Abstract
Purpose
Recent data suggest that intrinsic subtype and immune cell infiltration may predict response to trastuzumab-based therapy. We studied the interaction between these factors, changes in immune signatures following brief exposure to trastuzumab, and achievement of pathologic complete response (pCR) to subsequent preoperative trastuzumab and chemotherapy in HER2-positive breast cancer.
Experimental Design
In patients enrolled on two multicenter trials (03-311 and 211B), tumor core biopsies were obtained at baseline and after brief exposure to single-agent trastuzumab or nab-paclitaxel. Gene expression profiles were assessed to assign PAM50 subtypes, measure immune cell activation, and were correlated with response.
Results
The pCR rate was significantly higher in HER2-enriched tumors in the Discovery, 03-311 (36%, P = 0.043) dataset, as compared with other subtypes, which validated in 211B (50%, P = 0.048). Significant increases in a signature of immune cell admixture (Immune Index) were observed only following brief exposure to trastuzumab in HER2-enriched tumors (Discovery/03-311, P = 0.05; Validation/211B, P = 0.02). Increased Immune Index was predictive of response after brief exposure (03-311, P = 0.03; 211B, P = 0.04), but not at baseline, in addition to increased expression of a CD4+ follicular helper T-cell signature (03-311, P = 0.05; 211B, P = 0.04). Brief exposure to trastuzumab significantly increased gene expression of the T-cell marker PD-1 in HER2-enriched tumors (Discovery/03-311, P = 0.045) and PD-1 positivity by IHC (Validation/211B, P = 0.035).
Conclusions
Correlations between pCR rates, increases in Immune Index and markers of T-cell activity following brief exposure to trastuzumab in HER2-enriched tumors provide novel insights into the interaction between tumor biology, antitumor immunity, and response to treatment, and suggest potential clinically useful biomarkers in HER2+ breast cancers.
Introduction
The HER2 receptor is amplified and overexpressed in approximately 20% of all breast cancers and the addition of HER2-targeted therapy in the form of the mAb trastuzumab to chemotherapy has been shown to improve overall survival in both early stage and advanced disease (1–4). However, as many as 23% of patients with early-stage HER2-positive (HER2+) treated with adjuvant chemotherapy and trastuzumab may recur within 10 years (5), highlighting the importance of identifying which HER2+ patients respond to this treatment and those that do not. Alternative, novel HER2-targeted agents, including lapatinib, pertuzumab, and trastuzumabemtansine (T-DM1), administered either in place of or in combination with trastuzumab, have been developed and have demonstrated activity in trastuzumab-resistant cancers, but the challenge remains to try to identify patients who require addition of these agents upfront.
Genomic profiling of HER2+ tumors has shown that HER2+ breast cancer is clinically and biologically heterogeneous, and this heterogeneity complicates efforts to identify biomarkers predictive of response in the initial breast tumor biopsy (6). In previous analyses, we have found that brief exposure to trastuzumab may unmask potential biomarkers of response not apparent in pretreatment samples (7). This highlights the potential value of this approach to help to unravel the heterogeneity of the disease and understand mechanisms of response or resistance to trastuzumab in vivo.
There has been increasing evidence that the immune system plays a significant role in the therapeutic effects of HER2-targeted therapy (8). Trastuzumab has been shown to facilitate targeting of HER2-expressing tumor cells via antibody-dependent cellular cytotoxicity (ADCC), through recruitment of natural killer cells (9, 10). The presence of tumor-infiltrating lymphocytes (TIL) was predictive of improved outcomes with trastuzumab-containing therapy in a phase III adjuvant trial (11), suggesting that the evaluation of immune markers may be important. A recent study of mRNA expression in tumor samples collected from patients treated on NCCTG N9831 adjuvant trial identified a signature of immune function genes that was predictive of relapse-free survival in patients treated with trastuzumab plus chemotherapy but not in patients assigned to the chemotherapy-only control arm (12). While preclinical studies suggest that ADCC may be a critical component of trastuzumab benefit (13, 14), and subpopulations of immune cells have been associated with improved breast cancer survival (15), no study has yet correlated the in vivo effects of trastuzumab on immune markers with its impact on response to treatment. The brief exposure paradigm is an important approach to test the hypothesis that trastuzumab-induced changes in the tumor’s immune microenvironment might impact response to therapy, as we have previously shown for tumor-specific pathways (16).
In this study, the preoperative brief exposure paradigm was used to evaluate changes in the tumor and its microenvironment with a loading dose of trastuzumab. We utilized transcriptomic data from samples obtained from patients who enrolled on two prospective multicenter clinical trials (03-311 and BrUOG 211B) of stage II–III HER2+ breast cancers treated with preoperative trastuzumab-containing chemotherapy. HER2 intrinsic breast cancer subtypes and immune-related transcriptional signatures were evaluated in relation to pathologic complete response (pCR), a surrogate measure of long-term outcome. This approach may help us better understand the role of the innate immune response in mediating response to trastuzumab-containing chemotherapy in HER2+ early breast cancer.
Materials and Methods
Trial design and patient characteristics
Tissue samples were collected from patients enrolled on two independent multicenter phase II neoadjuvant trials (03-311, NCT00148668; and BrUOG 211B, NCT00617942) for clinical stage II–III HER2+ breast cancer, with HER2 positivity defined as either overexpression by immunohistochemical (IHC) stain (3+) or a FISH ratio for HER/CEP17 of >2.0.
The 03-311 trial was conducted at the Dana-Farber Cancer Institute and the Yale Comprehensive Cancer Center (YCCC). Once a patient signed informed consent, pretreatment tumor biopsies were obtained, and then she received a loading dose of trastuzumab (8 mg/kg). Repeat tumor biopsies were obtained approximately 14 days later, after which the patient started treatment with either vinorelbine 25 mg/m2 weekly × 12 weeks (NH) or docetaxel 75 mg/m2 and carboplatin AUC 6 every 3 weeks (TCH) with trastuzumab 2 mg/kg weekly for 12 weeks (TCH); the choice of chemotherapy regimen and subsequent surgical management of the breast and axilla were at the discretion of the treating physicians. A study detailing the long-term follow-up of patients in the 03-311 trial suggested an association between pCR and improved long-term recurrence-free survival (17). Supplementary Fig. S1 illustrates samples collected from patients enrolled on 03-311.
BrUOG 211B (211B) was conducted by the Brown University Oncology Group (BrUOG) at its participating hospitals and at the YCCC and the City of Hope Comprehensive Cancer Center (COHCCC). Enrolled patients underwent baseline tumor biopsies, then received “run-in” treatment determined by their institution, with patients enrolled at a BrUOG institution receiving a loading dose (6 mg/kg) of trastuzumab while patients enrolled at YCCC or COHCCC received two weekly doses of nab-paclitaxel 100 mg/m2. Repeat tumor biopsies were obtained between 10 and 14 days after trastuzumab or the first dose of nab-paclitaxel. All patients were then treated with carboplatin AUC 6 every 3 weeks and nab-paclitaxel 100 mg/m2 and trastuzumab 2 mg/kg weekly for 18 weeks, followed by surgical management of the breast and axilla at the discretion of their treating physicians. Supplementary Fig. S2 illustrates samples collected from patients enrolled on 211B.
On both studies clinical response was assessed during and at the completion of preoperative therapy, and pathologic response was scored by institutional pathologists, with pCR defined as the absence of residual invasive disease in both the breast and any sampled axillary nodes; for 03-311, patients who failed to achieve pCR were grouped as objective responders (OBJR) category, which included complete and partial response (CR + PR), and nonresponders (NOR) category including stable and progressive disease. For 211B, RECIST criteria were used but were not yet developed when the 03-311 trial was conducted. In addition, for 211B pathologists were asked to assess residual cancer burden (RCB; refs. 18, 19).
Tissue processing
Fresh core biopsy tissue was placed immediately into optimal cutting temperature (OCT) and frozen at −80°C. An H&E section was performed on each OCT block and the percentage of invasive tumor in the sample determined by a pathologist. Core biopsies containing 10% or greater invasive cancer were removed from the OCT and DNA and RNA were simultaneously extracted using the Qiagen AllPrep DNA/RNA Kit (cat# 80204). DNA and RNA concentration and 260/280 ratios were determined using a NanoDrop. The quality of the RNA was assessed using a 2100 Bioanalyzer (Agilent Technologies) to determine a RNA integrity number (RIN). Prior to sequencing, a more accurate concentration of the RNA was quantified using a fluorometric assay on the Qubit 2.0 Fluorometer (Life Technologies).
Microarray profiling
Samples from the 03-311 study were profiled using Illumina HT12v3 Beadchip microarrays. Briefly, the arrays were run on 200 ng RNA extracted from the tissue samples using standard processing protocols in the Yale Center for Genome Analysis. The raw microarray data were then processed for background correction using the Illumina Bead Studio. The Bead Studio output was then processed with the lumi package in Bioconductor using the variance stabilizing transform and quantile normalization. The resulting normalized log2 gene expression values were used for all subsequent downstream analyses. The microarray data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE76360 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=cnkjqomaxlclncv&acc=GSE76360).
Targeted RNA sequencing, data analysis, and quality assurance
A custom Targeted Amplicon RNA-Sequencing panel for 957 genes of interest [including PAM50 genes (20, 21), ESTIMATE Immune Index genes (22), and signatures of immune cell subsets B-cell, T-cell, macrophage, CD4+ Tfh, Th1, Treg, and CD8+ cytolytic genes (15, 23, 24)] as well as IPO8, ALAS1, GAPDH, and SDHA housekeeping genes was designed using Illumina’s Design Studio. For library preparation, the IlluminaTruSeq Targeted RNA Expression Kit was utilized. Briefly, 50 ng of input RNA was reverse transcribed. Next the custom oligo pool targeting the genes of interest was hybridized to the cDNA, followed by extension and ligation to enrich for the targets of interest. The enriched regions were then PCR-amplified and ligated to adapters and indices for multiplexing purposes. The libraries were normalized, pooled, and assessed for quality using quantitative real-time PCR. Samples were multiplexed in batches of 10–32 samples per 50 cycle MiSeq run (Illumina) and sequenced using the MiSeq Reagent Kit v3 (Illumina).
An average of 1.05 million paired-end reads were obtained per sample, over 4 different MiSeq runs. Given that each gene on the custom panel was assigned a single 50 bp validated gene-target probe based on Illumina’s Design Studio, the resulting reads per sample were aligned to the gene-target probe sequences using a banded Smith–Waterman alignment tailored for alignment across amplicon targets as low as 10 bp. This resulted in gene-level read-counts per sample for all of the 957 genes on the targeted RNASeq custom panel. Notably, as read-counts were derived using the number of reads aligned to the individual 50 bp gene-target probes, read-counts across genes are comparable without the need for transcript length normalization. As detailed in Illumina’s Targeted Amplicon RNA-Seq analysis pipeline, we then estimated the relative gene expression level for any given gene in a given sample by subtracting the median of the log2-transformed read-counts of four housekeeping genes (IPO8, ALAS1, GAPDH, and SDHA) in that sample from the log2-transformed read-count for the given gene. The resulting log2-transformed relative gene expression levels were used for all subsequent downstream analyses.
PAM50 subtyping
The log2-transformed gene expression values of the PAM50 genes were used to derive the PAM50-based subtyping assay (20) for all samples processed by either microarray or Targeted RNASeq. In either case, median subtraction was performed on the log2-transformed expression values of each of the PAM50 genes, followed by unsupervised hierarchical clustering using Pearson correlation for distance and average linkage. Major clusters were identified and assigned to HER2-luminal, HER2-basal, and HER2-enriched subgroups based on ER/PR IHC values and relative expression of the proliferation-associated genes within the PAM50 gene list (21). For each tumor, we assigned the subtype based on the membership of the baseline biopsy sample in the PAM50 clustering. In cases where no baseline sample was available for a given tumor, the subtype was assigned using the subtype membership of the post brief exposure sample.
Immune index estimation
The extent of immune cell admixture in the tumor samples was estimated using a previously published algorithm, ESTIMATE (22), which is derived using the expression levels of a 140-gene immune signature. Briefly, this published 140-gene immune signature uses a single-sample gene set enrichment methodology to derive an index (Immune Index) that corresponds to the level of infiltrating immune cells in a given sample (22). The resulting Immune Index allows for relative comparisons of the extent of immune admixture across samples profiled on the same platform. The Immune Index values for all samples in this study were estimated using the R-package associated with the published algorithm (22). The Immune Index values obtained for the 03-311 samples profiled using the Illumina HT12v3 Beadchip microarrays were log2-transformed and used for comparisons within subgroups of the 03-311 dataset. Similarly, the Immune Index values obtained for the 211B samples using gene expression levels derived from Targeted RNA-Sequencing were again compared within subgroups of the 211B dataset. Given the methodology and different profiling platforms, Immune Index values are not comparable across the two datasets, and therefore all of our Immune Index comparisons were performed within each dataset.
Signatures of immune cell subsets
B-cell, T-cell, and macrophage signatures
Previously published signatures related to specific immune cell subsets (23) including a 23-gene signature specific to B cells, an 83-gene signature specific to T cells and a 105 gene signature specific to macrophages were used as surrogate estimates of the individual immune cell subsets. For each sample in the Discovery 03-311 cohort, the specific immune cell subset signature index was estimated as the average value of the normalized log2 expression levels of the individual genes in the signature. Similarly, in the 211B cohort, the signature index was calculated as the average of the relative gene expression levels of each of the genes within the signatures.
CD4+ Tfh signature
An 8-gene follicular helper T-cell (Tfh) signature (CD200, CXCL13, FLJ37440, ICOS, SGPP2, SH2D1A, VSTM3, PDCD1) whose expression levels were previously shown to be associated with CD4+ follicular helper T cells (15) was used as a surrogate estimate of CD4+ Tfh cells. For each sample in the Discovery 03-311 cohort, the signature index was estimated as the signed average value of the normalized log2 expression levels of the 8 genes in this signature. Similarly, in the 211B cohort, the signature index was calculated as the signed average of the relative gene expression of each of the 8 genes with respect to the four housekeeping genes as detailed in the Targeted RNA Sequencing data analysis method section above.
CD4+ Th1 signature
A 12-gene Th1 signature (CCND2, CD38, CTLA4, RAB27A, RAB33A, SH2D2A, STAT1, TNFRSF4, IL12RB2, SAMD9L, CXCL9, IFNG) whose expression levels were shown to be significantly associated with CD4+ Th1 cells (15) was used as a surrogate measure of the extent of CD4+ Th1 infiltration in each sample. Similar to the Tfh signature above, the signed average expression level of the 12 Th1 genes was estimated per sample and used as a Th1 signature index in both the 03-311 and 211B cohorts.
CD4+ Treg signature
An 8-gene Treg signature (FOXP3, IL2, IL10, LGALS1, LGALS3, STAT5A, TGFB2, XCL1) whose expression levels were shown to be significantly associated with CD4+ T regulatory cells (Tregs; ref. 15) was used as a surrogate measure of the extent of Treg infiltration in each sample. Similar to the Tfh signature above, the signed average expression level of the 8 Treg genes was estimated per sample and used as a Treg signature index in both the 03-311 and 211B cohorts.
CD8+ T-cell cytolytic activity genes
Genes known to be upregulated upon CD8+ T-cell activation (PRF1, GZMK, GZMH, NKG7) were used as a surrogate of CD8+ T-cell cytolytic activity (24), both individually and as a signature. As above, the average expression level of the cytolytic activity genes was estimated per sample and used as a Cytolytic Activity signature index in both the 03-311 and 211B cohorts.
IHC
PD-1 expression in tumor biopsies derived from the 211B trial was assessed using IHC performed by the Immunohistochemistry Laboratory of University Hospitals Case Medical Center (Cleveland, OH). Briefly, unstained 4-μm sections of tissue were prepared from paraffin blocks and baked for 30 minutes at 60°C in a Boekel Lab oven. The slides were then processed using a BenchMark Ultra (Ventana Medical Systems) automated immunostainer. The slides were deparaffinized, antigen retrieved with standard Cell Conditioning 1 (Ventana Medical Systems), a Tris-based buffer pH 8.3 solution for 52 minutes at 95°C, then incubated at 37°C with the primary antibody PD-1 (clone NAT105 mouse mAb from Cell Marque) for 16 minutes and subsequently counterstained with Hematoxylin for 8 minutes. PD-1 scoring was performed blinded fashion, with the cutoffs chosen prior to association with other clinical/genomic data. PD-1 staining was scored as follows: tissues with no staining in any of the stroma-related tumor-infiltrating lymphocytes (TIL) or absence of TILs were scored as 0; tissues with less than 10% of TILs with PD-1 staining were scored as 1; tissues with 10%–50% of TILs staining for PD-1 were scored as 2; and tissues with >50% staining of TILs for PD-1 were scored as 3. We then categorized scores 0 and 1 as low, 2 as medium, and 3 as high levels of PD-1 staining. Published studies do not have a standardized method to score PD-1. Furthermore, artefactual tissue distortions, poor tissue quality limited our ability to provide precise counts of PD-1+ TILs in this dataset.
Results
Patient characteristics
The clinical characteristics of the patients from 03-311 and 211B trials are presented in Table 1. There were a total of 81 and 60 stage II–III patients enrolled in the 03-311 and 211B trials, respectively. No significant differences in tumor stage, tumor size, or hormonal status were observed between the two trials.
Table 1.
Clinical characteristics of patients enrolled in the 03-311 and BrUOG 211B trials and for whom gene expression data was available
| Clinical characteristics | DFCI 03-311, Number of patients (% of cohort)
|
BrUOG 211B, Number of patients (% of cohort)
|
|||
|---|---|---|---|---|---|
| Total patients (N = 81) |
Gene expression data available (N = 72) |
Total patients (N = 60) |
Gene expression data available (N = 41) |
||
| Receptor status | ER+ and/or PR+ | 34 (42%) | 31 (43%) | 32 (53%) | 25 (61%) |
| ER− and PR− | 47 (58%) | 41 (57%) | 28 (47%) | 16 (39%) | |
| Clinical stage | IIA | 26 (32%) | 22 (30%) | 20 (33%) | 15 (37%) |
| IIB | 28 (35%) | 24 (33%) | 19 (32%) | 15 (37%) | |
| IIIA | 16 (20%) | 15 (21%) | 11 (18%) | 6 (15%) | |
| IIIB | 5 (6%) | 5 (7%) | 7 (12%) | 3 (7%) | |
| IIIC | 3 (4%) | 3 (4%) | 3 (5%) | 2 (4%) | |
| Tumor size | T0/T1 | 3 (4%) | 2 (3%) | 5 (8%) | 3 (7%) |
| T2 | 46 (57%) | 40 (56%) | 36 (60%) | 27 (66%) | |
| T3 | 24 (30%) | 22 (31%) | 11 (18%) | 8 (20%) | |
| T4 | 6 (7%) | 6 (8%) | 8 (13%) | 3 (7%) | |
| Age | Mean (SD) | 48 (10.7) | 47 (9.7) | 52 (9.1) | 52 (9.0) |
HER2-enriched subtype exhibits the highest rate of pCR to trastuzumab-based preoperative therapy
We and others have shown that HER2 amplification may arise within Luminal and Basal lineages and have named these subtypes HER2-basal and HER2-luminal breast cancers (6, 25). The third subtype of HER2-positive breast cancer, which is typically ER/PR negative, is termed HER2-enriched, and has been shown to exhibit improved response to anti-HER2 therapy. In the context of the 03-311 and 211B trials, we have used this nomenclature. Using the log2-transformed normalized expression levels of the PAM50 genes, we performed hierarchical clustering (See Materials and Methods) of the baseline and post-brief exposure gene expression data from the 03-311 (Fig. 1A) and 211B (Fig. 1B) cohorts. We then assigned the PAM50 subtype calls to each of the major clusters using the ER/PR IHC status and the expression levels of proliferation genes included in the PAM50 signature (See Materials and Methods). The cluster of samples that were largely ER/PR negative by IHC and had low expression levels of basal-keratin genes (KRT14, KRT17, KRT5) were designated as HER2-enriched, whereas clusters containing samples largely ER/PR positive by IHC were designated as HER2-luminal, and clusters of ER/PR-negative samples that showed high expression of proliferation and basal-keratin genes were designated as HER2-basal. In addition to these major subtypes, both cohorts also revealed a cluster of samples that showed lower expression of most of the PAM50 genes, suggestive of low tumor content (Fig. 1A and B).
Figure 1.
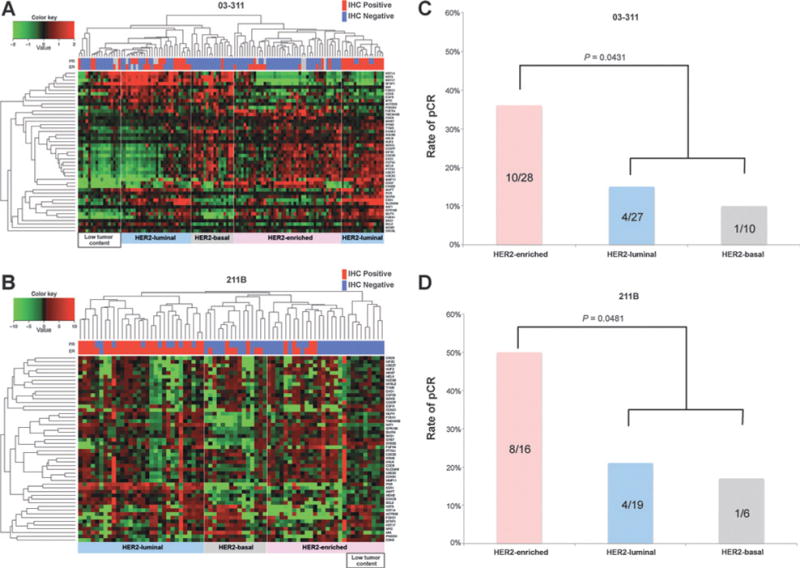
PAM50 subtyping. A, hierarchical clustering of microarray samples from the 03-311 cohort using the PAM50 genes. Three major clusters were identified, including Luminal, HER2-Basal, and HER2-enriched subtypes that correspond well with ER/PR IHC and the expression levels of proliferation genes. B, hierarchical clustering of samples from the 211B trial using targeted RNA sequencing of the PAM50 genes. Samples that showed poor expression of a majority of genes were considered to have low tumor content. C, rates of pCR across PAM50 subtypes in 03-311 cohort. Shown are the percentages of pCR within each subtype. Significance of the rate of pCR in the HER2-enriched subtype as compared with the remaining tumors was assessed using a two-sided Fisher exact test. D, rates of pCR across PAM50 subtypes in 211B cohort. Shown are the percentages of pCR within each subtype. The hypothesis that the HER2-enriched subtype exhibited the highest rate of pCR as compared with the rest was evaluated using a one-sided Fisher exact test.
We then proceeded to evaluate the association of PAM50 subtypes with rates of pCR with preoperative trastuzumab-containing therapy. We found that the HER2-enriched subtype had the highest rate of pCR in both the Discovery (03-311, Fig. 1C) cohort (36%, two-sided Fisher exact test, P = 0.0431) and the Validation (211B, Fig. 1D) cohort (50%, one-sided Fisher exact test, P = 0.0481). In the Discovery cohort (Fig. 1C), the HER2-luminal subtype had the next highest rate of pCR (15%), whereas the HER2-basal subtype had the lowest rate of pCR (10%). The Validation cohort (Fig. 1D) also mirrored the relative ranking among the subtypes in terms of the observed rates of pCR (21% in HER2-luminal and 17% in HER2-basal). Of note, the overall pCR rate was lower in 03-311 likely due to the shorter duration of chemotherapy compared with the BrUOG 211B. It is interesting to note that the HER-basal subtype had the lowest pCR rate, surprising in a typically ER-negative tumor, although the significance of this needs to be confirmed in larger studies, given the low number of HER2-basal samples in our trials. However it appears that the HER2-basal subtype may be more drug resistant than the HER2-enriched subtype based on studies from Carey and colleagues (26) and Harris and colleagues (6).
HER2-enriched subtype consistently shows a significant increase in immune index upon brief exposure to trastuzumab
We next hypothesized that the higher rate of pCR in HER2-enriched tumors may be due to immune activation by therapy in this subtype. We evaluated a publicly available gene expression signature of immune cell admixture (Immune Index) in the 03-311 and 211B cohorts (See Materials and Methods) to determine whether brief exposure to therapy results in changes in immune cell–based gene expression (22). Figure 2A shows the distribution of Immune Index estimates at baseline and post brief exposure to trastuzumab across each of the PAM50 subtypes in the 03-311 cohort. Consistent with our hypothesis, we found that the HER2-enriched subtype exhibited a significant (P = 0.05) increase in the median Immune Index following brief exposure to trastuzumab, while the other two major subtypes showed nonsignificant increases in Immune Index at the post-exposure timepoint. We then evaluated the Immune Index in 211B and found that the HER2-enriched subtype was again the only subtype showing a significant increase in median Immune Index (P = 0.05) following brief exposure to trastuzumab (Fig. 2B). Furthermore, in 211B, the HER2-enriched subtype showed a significant increase in Immune Index only upon brief exposure to trastuzumab, and not upon brief exposure to chemotherapy (P = 0.13; Supplementary Fig. S3). We note, however, that the small number of samples in the HER2-basal subtype in either trial precludes assessment of the significance of Immune Index changes in this subtype. Additional evaluation using ER/PR status revealed a significant increase in Immune Index only in the ER-negative subgroup within the 211B trial, whereas the ER-positive subgroup showed nonsignificant increases in both trials (Supplementary Figs. S4 and S5). Furthermore, we found no significant correlation between the Immune Index and tumor content of the biopsy samples in the two datasets at either the baseline or post brief exposure timepoints (Supplementary Fig. S6), suggesting that the observed associations are not related to biopsy-related changes.
Figure 2.
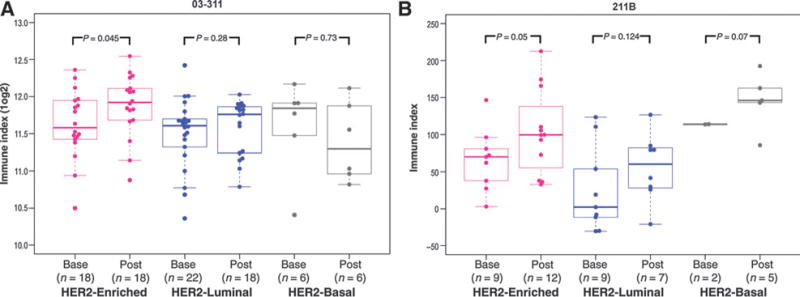
Comparisons of Immune Index following brief exposure to therapy across PAM50 subtypes. A, plotted are the Immune Index (log2) values in individual patient biopsies at baseline and after a single dose of trastuzumab in the 03-311 cohort across the HER2-enriched, luminal, and HER2-basal subtypes. Significance was calculated using a one-sided nonparametric Wilcoxon test. B, Immune Index values at baseline and following brief exposure to therapy (either trastuzumab or nab-paclitaxel) in the 211B cohort across the three subtypes.
In summary, our analysis showed that significant increases in Immune Index were observed only in the HER2-enriched subtype upon brief exposure to trastuzumab in both the 03-311 and 211B trials.
Immune signatures are discriminative of pCR at the post brief exposure timepoint
To test the hypothesis that the Immune Index signature was associated with response, we evaluated whether the Immune Index at either the baseline and/or the post brief exposure time-point could discriminate between responders and nonresponders. Figure 3A details the distribution of the Immune Index at baseline and after a single dose of trastuzumab in the Discovery cohort (03-311) across the three different response categories (pCR, OBJR, and NOR). We found that the median Immune Index was not significantly higher in the pCR as compared with OBJR (one-sided Wilcoxon, P = 0.63) or NOR (one-sided Wilcoxon, P = 0.48) at the baseline timepoint (Fig. 3A). However, after a single dose of trastuzumab, the median Immune Index was significantly higher in the pCR group as compared with the NOR group (one-sided Wilcoxon, P = 0.03, Fig. 3A).
Figure 3.
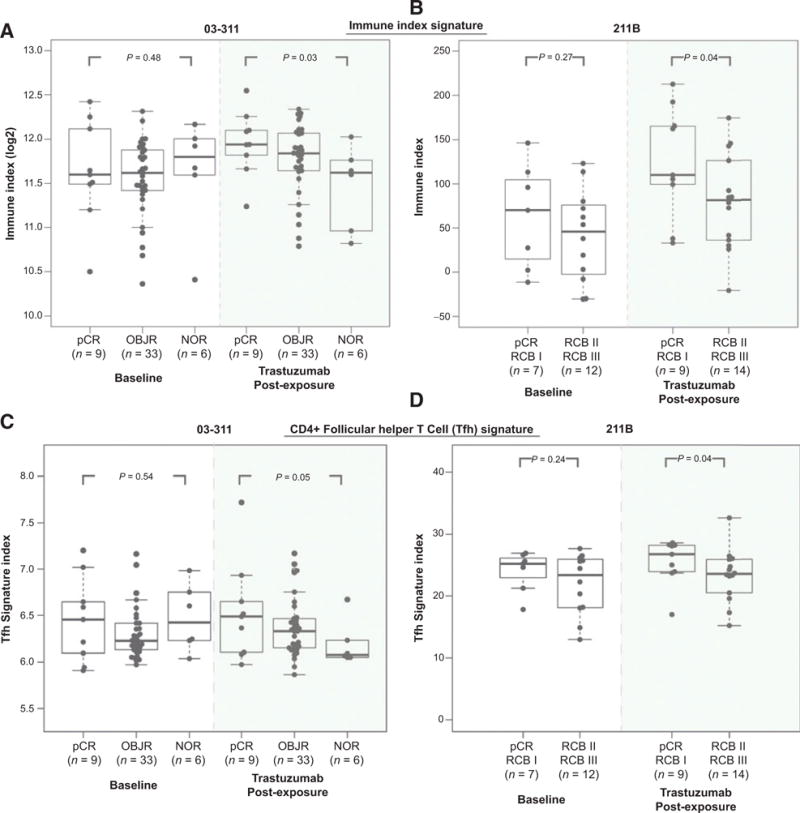
Association of immune signatures with response. A, shown are the Immune Index (log2) values at baseline and postexposure to trastuzumab across response groups in the 03-311 trial. B, Immune Index at baseline and post-exposure to trastuzumab across response groups in the 211B trial. Significance was assessed using a one-sided Wilcoxon test. C, plotted is the 8-gene signature index associated with CD4+ Tfh activity in the baseline and postexposure timepoint across response groups in the 03-311 trial. D, the Tfh signature index is plotted across response groups at baseline and postexposure to trastuzumab in the BrUOG 211B cohort.
We then evaluated whether this observation persisted in the 211B dataset (Fig. 3B). We found that the median Immune Index was not significantly higher (one-sided Wilcoxon, P = 0.27, Fig. 3B) at baseline in the tumors that had a complete or near-pCR to trastuzumab-containing therapy (pCR and RCB I) as compared with tumors with a suboptimal response to this therapy (RCB II-III). However, after a single-dose of trastuzumab, the median Immune Index was significantly higher (one-sided Wilcoxon P = 0.04, Fig. 3B) in the pCR and RCB1 as compared with the RCB II and III in the BrUOG cohort, validating the observation from the Discovery cohort. These findings suggest that a higher immune signature index upon brief exposure to trastuzumab may be predictive of response to trastuzumab-containing preoperative chemotherapy in HER2+ breast cancer.
The Immune Index results suggest a role of immune infiltration but do not provide a clear mechanistic explanation for these results. To gain insight into the potential mechanisms underlying the Immune Index findings, we evaluated the ability of individual genes belonging to the Immune Index signature to discriminate response after a single dose of trastuzumab across the two data-sets. We accordingly evaluated whether the expression level of each of the genes in the Immune Index signature were associated with response at either baseline and/or post brief exposure time-points, followed by q-value false discovery rate estimation to correct for multiple testing (27). We found that only two genes, GIMAP4 and ITGAL, were significantly (P < 0.05; FDR < 25%) discriminative of response at the postexposure timepoint in both the Discovery (03-311) and Validation (211B) datasets (Supplementary Table S1). GIMAP4 expression has been previously reported to be associated with CD4+ helper T-cell development (28), whereas ITGAL (CD11a) has been shown to be expressed in activated CD4+ T cells (29) suggesting that T-cell function may play an important role in response to trastuzumab-containing therapy.
We next evaluated predefined gene expression signatures associated with specific immune cell subsets such as B cells, T cells, and Macrophages (See Materials and Methods) to determine whether increases in specific immune cell subsets upon brief exposure to trastuzumab are associated with preoperative therapy response. Interestingly, we found that only the T-cell gene expression signature was significantly associated with response in both the 03-311 and 211B trials (Supplementary Table S2). In contrast, the B-cell signature was significantly associated with response only in the 211B trial at both baseline and post brief exposure timepoints, while the macrophage signature was significant only in the 03-311 trial at the post brief-exposure timepoint (Supplementary Table S2).
Given that the analysis of individual genes in the Immune Index signature (Supplementary Table S1) pointed to a the likely involvement of specific subsets of T cells such as the CD4+ immune T cells, we next evaluated predefined gene expression signatures specific to subsets of CD4+ immune infiltrating cells upon brief exposure to trastuzumab (See Materials and Methods). Using a publicly available 8-gene signature associated with CD4+ follicular helper T cells (Tfh; ref. 15), we found that a higher Tfh signature index was associated with pCR after post brief exposure to trastuzumab in the 03-311 Discovery cohort (Fig. 3C), and this finding was validated in the 211B cohort (Fig. 3D). To determine whether this is a nonspecific finding of tumor cell infiltration with T cells, we also evaluated signatures associated with CD4+ Th1 and CD4+ regulatory T cells (15), as well as genes associated with CD8+ T-cell cytolytic activity (ref. 24; See Materials and Methods). We found that higher expression of CD4+ Th1 and Treg signatures after a single dose of trastuzumab was discriminative of response in only one of the two datasets (Supplementary Fig. S7). Similarly, neither the individual genes associated with CD8+ T-cell cytolytic activity (See Materials and Methods) nor the 4-gene cytolytic signature were discriminative of response following brief exposure to therapy across both datasets (Supplementary Table S3).
Intriguingly, preclinical studies evaluating the effects of depletion of specific immune cell subsets on anti-neu antibody-mediated tumor regression have shown the essential role of CD4+ T cells while B cells were not required for the effectiveness of anti-neu therapy (30). These results, taken together with our observations across both trials, strongly suggest that higher T-cell activity, and specifically increased CD4+ follicular helper T cells, following brief exposure to trastuzumab, are associated with improved response to trastuzumab-containing preoperative therapy.
HER2-enriched subtype shows a significant increase in PD-1 expression upon brief exposure to trastuzumab
Given that PD-1 (PDCD1) is one of the genes in the Tfh signature associated with T-cell activity, and is of therapeutic interest, we evaluated PD-1 expression levels at baseline and post brief exposure to trastuzumab in the 03-311 trial. We found significant upregulation of PD-1 expression to be specific to the HER2-enriched subtype (P = 0.045) as opposed to the HER2-luminal (P = 0.738) and HER2-basal (P = 0.953) subtypes in the 03-311 trial (Fig. 4A). To validate these findings we assessed protein expression of PD-1 using IHC in samples from the 211B trial as unstained slides were not available from the 03-311 cohort. We categorized tissues into low, medium, and high levels of PD-1 staining (See Materials and Methods) and evaluated whether changes in PD-1 expression upon brief exposure to trastuzumab were associated with subtypes. Figure 4B shows the PD-1 IHC scores at baseline and postexposure to trastuzumab across the PAM50 subtypes. Notably, only the HER2-enriched subtype showed a significant increase in PD-1 expression (P = 0.035) upon brief exposure to trastuzumab (Fig. 4B), suggesting that both gene expression and protein expression of PD-1 are upregulated in this subtype of HER2 tumors.
Figure 4.
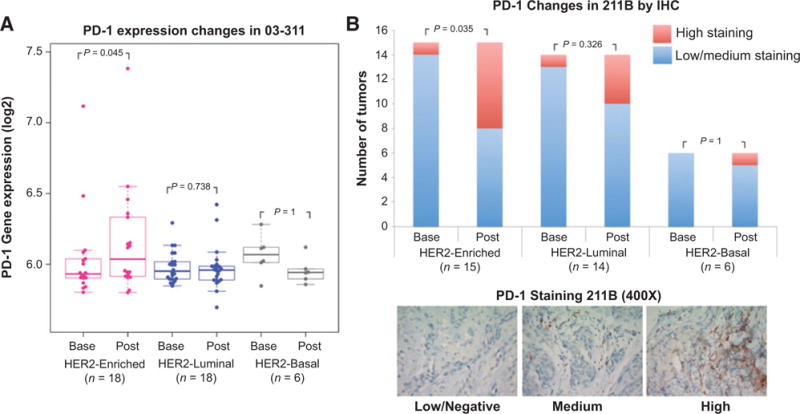
Modulation of PD-1 expression upon brief exposure to trastuzumab. A, plotted are the normalized log2 expression levels of PD1 in the 03-311 trial at baseline and post brief exposure to trastuzumab across the three PAM50 subtypes. The ladder plots indicate changes in expression in matched baseline and postexposure biopsy samples. Significance was assessed using a paired one-sided Wilcoxon test evaluating for increase in PD1 expression upon brief exposure to trastuzumab. B, the bar graphs plotted on top represent the number of tumors with low/medium (blue) or high (red) PD-1 protein expression by IHC at baseline and post brief exposure to trastuzumab across subtypes in the 211B trial. Significance was assessed using a Fisher exact test. Representative IHC stains of PD1 in the 211B trial in the low/negative, medium, and high categories at shown at 400× magnification in the bottom.
Discussion
In this study, we found that HER2+ tumors categorized as HER2-enriched by PAM50 genomic analysis exhibit the highest pCR rate to preoperative trastuzumab and chemotherapy in two prospective multicenter preoperative trials. In tumor samples from both trials, median Immune Index prior to treatment was not predictive of pathologic response in the baseline samples, but was after a single dose of trastuzumab, only in the HER2-enriched subset. This correlated with increased expression of a follicular helper T-cell signature and increased expression (by gene signature and IHC stain) of PD-1. This suggests that modulation of tumor immunity triggered by trastuzumab in this HER2-enriched subtype may mediate response to therapy.
Despite substantial efforts, reliable biomarkers that predict benefit from anti-HER2 therapy have yet to be identified in HER2-positive early breast cancer. While immune-related signatures (12) and extent of TILs (11) have been shown to be associated with response to trastuzumab-containing chemotherapy, their ability to predict benefit from specific HER2-targeted therapies and the in vivo effects of trastuzumab on immune response have not yet been elucidated. This study is the first report of a test and Validation set of preoperative trials in HER2+ breast cancer to show that immune changes in the tumor microenvironment following brief exposure to trastuzumab can generate useful predictive markers for response to combination trastuzumab and chemotherapy. We found that HER2-enriched tumors are more likely to upregulate T-cell–associated immune signatures in response to trastuzumab. In addition, we found that HER2-basal tumors are less likely to achieve pCR. Although the sample sizes were small in our study, this finding was recently confirmed in a preoperative trial of trastuzumab and lapatinib (26) in addition to our previous study (6).
Our study performed in vivo assessment of immune response with one dose of trastuzumab therapy and elaborates further on findings by Loi and colleagues that the immune system plays an important role in the benefit of trastuzumab (11). Specifically, we elucidated immune responses in molecular subtypes of HER2+ tumors and how this relates to response to therapy. By treating patients with one dose of trastuzumab therapy, we were able to demonstrate that immune cell function was significantly upregulated in the HER2-enriched subtype and that this upregulation was associated with response to subsequent preoperative therapy. These findings highlight the role of the innate and adaptive immune systems in mediating the antitumor effects of trastuzumab-containing chemotherapy and are consistent with multiple preclinical studies that have pointed to immune modulation as being an important mechanism of action of trastuzumab (13, 14). While CD4+ follicular helper T cells have previously been shown to be an important prognostic immune component (15), our study is the first to demonstrate that modulation of this immune component by trastuzumab is associated with response to preoperative therapy. In addition, our study is the first to show that a significant increase in PD-1 expression following brief exposure to trastuzumab is observed in the HER2-enriched subtype and appears to correlate with response. These findings not only help elucidate mechanisms of response to trastuzumab but suggest that the addition of immune checkpoint inhibitors to anti-HER2 therapy may be beneficial although which subtype would most likely benefit is unclear. While HER2-enriched tumors are a logical place to consider PD1/PDL1 blockade, our study suggests these patients are unlikely to need further therapy beyond trastuzumab and pertuzumab. However, it may be possible to use immune checkpoint inhibitors to stimulate adaptive immunity and potentiate T-cell activation in HER2-basal and HER2-lumimal tumors by removing negative feedback. These and other questions are optimally tested in clinical trials of HER2-targeted agents with anti-PD-1/PD-L1 therapy, many of which are ongoing.
An important aspect of our study is the utility of the brief exposure paradigm that has growing acceptance in the clinical research community (31, 32). Our findings show that this approach not only allows for the interrogation of immune modulation in the tumor by exposure to trastuzumab, but also unmasks biomarkers that can predict response after just a single dose of anti-HER2 therapy. These findings need to be validated in larger studies such as the NeoALTTO trial that incorporated a post brief exposure biopsy into the trial design and would allow investigators to compare the impact on immune activation of exposure to the HER family targeted tyrosine kinase inhibitor lapatinib with trastuzumab or the combination of both agents (31, 33). If validated, brief exposure biomarkers may help to identify patients who are likely to benefit from targeted therapy at an early timepoint, thus potentially enabling the identification of patients who may not need the addition of a second agent such as pertuzumab. Indeed, given the high costs involved in the use of pertuzumab (34), identifying patients who achieve optimal response to trastuzumab and chemotherapy alone could reduce overtreatment resulting in a more cost-effective therapeutic strategy.
One of the limitations of this study is the relative limited sample size in these two trials and the fact that adequate pretreatment and post brief exposure tumor samples were not available from all patients. Nevertheless, the subtype-specific and response-associated changes in immune signatures following brief exposure to trastuzumab in both the Discovery (03-311, N = 100 samples) and Validation (211B, N = 75 samples) datasets suggest that these findings are a robust observation. Another critique of this study relates to variation in tumor content in biopsy samples. Given that two independent datasets did not reveal significant correlation between tumor content and the Immune Index at either the baseline or post brief exposure timepoints (Supplementary Fig. S6), it is unlikely that these observations are a result of the vagaries of biopsy sampling. We also demonstrated in the 211B trial that the subtype-associated immune changes were specific to trastuzumab brief-exposure, with no significant changes being observed following treatment with nab-paclitaxel, again suggesting that tumor sampling issues, which should not be different by arm, are unlikely to be playing a major role. Another critique of the study could be differences in the chemotherapy regimens used between the two studies. However, given that both the trials had similar baseline patient characteristics and the consistent observation of increase in the Immune Index following brief exposure to trastuzumab in the HER2-enriched subtype, this effect seems to transcend any differences in regimen. This is encouraging and speaks to the likely generalizability of these observations.
Taken together, these data suggest that trastuzumab modulates activity of immune-specific transcriptional programs, and that this immune activation is significant only in the HER2-enriched subtype. Other subtypes, HER2-luminal and HER2-basal, do not show significant activation of these pathways and may be biologically quite different. These findings not only shed light on immune mechanisms at play in trastuzumab response but may identify patients whose tumors are likely to respond to trastuzumab thus not require additional therapy. In addition, this study identifies those subtypes that would potentially require additional therapy and/or alternative approaches (HER2-luminal, HER2-basal). If confirmed by others, “molecular triage” at an early timepoint may become an important clinical approach to identify which regimen a patient with HER2 disease should optimally receive as it appears that the baseline timepoint is unable to provide this information. In addition, these studies suggest the importance of genomic evaluation beyond HER2 IHC/FISH testing, as the subgroups can only be identified with gene expression. Finally, these studies suggest the use of new approaches such as immune checkpoint inhibitors to augment or trastuzumab in early stage disease, ideally in the preoperative setting. Only through clinical trials can we define the optimal approach for each patient, and the new paradigm of molecularly integrated studies must be undertaken in HER2 breast cancer to make this disease finally curable.
Supplementary Material
Translational Relevance.
HER2-positive breast cancer is biologically heterogeneous, with recent studies suggesting that intrinsic subtypes and immune infiltration estimates may predict response to trastuzumab-based therapy. However, no study has yet reported the in vivo effects of trastuzumab on the immune-related microenvironment following brief exposure to trastuzumab. We evaluated two multicenter, preoperative trials that used a run-in of trastuzumab prior to combination chemotherapy to discover and validate immune signature changes in vivo, that allowed the evaluation of subtype-specific immunogenic effects of trastuzumab. Our study demonstrates, for the first time, that immune signatures evaluated after a single dose of trastuzumab are predictive of response, possibly mediated by an induction of CD4+ helper T cells and PD-1–associated antitumor immunity within the HER-enriched subtype. These signatures, if further validated, have the potential to enable early evaluation of therapy response, thus providing an opportunity for triage to the most effective therapy for HER2+ early breast cancer patients.
Acknowledgments
V. Varadan reports receiving a commercial research grant from Philips Healthcare and is a consultant/advisory board member for Curis, Inc. I.E. Krop reports receiving other commercial research grants from Genentech. W. Sikov reports receiving a commercial research grant from Genentech. L.N. Harris reports receiving a commercial research grant from Philips Healthcare.
Grant Support
This work was supported by grants from Abraxis BioScience, Inc, now a subsidiary of Celgene Corporation, and Genentech for conduct of the clinical trial, the Breast Cancer Research Foundation for biospecimen collection and correlative science studies (to L.N. Harris). This research was also supported by pilot funding from the Case Comprehensive Cancer Center (P30 CA043703; V. Varadan), Career Development Program in Computational Genomic Epidemiology of Cancer (R25TCA094186; to V. Varadan), and the Rosalie and Morton Cohen Family Memorial Genomics Fund of University Hospitals (to V. Varadan).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Prior presentation: Oral presentation in the Predictive and Prognostic Biomarkers Minisymposium at AACR 2015.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: V. Varadan, H. Gilmore, K.L.S. Miskimen, A. Awa-dallah, E.P. Winer, V. Bossuyt, G. Somlo, W. Sikov, L.N. Harris
Development of methodology: V. Varadan, H. Gilmore, K.L.S. Miskimen, A. Awadallah, E.P. Winer, V. Bossuyt, L.N. Harris
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): H. Gilmore, K.L.S. Miskimen, A. Awadallah, V. Bossuyt, G. Somlo, M.M. Abu-Khalaf, M.A. Fenton, W. Sikov, L.N. Harris
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): V. Varadan, H. Gilmore, D. Tuck, S. Parsai, E.P. Winer, V. Bossuyt, W. Sikov, L.N. Harris
Writing, review, and/or revision of the manuscript: V. Varadan, H. Gilmore, K.L.S. Miskimen, D. Tuck, I.E. Krop, E.P. Winer, V. Bossuyt, G. Somlo, M.M. Abu-Khalaf, M.A. Fenton, W. Sikov, L.N. Harris
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): K.L.S. Miskimen, L.N. Harris
Study supervision: W. Sikov, L.N. Harris
References
- 1.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Singh JC, Jhaveri K, Esteva FJ. HER2-positive advanced breast cancer: optimizing patient outcomes and opportunities for drug development. Br J Cancer. 2014;111:1888–98. doi: 10.1038/bjc.2014.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 4.Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2012;9:16–32. doi: 10.1038/nrclinonc.2011.177. [DOI] [PubMed] [Google Scholar]
- 5.Perez EA, Romond EH, Suman VJ, Jeong JH, Sledge G, Geyer CE, Jr, et al. Trastuzumab plus adjuvant chemotherapy for human epidermal growth factor receptor 2-positive breast cancer: planned joint analysis of overall survival from NSABP B-31 and NCCTG N9831. J Clin Oncol. 2014;32:3744–52. doi: 10.1200/JCO.2014.55.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris LN, You F, Schnitt SJ, Witkiewicz A, Lu X, Sgroi D, et al. Predictors of resistance to preoperative trastuzumab and vinorelbine for HER2-positive early breast cancer. Clin Cancer Res. 2007;13:1198–207. doi: 10.1158/1078-0432.CCR-06-1304. [DOI] [PubMed] [Google Scholar]
- 7.Varadan V, Sandoval M, Harris LN. Biomarkers for predicting response to anti-HER2 agents. In: Stearns V, editor. Novel biomarkers in the continuum of breast cancer. New York: Springer; in press. [DOI] [PubMed] [Google Scholar]
- 8.Bianchini G, Gianni L. The immune system and response to HER2-targeted treatment in breast cancer. Lancet Oncol. 2014;15:e58–68. doi: 10.1016/S1470-2045(13)70477-7. [DOI] [PubMed] [Google Scholar]
- 9.Gennari R, Menard S, Fagnoni F, Ponchio L, Scelsi M, Tagliabue E, et al. Pilot study of the mechanism of action of preoperative trastuzumab in patients with primary operable breast tumors overexpressing HER2. Clin Cancer Res. 2004;10:5650–5. doi: 10.1158/1078-0432.CCR-04-0225. [DOI] [PubMed] [Google Scholar]
- 10.Denkert C, Darb-Esfahani S, Loibl S, Anagnostopoulos I, Johrens K. Anticancer immune response mechanisms in neoadjuvant and targeted therapy. Semin Immunopathol. 2011;33:341–51. doi: 10.1007/s00281-011-0261-0. [DOI] [PubMed] [Google Scholar]
- 11.Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol. 2014;25:1544–50. doi: 10.1093/annonc/mdu112. [DOI] [PubMed] [Google Scholar]
- 12.Perez EA, Thompson EA, Ballman KV, Anderson SK, Asmann YW, Kalari KR, et al. Genomic analysis reveals that immune function genes are strongly linked to clinical outcome in the North Central Cancer Treatment Group N9831 Adjuvant Trastuzumab Trial. J Clin Oncol. 2015;33:701–8. doi: 10.1200/JCO.2014.57.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barok M, Isola J, Palyi-Krekk Z, Nagy P, Juhasz I, Vereb G, et al. Trastuzumab causes antibody-dependent cellular cytotoxicity-mediated growth inhibition of submacroscopic JIMT-1 breast cancer xenografts despite intrinsic drug resistance. Mol Cancer Ther. 2007;6:2065–72. doi: 10.1158/1535-7163.MCT-06-0766. [DOI] [PubMed] [Google Scholar]
- 14.Kute TE, Savage L, Stehle JR, Jr, Kim-Shapiro JW, Blanks MJ, Wood J, et al. Breast tumor cells isolated from in vitro resistance to trastuzumab remain sensitive to trastuzumab anti-tumor effects in vivo and to ADCC killing. Cancer Immunol Immunother. 2009;58:1887–96. doi: 10.1007/s00262-009-0700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu-Trantien C, Loi S, Garaud S, Equeter C, Libin M, de Wind A, et al. CD4(+) follicular helper T cell infiltration predicts breast cancer survival. J Clin Invest. 2013;123:2873–92. doi: 10.1172/JCI67428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galanina N, Sprecher S, Bossuyt V, Sarkar S, Krop I, Winer E, et al. Evaluation of gene expression by RNA-seq after single dose of trastuzumab (T) reveals predictors of pathologic complete response (pCR) in HER2-positive early breast cancer. J Clin Oncol. 2012;30(suppl) abstr 10558. [Google Scholar]
- 17.Mayer EL, Gropper AB, Harris L, Gold JM, Parker L, Kuter I, et al. Long-term follow-up after preoperative trastuzumab and chemotherapy for HER2-overexpressing breast cancer. Clin Breast Cancer. 2015;15:24–30. doi: 10.1016/j.clbc.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Residual cancer burden calculator. 2013 Available from: http://www.mdanderson.org/education-and-research/resources-for-professionals/clinical-tools-and-resources/clinical-calculators/index.html.
- 19.Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007;25:4414–22. doi: 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 20.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16:5222–32. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iglesia MD, Vincent BG, Parker JS, Hoadley KA, Carey LA, Perou CM, et al. Prognostic B-cell signatures using mRNA-seq in patients with subtype-specific breast and ovarian cancer. Clin Cancer Res. 2014;20:3818–29. doi: 10.1158/1078-0432.CCR-13-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carey L, Barry WT, Pitcher B, Hoadley KA, Cheang MC, Anders CK, et al. Gene expression signatures in pre- and post-therapy (Rx) specimens from CALGB 40601 (Alliance), a neoadjuvant phase III trial of weekly paclitaxel and trastuzumab with or without lapatinib for HER2-positive breast cancer (BrCa) J Clin Oncol. 2014;32(suppl) abstr 506. [Google Scholar]
- 26.Carey LA, Berry DA, Cirrincione CT, Barry WT, Pitcher BN, Harris LN, et al. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol. 2016;34:542–9. doi: 10.1200/JCO.2015.62.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinonen MT, Kanduri K, Lahdesmaki HJ, Lahesmaa R, Henttinen TA. Tubulin- and actin-associating GIMAP4 is required for IFN-gamma secretion during Th cell differentiation. Immunol Cell Biol. 2015;93:158–66. doi: 10.1038/icb.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDermott DS, Varga SM. Quantifying antigen-specific CD4 T cells during a viral infection: CD4 T cell responses are larger than we think. J Immunol. 2011;187:5568–76. doi: 10.4049/jimmunol.1102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mortenson ED, Park S, Jiang Z, Wang S, Fu YX. Effective anti-neu-initiated antitumor responses require the complex role of CD4+ T cells. Clin Cancer Res. 2013;19:1476–86. doi: 10.1158/1078-0432.CCR-12-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baselga J, Bradbury I, Eidtmann H, Di Cosimo S, de Azambuja E, Aura C, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial. Lancet. 2012;379:633–40. doi: 10.1016/S0140-6736(11)61847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellis MJ, Suman VJ, Hoog J, Lin L, Snider J, Prat A, et al. Randomized phase II neoadjuvant comparison between letrozole, anastrozole, and exemestane for postmenopausal women with estrogen receptor-rich stage 2 to 3 breast cancer: clinical and biomarker outcomes and predictive value of the baseline PAM50-based intrinsic subtype–ACOSOG Z1031. J Clin Oncol. 2011;29:2342–9. doi: 10.1200/JCO.2010.31.6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Azambuja E, Holmes AP, Piccart-Gebhart M, Holmes E, Di Cosimo S, Swaby RF, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): survival outcomes of a randomised, open-label, multicentre, phase 3 trial and their association with pathological complete response. Lancet Oncol. 2014;15:1137–46. doi: 10.1016/S1470-2045(14)70320-1. [DOI] [PubMed] [Google Scholar]
- 34.Durkee BY, Qian Y, Pollom EL, King MT, Dudley SA, Shaffer JL, et al. Cost-effectiveness of pertuzumab in human epidermal growth factor receptor 2-positive metastatic breast cancer. J Clin Oncol. 2015 doi: 10.1200/JCO.2015.62.9105. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


