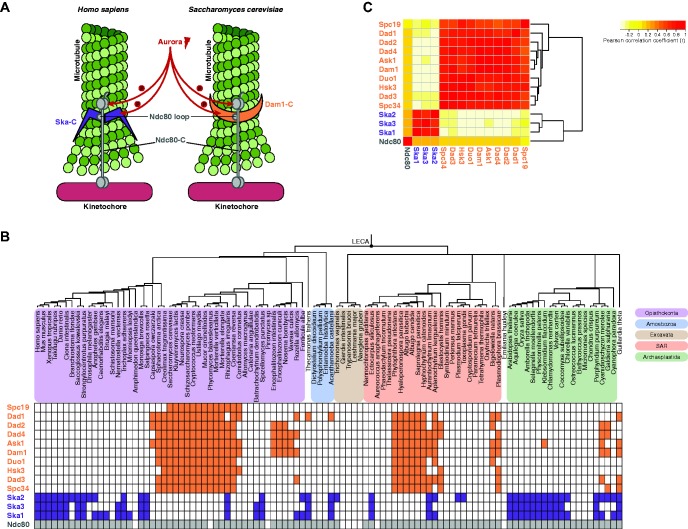
During eukaryotic cell division, duplicated sister chromatids are separated by the microtubules of the mitotic spindle. These microtubules connect to the sister chromatids via kinetochores, large protein structures that assemble onto the centromeric DNA (Cheeseman 2014). Microtubules depolymerize to pull sister chromatids apart, while maintaining their connection to the kinetochore. Kinetochores track these depolymerizing microtubules using the Ska complex (Ska-C, three subunits) in human and the Dam1 complex (Dam1-C, 10 subunits) in yeast (fig. 1A) (Cheeseman 2014). While the human and yeast kinetochores are largely homologous, these complexes instead seem analogous. This raises the question when and how these complexes were invented and whether kinetochores of other eukaryotic species may use homologous complexes to track microtubules.
Fig. 1.
—Presences and absences of the analogous Ska and Dam1 complexes. (A) Illustration of Dam1-C and Ska-C at the kinetochore-microtubule interface. Analogy between the complexes consists of their function in tracking dynamic, depolymerizing microtubules, their regulation by Aurora kinases (Aurora B in human, Ipl1 in budding yeast) and their interaction with Ndc80 via its internal loop region. Ndc80 is part of the four-subunit complex Ndc80-C. (B) Presences and absences (“phylogenetic profiles”) of the Ska-C and Dam1-C subunits and of Ndc80 across eukaryotes. The eukaryotic super groups are color-coded according to the legend. (C) Pairwise correlations between the phylogenetic profiles of all subunits.
To trace the evolutionary histories of Dam1-C and Ska-C, we determined the occurrences (“phylogenetic profiles”) of their subunits across the eukaryotic tree of life. We expected that microtubule-tracking complexes are broadly present in eukaryotic lineages, because microtubule-based chromosome segregation is conserved in eukaryotes (De Souza and Osmani 2007). Indeed, Ska-C subunits had been detected also in non-metazoan genomes and Dam1-C subunits had been detected in non-fungal ones (Cipriano 2013). To search for orthologs of Ska-C and Dam1-C subunits as well as of Ndc80, their interactor at kinetochores, we constructed a proteome database of 102 diverse eukaryotic species. This database was enriched for lineages reported to contain Dam1-C, in order to facilitate finding homologs of this apparently less abundant complex (see “Materials and Methods” section). Finding homologs of subunits of these complexes is complicated, because their sequences are highly divergent and because the Dam1-C subunit sequences are short. Therefore, we performed vigorous homology searches and de novo gene prediction (see “Materials and Methods” section). We detected Ska-C subunits in all and Dam-C subunits in four out of five eukaryotic supergroups (fig. 1B). Only the Dam1-C subunit Spc19 seems restricted to Fungi.
Within each complex the subunits had highly similar phylogenetic profiles. This similarity indicates that both complexes evolved each as a single evolutionary unit and reflects the interdependencies of their subunits (Kensche et al. 2008; Pellegrini 2012). Despite these similarities, various species lack subunits. These absences may be due to severe sequence divergence escaping our homology detection (i.e. false negatives: see supplementary text, Supplementary Material online), or they might indicate that functional complexes can consist of a subset of the subunits or have incorporated other proteins. Moreover, 19 species that contain an Ndc80 ortholog—suggesting they use microtubule-based chromosome segregation—lack Ska-C as well as Dam1-C subunits. Whether these species do not need a microtubule-tracking complex at the kinetochore or whether they contain yet other, non-homologous complexes is unknown but of great interest to further investigation.
Although most species have Ska-C or Dam1-C [74% (75/102), defined as at least one Ska-C subunit or at least three Dam1-C subunits], very few have both [7% (7/102)] (fig. 1B). To quantify how dissimilar the phylogenetic profiles of the complexes are, we calculated the Pearson correlation coefficient (r) between any two subunits (Wu et al. 2003). As expected, the intra-complex correlations were high (Ska-C: 0.72 < r < 0.81, Dam1-C: 0.51 < r < 0.91, supplementary table S1, Supplementary Material online). Strikingly, however, the inter-complex correlations were negative (−0.38 < r < −0.19) (fig. 1C). We estimated that such negative correlations are only found in 1.6% of all possible protein pairs in a genome-wide screen (see supplementary text, Supplementary Material online). This strong negative correlation suggests that Ska-C and Dam1-C are disfavored to co-occur in a species. It furthermore supports functional analogy of the complexes and predicts that their kinetochore functions are conserved across eukaryotes (Morett et al. 2003).
Ska-C and Dam1-C are distributed in a wide and scattered, yet inverse manner. Such distributions are rare in eukaryotes, but, as also indicated by our genome-wide screen, they are not unique: translation elongation factors eEF-1α and EFL form another example (Keeling and Inagaki 2004). Such distributions form a challenge for evolutionary reconstruction, because the reported low incidence of horizontal gene transfer (HGT) in eukaryotes argues for timing the origin of a gene in the last common ancestor of species carrying that gene (Keeling and Palmer 2008; Ku et al. 2015). Accordingly, Ska-C and Dam1-C would be inferred to have both been present in the LECA and in many other ancestral lineages, in contrast with what is observed in the majority of extant species. Subsequently, either Dam1-C or Ska-C was lost in most eukaryotic lineages. As such, this scenario would provide a unique case of parallel, reciprocal loss of non-homologous complexes. In another evolutionary scenario, one of the complexes was invented more recently than LECA and displaced the ancient complex, and subsequently spread to other clades of the eukaryotic tree of life via HGT, which would make this a unique case of eukaryote-to-eukaryote HGT and parallel gene displacement.
Ancient Gene Duplications Contributed to the Origin of Ska-C and Dam-C
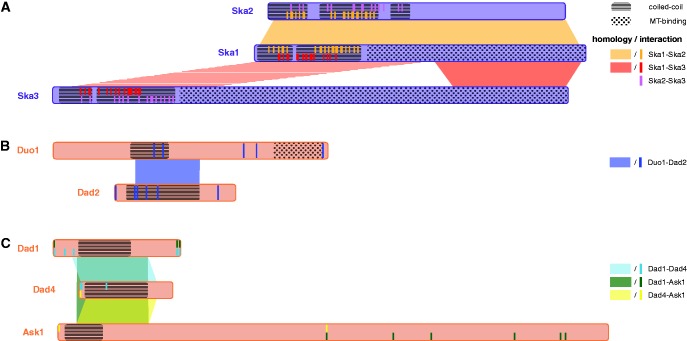
To shed light on the origins of Ska-C and Dam1-C, we searched for distant homologous protein families for each subunit in the Pfam database (Finn et al. 2014) using sensitive profile–profile comparisons. The Dam1-C subunits hit some prokaryotic families (supplementary table S2 and supplementary text, Supplementary Material online) but no homologous prokaryotic complex was identified. Strikingly, all Ska-C and three Dam1-C subunits hit another subunit of the same complex, indicating that these subunits are homologs. More significant intra-complex hits were found after adding the query profiles (constructed from the multiple sequence alignments of the orthologous groups) to the Pfam database (see “Materials and Methods” section). This latter search suggested that within Ska-C, all three subunits are homologous to one another, and that within Dam1-C, two sets of homologous subunits (Duo1-Dad2 and Dad1-Dad4-Ask1) exist (fig. 2 and supplementary fig. S1 and table S3, Supplementary Material online). Although Ska2 and Ska3 hit each other insignificantly (E-value = 13 or 22, dependent on the query profile), their common ancestry is implied by the transitive nature of homology.
Fig. 2.
—Homologous regions among Ska-C and Dam1-C subunits. Regions of homology, microtubule-binding (“MT-binding”), coiled-coils and pairwise interactions in the homologous clusters Ska1-Ska2-Ska3 (A), Duo1-Dad2 (B) and Dad1-Dad4-Ask1 (C). For the Ska-C subunits, the coiled-coil and interaction regions are based on the structure of this complex in H. sapiens (Jeyaprakash et al. 2012), for the Dam1-C subunits the coiled-coil regions are based on predictions and the interaction sites were derived from published cross-linking/mass spectrometry analyses in S. cerevisiae (Zelter et al. 2015). Although not found by profile–profile searches, the red striped region in (A) is proposed to be homologous to Ska1/2 coiled coil region based on structural similarities.
This intra-complex homology reveals that gene duplications contributed to the invention of Dam1-C and Ska-C. Dam1-C and Ska-C share duplication as a mode of invention with other protein complexes (Pereira-Leal et al. 2007). One mechanism explaining this phenomenon is that if a homodimer-forming protein duplicates, the interaction interface might be conserved, hence a heterodimer arises (Pereira-Leal et al. 2007). Such a scenario could apply to Ska-C because the interaction interfaces (the subunits’ N-terminal coiled-coils; Jeyaprakash et al. 2012) overlap with the homologous regions in at least Ska1 and Ska2. Since Ska3 also interacts with the other subunits via an N-terminal coiled-coil, we asked if the N-termini of Ska1 and Ska2 are homologous to that of Ska3. In support of this, the Ska3 profile hits Ska1 sequences in their N-terminus. Hence we hypothesize that the Ska-C subunits are homologous along their full lengths (fig. 2A, striped area indicates hit with human Ska1, supplementary fig. S1, Supplementary Material online). For Dam1-C, the interaction interfaces are less well specified, because no crystal structure is available (fig. 2B and C) (Zelter et al. 2015). Similar to Ska-C, the homologous regions overlapped with (predicted) coiled-coil regions, suggesting this structure is an ancient and important feature of both complexes (see supplementary text, Supplementary Material online).
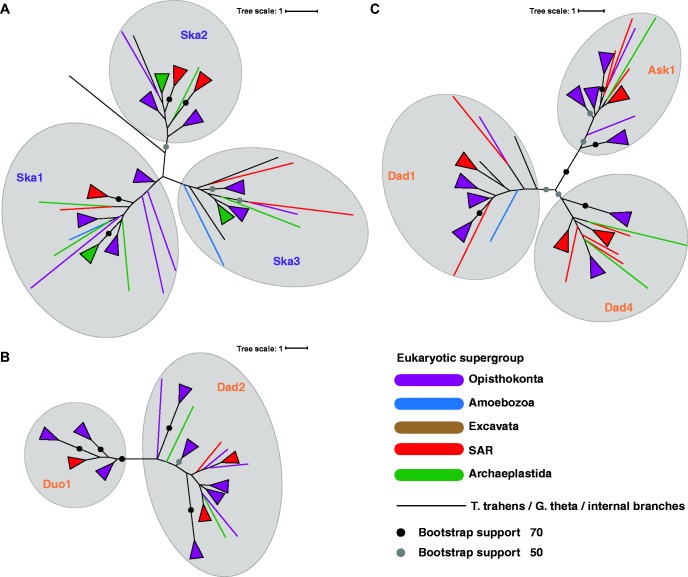
For the three homologous clusters, we estimated gene trees in an attempt to elucidate the evolutionary histories of Dam1-C and Ska-C. We generated multiple sequence alignments of the combined orthologs of a homology cluster (Ska1-Ska2-Ska3, Duo1-Dad2, Dad1-Dad4-Ask1, supplementary fig. S1, Supplementary Material online) to build the trees. The trees clearly separate the orthologous groups containing the Dam1-C/Ska-C subunits (fig. 3). Hence, the gene phylogenies confirm the phylogenetic profiles of the subunits (fig. 1B). In these phylogenies, the duplication node that unites the different orthologous groups indicates the origin of the subunits. Since all orthologous groups contain sequences from a wide range of species, and no pre-duplication sequences seem to exist, the duplications preceded the propagation of the complexes. In both the Ska-C and the Dam1-C trees many sequences have positions incongruent with the species phylogeny, which could indicate HGTs (fig. 3 and supplementary fig. S2, Supplementary Material online). However, because nodes uniting sequences from unrelated lineages have low support values and because the topologies of the subunit clusters differ within a complex, these gene trees do not provide sufficient evidence for HGT of either Ska-C or Dam1-C.
Fig. 3.
—Gene trees of Ska-C and Dam1-C homologous subunits. Maximum-likelihood gene trees of the combined orthologs of Ska1, Ska2 and Ska3 (A), Duo1 and Dad2 (B) and Dad1, Dad4 and Ask1 (C). Triangles denote a collection of branches from the same eukaryotic supergroup.
Apparently, the protein sequences of these subunits contained too little information to uncover their evolution. This lack of information is likely caused by the sequences diverging rapidly, and for Dam1-C subunits also by their short lengths. To increase the information content, we also made gene phylogenies from the concatenated alignments of Dam1-C and Ska-C subunits, assuming that for each complex the subunits evolved as a single evolutionary unit (supplementary fig. S3, Materials and Methods and supplementary text, Supplementary Material online). These trees were better supported and more congruent with the tree of life. They did not, however, allow for the identification of the transmission mechanism because there is no information to decide where these trees should be rooted.
Comparing Two Evolutionary Scenarios: “Both in LECA” versus “HGT of Dam1-C”
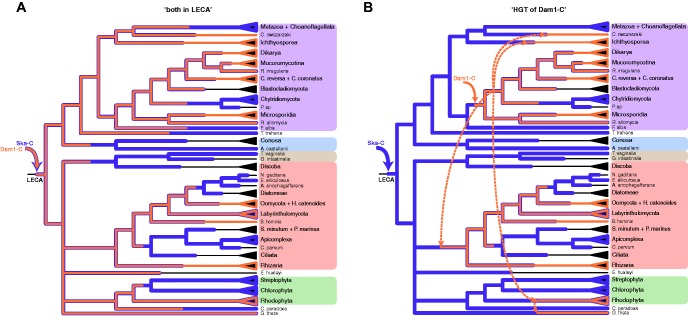
In an attempt to explain the inverse presences of Ska-C and Dam1-C in eukaryotes, we compared two evolutionary scenarios to assess which is more parsimonious. One scenario poses that LECA contained both Dam1-C and Ska-C and that no HGT events occurred, while another poses that one of the complexes was invented after LECA and spread to other eukaryotic clades by HGT. We do not consider a “both novel” scenario, because we assume that LECA had a microtubule-tracking complex to enable microtubule-based chromosome segregation. The first scenario (“both in LECA”) involves both Ska-C and Dam1-C being invented in the lineage leading to LECA, partially via the duplications reported above (fig. 4A). In the second scenario (“HGT of Dam1-C”) we favor Dam1-C being invented post-LECA rather than Ska-C because Dam1-C is present in fewer species (47 vs. 35 in a database enriched for Dam1-C-containing species, 47 vs. 27 in a “backbone” database, representing eukaryotic diversity—see “Materials and Methods” section) and in fewer supergroups (5 vs. 3) compared with Ska-C. For this “HGT of Dam1-C” scenario we specifically propose that Dam1-C was invented in a fungal ancestor, because this complex is most ubiquitous in fungi, and that it subsequently was horizontally transferred towards SAR, Ichthyosporea, the lineage of Capsaspora owczarzaki and Rhodophyta (fig. 4B). Dam1-C in Guillardia theta might be derived from this species’ secondary endosymbiont; a red alga (Douglas and Penny 1999). Please note that we here assume that all Dam1-C subunits were transferred together, as a single event, which we discuss in more detail below. Of course, when allowing for HGT many alternative scenarios can be envisioned (e.g. transfer of Dam1-C from the SAR group to the Fungi, or HGT of Ska-C, or combinations thereof), but for reasons of feasibility we here only examined one. We compared this “HGT of Dam1-C” scenario to the “both in LECA” scenario. In the latter scenario, 26% of the ancestors (the internal nodes in the species tree in fig. 1B) would have had both Dam1-C and Ska-C, compared with 7% of current-day species. In the “HGT of Dam1-C” scenario, only 14% of the ancestors would have had both complexes. We thus conclude that this scenario is more parsimonious in relation to the observed inverse presences of the complexes. In addition, while both scenarios entail 25 losses of Ska-C, “both in LECA” infers 13 losses of Dam1-C whereas “HGT of Dam1-C” infers only 4. Indeed, other HGT scenarios would likewise reduce the number of ancestral co-occurrences and losses relative to “both in LECA”.
Fig. 4.
—Two evolutionary scenarios for Ska-C and Dam1-C. Pruned species phylogeny of the eukaryotes in figure 1. Depicted are the “both in LECA” (A) and “HGT of Dam1-C” (B) scenarios, including invention (solid arrows), HGT (dashed arrows), conservation and loss of Ska-C and Dam1-C. Recent, lineage-specific losses are not shown.
The “HGT of Dam1-C” scenario is also more likely than the “both in LECA” scenario when considering their respective implications for the complexes’ functions. For Ska-C and Dam1-C to have co-existed in LECA and for long periods thereafter, their functions should most likely have been non-redundant. One complex might have had a non-kinetochore function or the complexes fulfilled the same kinetochore function in different life cycle stages. Subsequently, during post-LECA evolution, their functions became redundant in multiple independent lineages. One of the complexes would have recurrently taken over all ancestral functions previously performed by the distinct complexes, due to which the other complex got lost. In some lineages, the dominant complex would become Ska-C, while in others it would be Dam1-C. In other words, Dam-C and Ska-C evolved towards each other functionally, and this convergent evolution should have occurred in a parallel fashion in most eukaryotic lineages, with the exception of those encompassing species that still contain both complexes. Moreover, this scenario suggests that Dam1-C and/or Ska-C has a secondary, yet unknown “moonlighting” function.
In our “HGT of Dam1-C” scenario, Ska-C had a single microtubule-tracking function in LECA. Dam1-C, functionally analogous to Ska-C, was invented in an early fungal ancestor, so Ska-C was lost due to redundancy. Likewise, after Dam1-C was horizontally transferred to other eukaryotic lineages, it displaced Ska-C in these lineages, with the exception of some species that still contain both complexes. In those lineages, the complexes might have differentiated their functions recently, or one of the complexes might actually becoming displaced at present.
In summary, while both scenarios present unique and complex evolutionary trajectories, we think the “both in LECA” scenario is less likely given that it requires functions of two different complexes to converge, and to do so in an alternating (either Dam1-C or Ska-C “took over”) and independent manner in different lineages. Applying a similar reasoning, eukaryote-to-eukaryote HTG was proposed in the case of the two inversely present translation elongation factors EFL and eEF-1α (Keeling and Inagaki 2004).
Potential Mechanisms and Drivers of Dam1-C HGT
Various mechanism for eukaryotic HGT have been proposed, for example direct transformation, via viral vectors or transposable elements or via endosymbionts (Schönknecht et al. 2014). The latter might have played a role in HGT of Dam1-C to G. theta, as it contains a plastid derived from a secondary endosymbiosis of a red alga (Douglas and Penny 1999). As for other Dam1-C-containing species: Many have a “fungi-like”, osmotrophic lifestyle, viz. oomycetes and Hyphochytrium catenoides, the Labyrinthulomycota, Plasmodiophora brassicae (Rhizaria) and the Ichthyosporea (figs. 1 and 4). Osmotrophy might facilitate HGT, and moreover, some of these species form hyphae or other filamentous structures, which may fuse (anastomosis) and thereby might mediate HGT (Soanes and Richards 2014). Moreover, a shared lifestyle makes the donor and recipient species more likely to occupy similar niches and hence to physically co-localize. Interestingly, HGTs from fungi to oomycetes has been reported, and these occurred after oomycetes acquired osmotrophy and hyphae formation (Richards et al. 2011). Regardless of the exact mechanism, if HGT of Dam1-C (or Ska-C) occurred, it likely occurred to all subunit-encoding genes simultaneously: A single HGT event minimizes the number of HGT events and increases the probability that the genes are retained in the recipient species. A single HGT could have been accommodated by endosymbiosis or by genomic clustering of the subunits. In fungi, genomic clusters of functionally related genes exist, for example in secondary metabolism pathways, and some of these indeed have been horizontally transferred (reviewed in Soanes and Richards 2014).
What could have driven displacement of Ska-C by Dam1-C? Although they are considered analogous in their kinetochore function, they might differ slightly in their mechanisms of action (e.g. microtubule-tracking features, kinetochore localization, regulation). Such differences may have caused a preference for Dam1-C over Ska-C and vice versa in certain lineages. Maybe the common osmotrophic lifestyle shared by various Dam1-C containing species not only facilitated HGT, but also favored certain mechanistic alterations to the mitotic machinery. Studying mitosis in such species might yield a common theme that helps to explain the striking patterns of occurrence of Dam1-C versus Ska-C in eukaryotic species.
Materials and Methods
Compiling the Proteome Database
For studying the presences and absences of subunits of Dam1-C, Ska-C and of Ndc80 across the eukaryotic tree of life, we compiled a backbone database containing the protein sequences of 94 eukaryotic species. These species were selected in order to represent eukaryotic diversity. In order to avoid adding proteomes that relatively incomplete (containing many erroneously unpredicted genes)—which could lead to false absences in our ortholog detection—we assessed the completeness of candidate proteomes by the percentage of core KOGs present (248 core eukaryotic orthologous groups; Parra et al. 2009). If multiple annotations of the genome of a given species were available, we chose the annotation containing the highest number of KOGs. This also applies to situations in which multiple strains of a given species are sequenced. After initial searches for orthologs in the UniProtKB database (Boutet et al. 2016), this proteome database was supplemented with seven other species’ proteomes putatively having orthologs of Dam1-C subunits, in order to facilitate phylogenetic analyses (and later with H.catenoides homologs of the proteins of interest, for which we did not include the full predicted proteome—see below). The versions and sources of the selected proteomes can be found in supplementary table S4, Supplementary Material online.
Ortholog Detection
To find orthologs of Dam1-C subunits, Ska-C subunits and Ndc80, we started BLASTp homology searches with protein sequences form S. cerevisiae (Dam1-C subunits) and H. sapiens (Ska-C subunits, Ndc80) using BLASTp online (Johnson et al. 2008) and non-redundant protein sequences (nr) as a database. We aligned the resulting sequences with MAFFT (Katoh et al. 2002) (version v7.149b, option linsi, used for all other multiple sequence alignments in this study), and constructed a profile HMM. This HMM was used to initially check our local database for homologs, and it was submitted to jackhmmer online (Finn et al. 2015) versus UniProtKB (Boutet et al. 2016). Based on these results, interesting putative Dam1-C-containing species were added to our local database. Moreover, interesting hits from novel taxa, such as early-branching fungi and non-fungal lineages for Dam1-C subunits or plants for Ska-C subunits, were selected to serve as a query sequence for reciprocal homology searches, using either jackhmmer or psi-BLAST. The combined results of these homology searches were aligned to generate another profile HMM, which was used to create the initial set of orthologous sequences in the local proteome database. This HMM was required to converge on this initial set of orthologous sequences: if making an HMM profile from the obtained initial set, this second HMM should hit the sequences it was constructed from. This set was expanded by BLASTp searches versus the predicted genome of H. catenoides (which we were kindly provided access to by Thomas Richards, University of Exeter), using an oomycete query sequence. After addition of homologs not present in the predicted proteome (but present on the DNA—see “Gene Prediction of Putative Homologs” section), the HMMs derived from this sequence orthologous set was again used to search the local database, thereby confirming convergence of the orthologous set. Moreover, for proteins for which we already observed that non-orthologous sequences were hit (e.g. Ska3 sequences by Ska1, these proteins correspond to the homologous clusters in fig. 2), indicating paralogy, we confirmed the orthologous groups by generating gene trees of the multiple sequence alignments of the combined orthologous sequences. The alignments were trimmed using trimAl (Capella-Gutiérrez et al. 2009) with variable gt settings. RAxML was used to build the maximum-likelihood gene tree (Stamatakis 2014) (version 8.0.20, automatic model detection with GAMMA model of rate heterogeneity, rapid bootstrap analysis of 100 replicates—settings used throughout this study). Sequences of the orthologous groups can be found in supplementary files S1–14, Supplementary Material online, in which newly predicted genes are labeled “_p”.
Gene Prediction of Putative Homologs
To avoid false negatives due to improper gene prediction, we scanned the translated DNA sequences of the genomes with spurious absences. These spurious absences were selected based on the presence of the complex of interest (Ska-C: ≥1 subunits, Dam1-C: ≥3 subunits), except for Ndc80, for which we checked all absences. In these cases, the profile HMM of the orthologous set was used to search against the translated DNA sequences. If a hit was found in the DNA sequence, this hit was verified by searching with the hit region in the nr database using BLASTp. After confirmation, the corresponding gene was predicted by selecting the region (−5000 bp, +5000 bp) neighboring the hit and submitting this region to the AUGUSTUS web interface (Stanke and Morgenstern 2005) (multiple runs with various trained species, both strands, alternative transcripts: middle). In a few cases, no gene was predicted in the hit region, and we added the translated hit region to the orthologous group. In other cases the protein sequence of the predicted gene was added. This approach returned 24 additional homologs of Dam1-C subunits, one of a Ska-C subunit and an Ndc80 homolog.
Calculating Correlations between Phylogenetic Profiles
For the Dam1-C subunits, Ska-C subunits and Ndc80, we derived a phylogenetic profile (presences and absences) across our set of 102 eukaryotic genomes (genomes in supplementary table S4 + H. catenoides, Supplementary Material online). For each protein, this results in a string containing a “1” if it is present in a particular species (either single- or multi-copy), and a “0” if it is absent. For each possible pair of proteins, we measured to what extend the profiles correlate using Pearson correlation coefficient (Wu et al. 2003). The correlation coefficients were converted into distances (d = 1−r) and the proteins were clustered based on their phylogenetic profiles using average linkage.
Detecting Distant Homologs Using Profile–Profile Searches
In order to detect distantly related homologs of the Dam1-C and Ska-C subunits, HMM-HMM searches were performed using PRC (Madera 2008). As input, the profile HMMs of the Dam1-C and Ska-C subunit orthologous groups in our local database were used, derived from the trimmed (gt 0.1) multiple sequence alignments. The search database consisted of Pfam version 29.0 (Finn et al. 2014). Standard options for PRC were used, except for the maximum E-value (set to 100). For inferring homology between subunits of the same or the alternative complex, the search database was enriched with the query HMMs. We considered two subunits to be homologous if 1) they are each other’s best hit (or if there are no intervening hits except for within the same complex) and 2) the hit has an E-value < 10. Although the second criterion is usually considered to be too inclusive, hence yielding false positives, because of the first criterion and because of the apparent rapid sequence evolution of the subunits, we think it is appropriate here.
The homologous regions in figure 2 represent the hit regions within the respective profiles. Additional data were projected onto the illustrations of the proteins HMMs. The microtubule-interacting regions were based on studies in human (Jeyaprakash et al. 2012) and budding yeast (Zelter et al. 2015). The coiled-coil regions were based on structural information of the human Ska-C (Jeyaprakash et al. 2012) and on predictions for the budding yeast sequences using Pcoils ((Gruber et al. 2006) input is alignment of orthologous sequences, settings: apply weighting, MTIKK matrix, probability > 0.5, window size 28). The interacting residues were based on the complex structure of the human Ska-C (Jeyaprakash et al. 2012) and on cross-linking residues in Dam1-C (Zelter et al. 2015).
Phylogenetic Analyses
The identification of homology between various subunits of Dam1-C and Ska-C allowed for the construction of multiple sequence alignments of all homologous sequences consisting of multiple orthologous groups. For the well-supported homologous clusters Ska1-Ska2-Ska3, Duo1-Dad2 and Dad1-Dad4-Ask1, we aligned the sequences of the combined orthologous groups per cluster, and trimmed these alignments using trimAl ((Capella-Gutiérrez et al. 2009) gt 0.7, 0.7, 0.3, respectively), keeping only the homologous regions. From these regions, gene phylogenies were inferred. In addition, multiple sequence alignments were derived for each orthologous group separately, selecting only sequences from species having a certain complex (Ska-C: ≥1 subunits, Dam1-C: ≥3 subunits). If a species had multiple copies of a given orthologous group, one was randomly chosen, given that these are all recent duplicates and showed little divergence. The resulting alignments were concatenated, resulting in a single sequence per Dam1-C- or Ska-C-containing species. For Dam1-C, the Spc19 subunit was excluded because of its limited phylogenetic profile. The concatenated alignments were trimmed (gt 0.3 for Ska-C, gt 0.5 for Dam1-C) and the complex phylogenies were made. The resulting topologies of the maximum-likelihood phylogenies were tested for the significance of their likelihoods compared with the species phylogeny, a pruned version of figure 1, using the SH-test as recommended (Goldman et al. 2000) provided by IQ-TREE (Nguyen et al. 2015).
Inferring Ancestral States
For the Dam1-C and Ska-C, we inferred the evolutionary histories along the species phylogeny in figure 1 by applying Dollo parsimony, which allows for a single invention only. As input, the phylogenetic profiles of the full (Ska-C: ≥1 subunits, Dam1-C: ≥3 subunits) in current-day species were taken. All internal nodes were labeled by their inferred status (having/lacking) Dam1-C and Ska-C. From these, co-occurrence analysis of the complexes in these internal nodes could be calculated. This procedure was repeated for the alternative scenario, where internal nodes were now labeled in a parsimonious manner except for six instances of Dam1-C invention, which indicate the proposed HTGs.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Author Contributions
J.H. defined orthologs and performed phylogenetic analyses. B.S. and G.K. conceived and managed the project. J.H., B.S. and G.K. wrote the manuscript.
Supplementary Material
Acknowledgments
The authors thank Leny van Wijk en John van Dam for providing the eukaryotic proteome set. We thank Thomas Richards for providing permission to the genome data of H. catenoides and Eelco Tromer for extensive discussions. This work was supported by the UMC Utrecht and is part of the VICI research programme with project number 016.160.638 of the Netherlands Organisation for Scientific Research (NWO).
Literature Cited
- Boutet E, et al. 2016. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view In: Edwards D, editor. Plant bioinformatics: methods and protocols. New York (NY: ): Springer New York; p. 23-54. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM. 2014. The kinetochore. Cold Spring Harb Perspect Biol. 6:a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriano MJ. 2013. An analysis of kinetochore proteins in a wide range of eukaryotes and the kinetochore of Giardia lamblia: Davis: University of California. [Google Scholar]
- De Souza CPC, Osmani SA.. 2007. Mitosis, not just open or closed. Eukaryot Cell 6:1521–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas ES, Penny LS.. 1999. The plastid genome of the cryptophyte alga, Guillardia theta: complete sequence and conserved synteny groups confirm its common ancestry with red algae. J Mol Evol. 48:236–244. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2015. HMMER web server: 2015 update. Nucleic Acids Res. 43:W30–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Anderson JP, Rodrigo AG.. 2000. Likelihood-based tests of topologies in phylogenetics. Syst Biol. 49:652–670. [DOI] [PubMed] [Google Scholar]
- Gruber M, Söding J, Lupas AN.. 2006. Comparative analysis of coiled-coil prediction methods. J Struct Biol. 155:140–145. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash AA, et al. 2012. Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol Cell 46:274–286. [DOI] [PubMed] [Google Scholar]
- Johnson M, et al. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res. 36:W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Inagaki Y.. 2004. A class of eukaryotic GTPase with a punctate distribution suggesting multiple functional replacements of translation elongation factor 1α. Proc Natl Acad Sci U S A. 101:15380–15385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD.. 2008. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 9:605–618. [DOI] [PubMed] [Google Scholar]
- Kensche PR, van Noort V, Dutilh BE, Huynen MA.. 2008. Practical and theoretical advances in predicting the function of a protein by its phylogenetic distribution. J R Soc Interface 5:151–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku C, et al. 2015. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524:427–432. [DOI] [PubMed] [Google Scholar]
- Madera M. 2008. Profile Comparer: a program for scoring and aligning profile hidden Markov models. Bioinformatics 24:2630–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morett E, et al. 2003. Systematic discovery of analogous enzymes in thiamin biosynthesis. Nat Biotechnol. 21:790–795. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Ning Z, Keane T, Korf I.. 2009. Assessing the gene space in draft genomes. Nucleic Acids Res. 37:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M. 2012. Using phylogenetic profiles to predict functional relationships In: van Helden J, Toussaint A, Thieffry D, editors. Bacterial molecular networks: methods and protocols. New York (NY: ): Springer New York; p. 167-177. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Levy ED, Kamp C, Teichmann SA.. 2007. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA, et al. 2011. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc Natl Acad Sci. 108:15258–15263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönknecht G, Weber AP, Lercher MJ.. 2014. Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. BioEssays 36:9–20. [DOI] [PubMed] [Google Scholar]
- Soanes D, Richards TA.. 2014. Horizontal gene transfer in eukaryotic plant pathogens. Annu Rev Phytopathol. 52:583–614. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B.. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33:W465–W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Kasif S, DeLisi C.. 2003. Identification of functional links between genes using phylogenetic profiles. Bioinformatics 19:1524–1530. [DOI] [PubMed] [Google Scholar]
- Zelter A, et al. 2015. The molecular architecture of the Dam1 kinetochore complex is defined by cross-linking based structural modelling. Nat Commun. 6:8673.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.