Summary
The development of sensitive and non-invasive “liquid biopsies” presents new opportunities for longitudinal monitoring of tumor dissemination and clonal evolution. The number of circulating tumor cells (CTCs) is prognostic in multiple myeloma (MM), but there is little information on their genetic features. Here, we have analyzed the genomic landscape of CTCs from 29 MM patients, including eight cases with matched/paired bone marrow (BM) tumor cells. Our results show that 100% of clonal mutations in patient BM were detected in CTCs and that 99% of clonal mutations in CTCs were present in BM MM. These include typical driver mutations in MM such as in KRAS, NRAS, or BRAF. These data suggest that BM and CTC samples have similar clonal structures, as discordances between the two were restricted to subclonal mutations. Accordingly, our results pave the way for potentially less invasive mutation screening of MM patients through characterization of CTCs.
Introduction
The development of “liquid biopsies” presents new opportunities for non-invasive monitoring of clonal heterogeneity (Lawrence et al., 2014). Multiple myeloma (MM) is a plasma cell (PC) malignancy characterized by patchy bone marrow (BM) infiltration. Recent studies of massive parallel sequencing of tumor cells obtained from the BM of patients with MM have demonstrated significant clonal heterogeneity in MM with a median of five clones present in each sample (Lohr et al., 2014b; Bolli et al., 2014; Corre et al., 2015). Despite this remarkable clonal heterogeneity, it could be envisioned that such clonal diversity may be even higher since single BM samples only represent a small fraction of the whole BM compartment, and the pattern of BM infiltration in MM is typically patchy. In addition, BM biopsies are painful and cannot be repeated multiple times during the course of therapy, indicating a need for less invasive methods to molecularly characterize MM patients and monitor disease progression during the therapy. Thus, optimal characterization of circulating tumor cells (CTCs) may represent a non-invasive method to capture relevant mutations present in PC clones. However, it is presently unknown whether using liquid biopsies (i.e., patients’ genetic characterization performed in peripheral blood [PB] samples) can provide a more complete profile of MM clonal diversity. Unlike other hematological malignancies (e.g., leukemia), MM does not have a substantial numbers of CTCs burden except in late stages of disease progression such as in PC leukemia. Of note, standard exome sequencing has recently been performed in single CTCs in prostate cancer, demonstrating that 70% of CTC mutations were present in matched tumor tissue (Lohr et al., 2014a).
Here, we used sensitive multiparameter flow cytometry (MFC) to detect and isolate CTCs in the PB of MM patients. We performed whole-exome sequencing in sorted CTCs and compared their mutational profile to that of patient-paired BM clonal PCs. Confirmatory studies using a targeted sequencing panel demonstrate fidelity of the mutational profile observed in those with matched BM and CTC samples. Thus, our results reveal that CTCs can potentially be used as a non-invasive biomarker to perform mutational profiling of MM patients.
Results
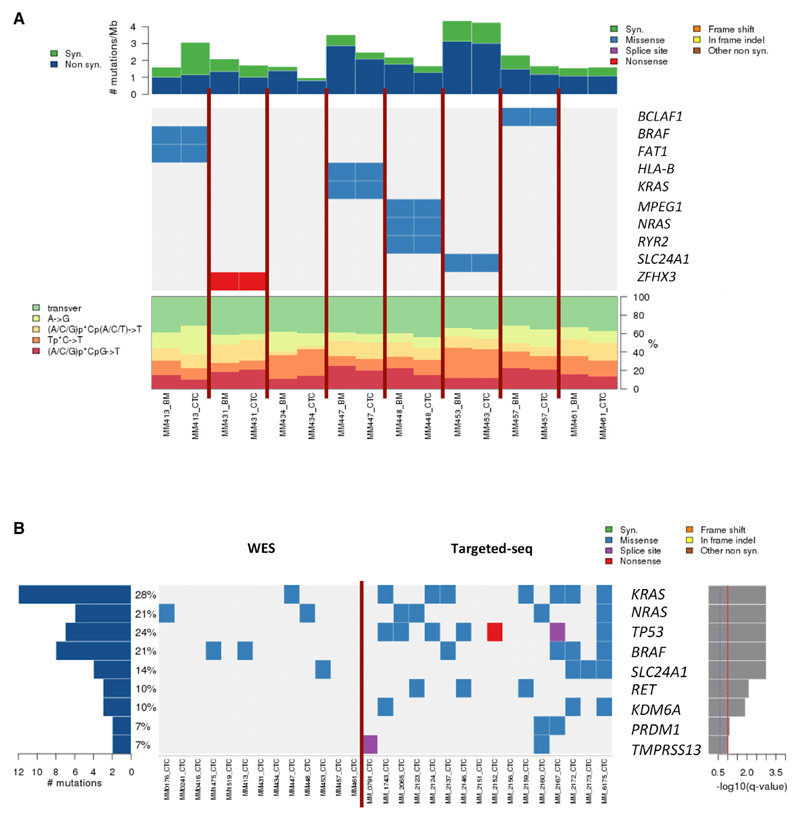
We first determined the mutational profile of CTCs of patients with MM of eight matched CTC samples compared to paired BM tumor cells and germline non-tumor cell DNA from PB Tlymphocytes. We identified 658 and 572 coding somatic single-nucleotide variants (SSNVs) in patient-paired BM clonal PCs and CTCs, respectively. Overall, 90% of CTC mutations were present in BM tumors and 93% of BM mutations were present in CTC samples, and, upon analyzing the mutational variants by nucleotide change, we found that the percentages of each change in BM myeloma PCs and CTCs were concordant. We then focused on the variants that are well known to be driver mutations in myeloma and other cancers, to define/investigate the role of CTCs in being a good surrogate for the most relevant variants observed in BM samples. Among 70 MM-related genes and 246 pan-cancer driver genes (Weinstein et al., 2013; Omberg et al., 2013), a total of 18 somatic single nucleotide variants (SSNVs) in 13 genes were identified in our cohort, and ten out of the 13 genes were matched between BM clonal PCs and CTCs. It is noteworthy that the genes with the highest frequency in MM, such as KRAS, NRAS, and BRAF, were present in these samples and were shared between patient-paired BM clonal PCs and CTCs (Figure 1A).
Figure 1. Concordance of SSNVs Found in Matched BM Clonal PCs and CTCs.
(A) Rate of synonymous and nonsynonymous mutations are expressed in number of mutations per megabase. Heatmap representation of individual mutations is present in a series of eight paired BM and CTC samples. Breakdown of individual base-substitution rates is shown for each sample as well.
(B) Heatmap representation of individual mutations is present in 13 WES CTC samples and 16 targeted sequencing CTC samples. Percentages represent the fraction of tumors harboring at least one mutation in specified genes.
The frequency of mutated genes in MM patients and pan-cancer driver genes in CTCs was further investigated in a subsequent analysis of additional samples. Therefore, we analyzed five CTC samples by whole-exome sequencing (without whole-exome amplification) and 16 CTC samples by deep-targeted sequencing (∼900×) using a custom-developed panel. These additional 21 patients (without available paired BM tumor cells) showed similar mutational profiles (Figure 1B) as compared to the first cohort with matched BM tumor cells and CTCs, as well as previous reports on mutation analyses from BM tumor cells (Bolli et al., 2014; Lohr et al., 2014b; Walker et al., 2015). The higher frequency of mutations detected in the later 16 patients was related to the deeper coverage of the targeted sequencing approach.
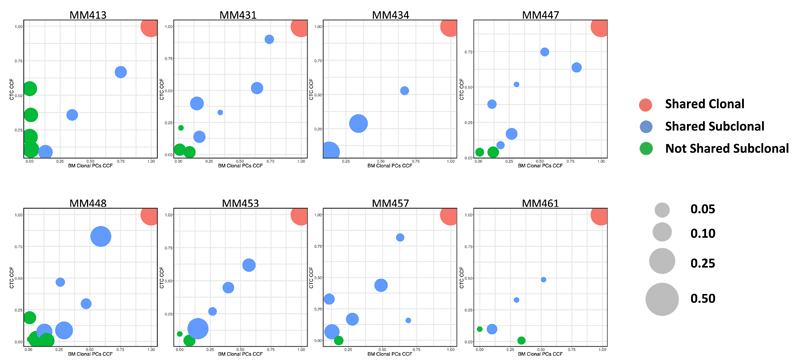
After showing that CTCs were a representative tumor compartment for mutation screening in MM, we then investigated the clonal distribution between BM myeloma clonal PCs and CTCs in order to compare clonal and subclonal architectures between each tissue-specific clone (Figure 2). Overall, 100% of clonal mutations in BM were confirmed in CTC samples, and 99% of clonal mutations in CTCs were present in BM tumors. On the other hand, 88% of subclonal mutations in BM were confirmed in CTC samples, and 81% of subclonal mutations in CTCs were confirmed in BM clonal PC samples. In other words, we observed that 84% of these clonal and subclonal SSNVs were shared between BM and CTC samples (i.e., cancer cell fraction [CCF] >0.05 in both samples), 42% (range 22%–73%) of SSNVs were clonal (i.e., cancer cell fraction, CCF ≥0.9), and 42% (range 18%–65%) of SSNVs were found to be subclonal (i.e., CCF <0.9). Conversely, 16% (range 0%–50%) of SSNVs were subclonal and not shared between BM tumor PCs and CTCs (i.e., CCF <0.05 in either BM clonal PCs or CTCs).
Figure 2. Clustering Analysis of Clonal and Subclonal SSNVs between Matched BM Clonal PCs and CTCs.
Clustering analysis of CCF for SSNVs between matched BM clonal PCs and CTCs is shown in this figure. Shared clonal SSNVs were defined as events having ≥0.9 CCF in both samples (red). Shared subclonal SSNVs were identified as events having ≥0.05 CCF in both samples (blue). Not shared subclonal SSNVs were defined as events having <0.05 CCF in either BM or CTC samples (green). The size of each cluster indicates the frequency of SSNVs within the same sample.
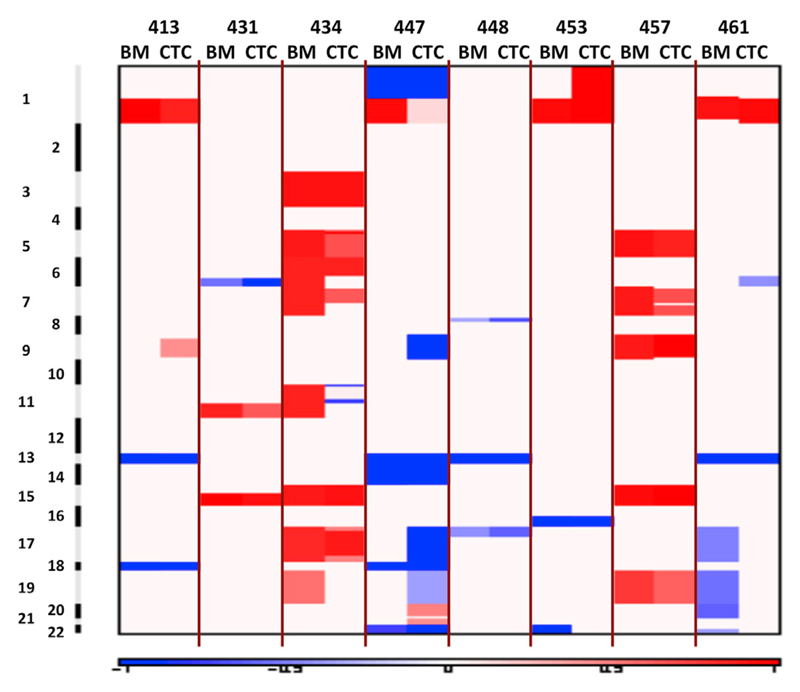
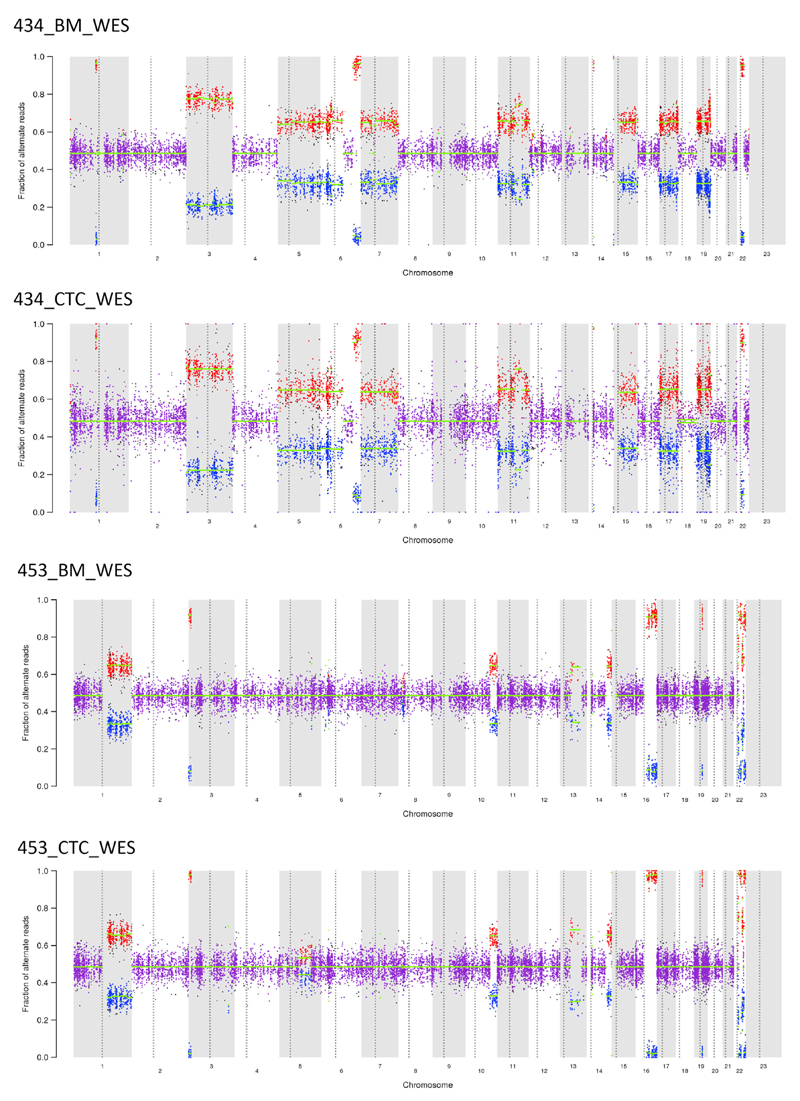
Since specific cytogenetic abnormalities are of major importance for risk stratification in MM, we further evaluated somatic copy-number alterations (SCNAs) and compared them between matched BM and PB tumor cells across paired samples (Figures 3 and 4). A concordance between BM clonal PCs and CTCs was observed in ∼92% (range 77%–100%) of arm level SCNAs. Classic MM-related SCNAs, such as 1q21 amplification and 13q deletion, were present both on BM clonal PCs and CTCs from some patients (Figure 3) (Corre et al., 2015; Walker et al., 2010; Mohamed et al., 2007; Jenner et al., 2007). Two examples of the concordance observed between BM and CTC SCNAs are provided in Figure 4, in which we were able to detect, in individual patients, concordant patterns of SCNAs between BM and CTC samples.
Figure 3. Concordance of Somatic Copy-Number Alterations Found in Matched BM Clonal PCs and CTCs.
Arm level of somatic copy-number alterations are compared within the same patient (left, BM; right, CTC). Red indicates amplification, and blue indicates deletion.
Figure 4. Representative Example of Somatic Copy-Number Alterations Found in Matched BM Clonal PCs and CTCs.
Allelic copy number ratios are shown for both BM (top) and CTC (bottom) samples within the same patient (434 and 453, respectively).
Discussion
In MM, there is a marked fluctuation of different clones throughout patients’ clinical course, implying that multiple BM aspirates are needed to determine the genomic profile of patients, specifically with the development of new targeted thera pies for actionable mutations (e.g., BRAF inhibitors for patients with BRAF mutations). Accordingly, the primary objective of our study was to determine the feasibility of performing genomic characterization of MM patients non-invasively and define whether the mutation profile of CTCs reflected that of patient-paired BM clonal PCs.
The field of liquid biopsies is one of the more intensively investigated areas of research in oncology at this time. We felt that this should also be a priority in MM, a disease in which patients go through a large number of BM aspirates to determine their genomic profile before starting a new line of therapy. In the present study, we demonstrated that whole-exome sequence of MM CTCs is feasible by combining highly sensitive multicolor flow cytometry that enabled us to detect and collect a sufficient number of purified CTCs with optimized molecular approaches. Regarding the frequency of patients with sufficient CTCs for genetic studies in PB, our recent experience using ultra-sensitive next-generation flow cytometry is that CTCs are detectable in 61% of monoclonal gammopathy of undetermined significance (MGUS) and 100% of smoldering and active MM (newly diagnosed and relapsed). The median number of CTCs per microliter in the blood of patients with MGUS, smoldering MM, and active MM are 0.011, 0.14, and 2.01, respectively (Sanoja et al., 2015). Accordingly, in patients with active MM as much as 15–20 mL of blood would suffice to sort 30,000 CTCs in a significant fraction of patients. A recent publication by Lohr et al. describes a method that allows for the isolation and genomic characterization of single MM CTCs by targeted sequence (Lohr et al., 2016). Meanwhile, whole-exome sequencing has great advantages against targeted sequencing approach, which includes detection of non-recurrent but potential driver gene mutations, deconvolution of copy-number alteration, and analysis of clonal structure of the patient’s tumor. Indeed, we demonstrated in this study that similar mutation patterns and SCNA are typically observed for clonal as well as subclonal mutations. However, discordant genomic alterations between matched BM clonal PCs and CTCs were also detected. Further studies in larger series are warranted to address whether SSNVs present in CTCs but not in BM tumor cells from some patients are a proof of concept that single BM biopsies afford limited information due to patchy infiltration and extramedullary disease. Conversely, the fact that in other patients BM clonal PCs show higher numbers of SSNVs reopens the question of whether CTCs are more immature and therefore display lower subclonal mutations, or whether we are witnessing unknown levels of spatial heterogeneity in which tumor cells from the same patient but isolated from different tissues will consistently show differences in subclonal mutations (Gerlinger et al., 2012). Alternatively, differences in total number of mutations observed between BM and CTC samples may also be attributable to differences in detection sensitivity for subclonal mutations. Most of the discrepant variants were subclonal mutations, and all clonal mutations were shared between BM and CTC samples. Even though intertumoral heterogeneity has been already described when sequencing two tumor biopsies from the same patient (Gerlinger et al., 2012), we believe that the discrepancy could also be driven by the effect of whole-genome amplification in some of the samples. We attempted to be strict in calling variants in the samples that underwent whole-genome amplification because we took only the shared variants identified in two parallel libraries constructed by two different WGA reactions to eliminate random errors caused by whole-genome amplification, and, by doing that, we had a smaller number of variants called in the CTC samples. Indeed, these results are similar to studies that have shown that the false-positive rate of WGA can be overcome by taking the consensus of two independent WGA reactions (Lohr et al., 2014a; Zhang et al., 2015).
With the development of better technologies and sequencing of longitudinal CTCs, we may be able to use PB samples to determine the mutational landscape of MM patients during disease presentation and progression. This approach could eliminate the need for multiple invasive BM aspirates to determine genomic alterations and monitor clonal evolution during disease progression and after therapeutic interventions. Additional efforts are warranted to define, in large series of patients, the level of concordance for SCNA and translocations between (whole genome/exome or targeted) sequencing versus the gold standard fluorescence in situ hybridization (FISH). An additional question to be addressed is the potential association between the mutation profile of CTCs and clinical outcomes; however, it should be noted that the prognostic significance of mutation profiling in MM remains largely unknown, and, perhaps, its major application could be to personalize patients’ treatment rather than prognostication. Together, this study defines a new role for CTCs in the prognostic and molecular profiling of MM patients.
Experimental Procedures
Patient Sample Collection and Study Approval
We prospectively collected samples from patients seen in the clinic at Dana-Farber Cancer Institute (DFCI) or Clinica Universidad de Navarra from 2011 to 2015. Among 29 unique patients with MM, we obtained eight samples of newly diagnosed untreated patients whose bone marrow, CTC, and germline T lymphocytes were available and selected for paired exome sequencing. Additional whole-exome sequencing studies were performed in five patients with flow-sorted CTCs but without available BM clonal PCs.
The review boards of participating centers approved the study, which was conducted according to the Declaration of Helsinki and International Conference on Harmonization Guidelines for Good Clinical Practice. All patients provided written informed consent. The study approval numbers are 07-150 (DFCI) and 073/2015 (Clinica Universidad de Navarra).
Whole-Exome Sequencing
BM myeloma PCs and CTCs were sorted from paired BM and PB from eight patients with symptomatic MM using a fluorescence-activated cell sorting FACSAria IIb sorter (BD Biosciences). Both tumor fractions were sorted according to the individual patient-specific aberrant phenotypes, and PB T lymphocytes were simultaneously collected for germline control. Genomic DNA was extracted using QIAamp DNA micro kit (QIAGEN) according to the manufacturer’s protocols, and double-stranded DNA concentration was quantified using PicoGreen dsDNA Assay kit (Life Technologies). The cell number of CTCs used and total amount of genomic DNA obtained are shown in Table S1.
For cases in which the total amount of DNA extracted from BM myeloma PCs (n = 1) and CTCs (n = 7) was limited, the genomic DNA was amplified using GenomePlex Whole Genome Amplification (WGA) Kits (Sigma-Aldrich) according to the manufacturer’s instructions. To capture the coding regions, we used the SureSelectQXT Target Enrichment kit (Agilent). All sequencing was performed on the Illumina HiSeq 2000 platform (Illumina) at the New York Genome Center or at the Broad Institute.
A detailed description of data processing is provided in the Supplemental Experimental Procedures.
Quality Control of Sequencing Data
For details, see Supplemental Experimental Procedures.
Curation of MM and Pan-Cancer Driver Genes
For details, see Supplemental Experimental Procedures.
Targeted Sequencing of CTCs
CTCs were magnetically enriched with anti-human CD138 antibody conjugated with microbeads (Miltenyi Biotec) from 10 mL of PB of patients with symptomatic MM.
A detailed description of the procedure is provided in the Supplemental Experimental Procedures.
Statistical and Bioinformatics Analysis
Characterizing the Shared and Unique SNVs in BM Clonal Cells and CTCs
Single-nucleotide variants (SNVs) were called by MuTect (Cibulskis et al., 2013) by comparing tumor samples to matched normal samples using default parameters, with an additional filter that requires at least three high-quality reads supporting alternative variants.
Estimation of SSNV Cancer Cell Fraction and Clonal Dynamic
The algorithm ABSOLUTE was applied to estimate sample purity, ploidy, and absolute somatic copy numbers (Carter et al., 2012; Landau et al., 2013). These were used to calculate and compare the cancer cell fraction of SSNVs that were identified from MuTect and manually reviewed calling in both BM clonal PCs and CTCs via PHYLOGIC (Brastianos et al., 2015; Stachler et al., 2015). Clonal SSNVs were defined with a CCF >0.9. Shared subclonal SSNVs were defined with a CCF >0.05 in both BM and CTC samples. We defined non-shared subclonal SSNVs in either BM or CTC samples as those with a CCF lower than 0.05.
Somatic Copy-Number Alteration Identification
To estimate somatic copy-number alteration, we used ReCapSeg (http://gatkforums.broadinstitute.org/gatk/categories/recapseg-documentation), which calculated proportional coverage for each target region and then normalized each segment using the median proportional coverage in a panel of normal (PON) samples sequenced with the same capture technology. The sample was projected to a hyperplane defined by the PON, and the tumor copy ratio was estimated. These copy-ratio profiles were segmented with circular binary segmentation (CBS) (Venkatraman and Olshen, 2007). To estimate allelic copy number, germline heterozygous sites in the normal sample were called via GATK Haplotype Caller. Then, the contribution of each homologous chromosome was assessed via reference and alternate read counts at the germline heterozygous sites. Finally, we segmented the allele specific copy ratios using R package PSCBS. After accounting for purity and ploidy of each sample, we identified significant somatic copy-number alterations across the samples via GISTIC 2.0 (Mermel et al., 2011).
Supplemental Information
Highlights.
MM patient-derived CTCs mirror the mutational profile obtained from BM biopsy
Somatic copy-number alteration of CTCs was highly concordant with paired BM tumors
We suggest roles for CTCs in the prognostic and molecular profiling of MM patients
Acknowledgments
This work was supported in part by the Leukemia and Lympoma Society Specialized Center of Research (SCOR) grant and NIH R01 CA181683-01A1. This study was also supported by the Cooperative Research Thematic Network grants RD12/0036/0058 (CIBERONC); Instituto de Salud Carlos III, Spain, Instituto de Salud Carlos III/Subdireccio´ n General de Investigacio´ n Sanitaria (FIS: PI060339; 06/1354; 02/0905; 01/0089/01-02; PS09/01897/01370; G03/136; Sara Borrell: CD13/00340); and Asociacio´ n Espan˜ ola Contra el Ca´ ncer (GCB120981SAN). The study was also supported internationally by the International Myeloma Foundation Junior Grant Proposal, the Multiple Myeloma Research Foundation research fellow award, the American Association for Cancer Research (15-40-38-PAIV), and a European Research Council Starting Grant (680200-MYELOMANEXT).
Footnotes
Author Contributions
Y.M., B.P., J.S., J.P., S.M., A.D., J.F.S.M., F.M., and I.M.G. collected the samples, analyzed the data, performed the sequencing studies, and performed bioinformatics studies and wrote the manuscript. B.P., D.A., M.M., A.P.-G., Y.A., S.T., D.H., A.M.R., and A.S. collected samples and clinical data and performed flow cytometry. M.C., J.L., G.H., S.F., E.M.V.A., and V.D. performed sequencing and bioinformatics studies. F.P. and J.F.S.M. collected patient samples in Spanish centers. K.C.A. and N.C.M. collected patient samples at DFCI.
Accession Numbers
The accession number for the BAM files reported in this paper is NCBI dbGaP: phs001323.
References
- Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martin corena I, Dawson KJ, Iorio F, Nik-Zainal S, Bignell GR, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS, Horowitz PM, Cibulskis K, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5:1164–1177. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corre J, Munshi N, Avet-Loiseau H. Genetics of multiple myeloma: Another heterogeneity level? Blood. 2015;125:1870–1876. doi: 10.1182/blood-2014-10-567370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner MW, Leone PE, Walker BA, Ross FM, Johnson DC, Gonzalez D, Chiecchio L, Dachs Cabanas E, Dagrada GP, Nightingale M, et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood. 2007;110:3291–3300. doi: 10.1182/blood-2007-02-075069. [DOI] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014a;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, Sougnez C, Knoechel B, Gould J, Saksena G, et al. Multiple Myeloma Research Consortium Widespread genetic heterogeneity in multiple myeloma: Implications for targeted therapy. Cancer Cell. 2014b;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Kim S, Gould J, Knoechel B, Drier Y, Cotton MJ, Gray D, Birrer N, Wong B, Ha G, et al. Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. Sci Transl Med. 2016;8:363ra147. doi: 10.1126/scitranslmed.aac7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed AN, Bentley G, Bonnett ML, Zonder J, Al-Katib A. Chromosome aberrations in a series of 120 multiple myeloma cases with abnormal karyotypes. Am J Hematol. 2007;82:1080–1087. doi: 10.1002/ajh.20998. [DOI] [PubMed] [Google Scholar]
- Omberg L, Ellrott K, Yuan Y, Kandoth C, Wong C, Kellen MR, Friend SH, Stuart J, Liang H, Margolin AA. Enabling transparent and collaborative computational analysis of 12 tumor types within The Cancer Genome Atlas. Nat Genet. 2013;45:1121–1126. doi: 10.1038/ng.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanoja L, Paiva B, Flores-Montero J, Puig N, Burgos L, García O, Prósper F, Merino J, Vidriales MB, Mateos MV, et al. Next generation flow (NGF): A high sensitive technique to detect circulating peripheral blood (PB) clonal plasma cells (cPC) in patients with newly diagnosed of plasma cell neoplasms (PCN) Blood (Proceedings of the 57th ASH Annual Meeting) 2015;126:4180. [Google Scholar]
- Stachler MD, Taylor-Weiner A, Peng S, McKenna A, Agoston AT, Odze RD, Davison JM, Nason KS, Loda M, Leshchiner I, et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat Genet. 2015;47:1047–1055. doi: 10.1038/ng.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD, Johnson DC, Gonzalez D, Dagrada GP, Protheroe RK, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010;116:e56–e65. doi: 10.1182/blood-2010-04-279596. [DOI] [PubMed] [Google Scholar]
- Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, Proszek PZ, Johnson DC, Kaiser MF, Melchor L, et al. Mutational spectrum, copy number changes, and outcome: Results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015;33:3911–3920. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, Cancer Genome Atlas Research Network The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Adalsteinsson VA, Francis J, Cornils H, Jung J, Maire C, Ligon KL, Meyerson M, Love JC. Calibrating genomic and allelic coverage bias in single-cell sequencing. Nat Commun. 2015;6:6822. doi: 10.1038/ncomms7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.