Abstract
The ‘quenda genotype’ of Giardia was first identified in quenda (syn. southern brown bandicoots, Isoodon obesulus) in Western Australia in 2004. We aimed to formally describe this genotype as a species of Giardia, Giardia peramelis. Seventy five faecal samples positive for G. peramelis were obtained from quenda within the Statistical Division of Perth, Western Australia. These samples were used in morphological and molecular characterisation of G. peramelis. PCR amplification and sequencing was most successful at the 18S rRNA and ITS1-5.8s-ITS2 loci. Phylogenetic analyses placed G. peramelis external to the ‘Giardia duodenalis species complex’ and Giardia microti. This confirmed the uniqueness of G. peramelis, warranting classification as a separate species of Giardia. Study findings suggest quenda are a natural host for G. peramelis.
Keywords: Giardia, Quenda, Bandicoot, Marsupial, Parasite, G. peramelis
Graphical abstract
Highlights
-
•
Giardia peramelis is formally described.
-
•
PCR techniques for G. peramelis are outlined.
-
•
G. peramelis phylogenetic trees are presented.
1. Introduction
A novel genotype of Giardia was isolated and described from a quenda (syn. southern brown bandicoot, Isoodon obesulus) in southwest Western Australia (Adams et al., 2004). This was believed to be a distinct species of Giardia, but the lack of samples from additional infected animals precluded formal description of this genotype as a separate species at that time.
Since this initial description, the ‘quenda genotype’ of Giardia has been documented in quenda in several other locations in Western Australia (Thompson et al., 2010). It has also been identified using PCR from a calf in Western Australia, though it is unclear whether this reflected infection or cysts passing through the calf gut after ingestion of contaminated pasture (Ng et al., 2011a). The ‘quenda genotype’ of Giardia has not been isolated from other Australian marsupial species surveyed for Giardia spp. (McCarthy et al., 2008, Thompson et al., 2008, Thompson et al., 2010, Ng et al., 2011b, Thomasz, 2014, Vermeulen et al., 2015).
We undertook a parasitological survey of quenda in the greater Perth region, investigating the epidemiology of Giardia spp. infections in this species, and found infection with the ‘quenda genotype’ of Giardia to be common. We aimed to formally describe the ‘quenda genotype’ of Giardia as a separate species, Giardia peramelis, by describing the morphology of cysts and trophozoites and genetically characterising the parasite. It is recognised that for any parasite, once adequate data are available the names should be formalised at the species level (Brooks and Hoberg, 2000). This provides stability and is essential for effective communication. More specifically, we considered formal description of this parasite important to expand knowledge of the phylogenetic range of the genus Giardia; and in consideration of the public health significance of Giardia spp. in Australian marsupials, in differentiating zoonotic and non-zoonotic ‘strains’ of the parasite.
2. Materials and methods
2.1. Obtaining G. peramelis specimens
Faecal samples were collected from quenda trapped across 51 locations in the Statistical Division of Perth, Western Australia. Faecal material was also collected from the large intestine of quenda carcasses, obtained opportunistically from the same area. All samples were obtained under Murdoch University Animal Ethics Permit (R2530/12), and Department of Parks Wildlife Regulation 17 (SF009640) and Regulation 4 (CE004287) permits.
From each quenda, 2 mL faeces were thoroughly mixed in to 8 mL 10% buffered formalin, and 1 mL faeces were thoroughly mixed in to 8 mL 70% ethanol. Preserved faecal samples were stored at 4 °C until analysis.
The formalin-preserved faecal samples were screened for Giardia spp. cysts using immunofluorescence microscopy. Merifluor Cryptosporidium/Giardia kits (Meridian Bioscience, Inc. USA) were used according to manufacturer's directions for unconcentrated faecal samples. Slides were examined at 200X magnification. Samples positive for Giardia spp. were differentiated to a species level via PCR and sequencing (methodology in section 2.4). In addition, ten immunofluorescence negative samples from trapped quenda were randomly selected and subject to the same PCR and sequencing protocols as the immunofluorescence positive samples.
2.2. G. peramelis morphological description-trophozoites
Wet mounts were prepared from the small intestinal mucosa of two quenda carcasses, which were positive for G. peramelis on faecal testing (and were not positive for any other species of Giardia), and were considered sufficiently fresh, with minimal mucosal autolysis, for detection of trophozoites. The first third of the small intestine was gently scraped and the scrapings mounted on a microscope slide. The slides were examined for trophozoites using an Olympus BX50 microscope. A sample of mucosal scrapings from each quenda was also used to seed flasks containing Giardia media (section 2.2, below), and cultures were monitored regularly for the appearance of trophozoites.
Excystation of G. peramelis cysts was attempted three times, using faecal samples from three quenda that were positive for G. peramelis by immunofluorescence microscopy and PCR (and not positive for any other species of Giardia).
To purify G. peramelis cysts, 1 g of fresh faeces containing G. peramelis was homogenised in 1X PBS, passed through layers of gauze, and centrifuged at 0.6 G. Two wash steps were carried out, where the supernatant was removed, the pellet was resuspended in 1X PBS, and the sample was centrifuged at 0.6 G. Two sucrose samples were made-one to a specific gravity of 1.18, and another made up to 0.8 M. The 0.8 M solution was layered on top of the SG 1.18 solution, and the purified sample was layered on top. The sample was centrifuged at 0.2 G, and cysts were collected at the water/sucrose interphase. 1X PBS was added to this isolation and it was centrifuged at 0.6 G, with the supernatant removed subsequently. The cysts were resuspended in a final volume of 1 ml 1X PBS, and examined under the microscope.
For excystment, cysts were resuspended in 10 mL of acidified Hanks Balanced Salt Solution (HBSS) (30 g/L biosate peptone (BD, Annapolis, USA), 10 g/L glucose (Sigma–Aldrich, St. Louis, USA), 2 g/L sodium chloride (Chem-Supply, Adelaide, Australia), 2 g/L cysteine (Sigma–Aldrich, St. Louis, USA), 1 g/L K2HPO4 (Chem-Supply, Adelaide, Australia), 0.6 g/L KH2PO4 (Merck, Melbourne, Australia), 0.01 g/L ferric ammonium citrate (Sigma–Aldrich, St. Louis, USA), 0.2 g/L ascorbic acid (Sigma–Aldrich, St. Louis, USA), 0.5 g/L bovine bile (Sigma–Aldrich, St. Louis, USA), 100 ml/L newborn calf serum (SAFC Biosciences, Lenexa, USA) and 10 ml/L penicillin/streptomycin (Sigma–Aldrich, St. Louis, USA), pH 2) and incubated at 37 °C for 30 min. Cysts were then centrifuged at 0.2 g and washed twice in HBSS (pH7.2), and finally resuspended in HBSS (pH 7.2) medium and incubated at 37 °C. Cysts were monitored daily for excystment, for one week.
2.3. G. peramelis morphological description-cysts
To describe the morphology of G. peramelis cysts, faecal smears were prepared using formalin-preserved faecal samples from eight quenda that were positive for G. peramelis by PCR with sequencing (and not positive for any other species of Giardia). Smears were examined by bright field and Nomarski differential interference microscopy, using an Olympus BX50 microscope. G. peramelis cysts were photographed at 1000X magnification. ImageJ software (US National Institute of Health, Bethesda, Maryland), was used to measure cyst length and width.
2.4. G. peramelis molecular characterisation
2.4.1. DNA extraction and PCR amplification
Amplification by PCR was attempted on all immunofluorescence microscopy positive faecal samples and the ten randomly selected immunofluorescence negative samples. Ethanol-preserved faecal samples were centrifuged to separate ethanol from faeces, with the ethanol supernatant discarded. Samples were then twice re-homogenised in distilled water, centrifuged and supernatant discarded. DNA extraction was then conducted using the Maxwell® 16 Instrument (Promega, Madison, USA) as per manufacturer's instruction.
Amplification by PCR was attempted at three loci: 18S rRNA, ITS1-5.8s-ITS2 and glutamate dehydrogenase (gdh). Initially, a semi-nested PCR protocol was employed to amplify a 130 bp product of the 18s rRNA, with primers RH11/RH4 and RH11/GiarR (Hopkins et al., 1997, Read et al., 2002). The PCR reaction was performed in 25 μl volumes, consisting of 1–2 μl of extracted DNA, 2.0 mM MgCl2, 1 × reaction buffer (20 mM Tris-HCL, pH 8.5 at 25 °C, 50 mM KCl), 400 μM of each dNTP, 0.4 μM of each primer, 0.5 units of Taq DNA polymerase (Fisher Biotec, Perth, Australia), and DMSO 5%. Amplification conditions were modified from Hopkins et al. (1997), and involved a denaturing step of 95 °C for 6 min, then 40 cycles of 95 °C for 30 s, 53 °C for 30 s (56 °C in the secondary round) and 72 °C for 30 s, followed by a final extension of 72 °C for 7 min.
A nested PCR protocol was conducted to amplify a 330 bp product of the ITS1-5.8S-ITS2 region of the ribosomal gene, with primers developed by Caccio et al. (2010). The PCR reactions were the same as those used for 18s rRNA, but performed in 50 μl volumes. Conditions for amplifications were modified from Caccio et al. (2010), and involved an initial denaturing step of 95 °C for 5 min, then 40 cycles of 95 °C for 45 s, 59 °C for 30 s and 72 °C for 30 s, followed by a final extension of 72 °C for 7 min.
Finally, for gdh, a nested PCR protocol was used to amplify a 530 bp product, using the primer pairs Gdh1/Gdh2 and Gdh3/Gdh4 for the primary and secondary rounds respectively, as per Caccio and Ryan (2008). The PCR reaction was performed in 25 μl volumes, consisting of 2 μl of extracted DNA, 1.5 mM MgCl2, 1 × reaction buffer, 200 μM of each dNTP, 0.4 μM of each primer, 1 unit of Taq DNA polymerase (Fisher Biotec, Perth, Australia) and DMSO 5%. Conditions for amplifications were the same for both rounds, and involved an initial denaturing step of 94 °C for 2 min, then 35 cycles of 94 °C for 30 s, 50 °C for 30 s and 72 °C for 60 s, followed by a final extension of 72 °C for 7 min.
2.4.2. Sequencing of amplified product
PCR products were purified using an Agencourt AMPure XP system (Beckman coulter, Beverly, USA). Sequence reactions were performed using the Big Dye Terminator Version 3.1 cycle sequencing kit (Applied Biosystems), according to the manufacturer's instructions. Reactions were electrophoresed on an ABI 3730 48 capillary machine. Amplicons were sequenced in both directions, with resultant nucleotide sequences compared with published sequences on NCBI GenBank® using the basic alignment search tool (BLAST). Further sequence analysis was conducted using the sequence alignment program Sequencher™ 4.8 (Gene Codes, Ann Arbour, MI, USA).
Nucleotide sequence data reported in this paper are available in the NCBI GenBank® database under accession numbers KU306911, KU306912, KU306913, KU306914 and KU306915.
2.4.3. Phylogenetic analyses
Phylogenetic analyses of sequences obtained in this study were conducted using the programme MEGA6 (Tamura et al., 2013). Phylogenetic trees were inferred with the neighbour-joining method, with a bootstrapping of 1000 replicates. In addition, analyses were conducted using the maximum likelihood and maximum parsimony methods. Evolutionary distances were calculated using the Kimura 2-parameter method (Kimura, 1980). Published sequences representing Giardia muris, Giardia ardeae, Giardia microti and all assemblages within the ‘Giardia duodenalis species complex’ (G. duodenalis, Giardia enterica, G. canis, Giardia bovis, G. cati, Giardia simondi and the ‘pinniped genotype’) were retrieved from GenBank® (accession numbers for 18S rRNA: X65063, Z17120, AF006676, X52949, AF199446, DQ100287, AF199447, U09491, AF199443, AF199449, AF199448, AF199444, AF199450 and AY309064; accession numbers for ITS1-5.8s-ITS2: GU126450, X65063, X58290, M73684, GU124448, X52949, GU126432/33/34/35, AF239840, U09491, GU126437/38/40/43/44/45).
3. Results
Faecal samples positive for Giardia spp. by immunofluorescence microscopy were obtained from 99 trapped quenda, and 11 quenda carcasses. Of the immunofluorescence positive samples, 63/99 trapped quenda and 11/11 quenda carcasses were confirmed to be infected with G. peramelis by PCR and sequencing at one or more loci. Thirty six immunofluorescence positive faecal samples from trapped animals either did not amplify by PCR, or amplified product failed to give a readable sequence.
Of the ten randomly selected immunofluorescence microscopy negative samples tested by PCR and sequencing, one was positive for G. peramelis by PCR and sequencing at one locus.
3.1. G. peramelis morphology
G. peramelis trophozoites were not detected in intestinal scrapings of the two tested quenda carcasses, or in the cultures seeded with mucosal scrapings from the two quenda, and all attempts at G. peramelis excystation failed. This precluded morphological description of G. peramelis trophozoites.
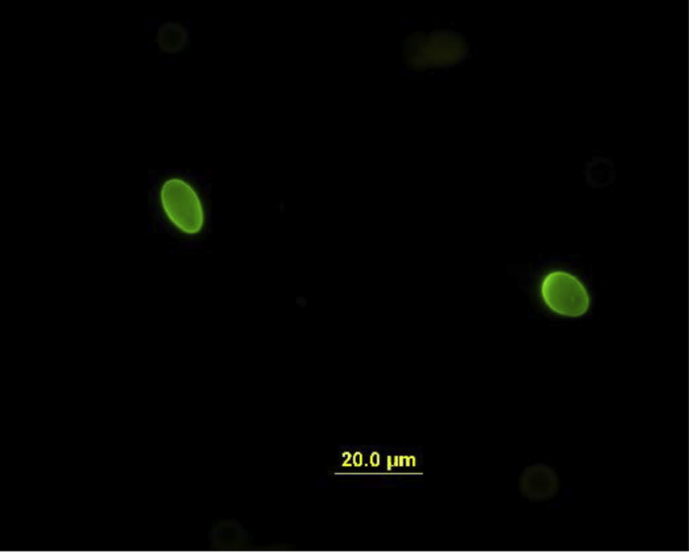
G. peramelis cysts are morphologically indistinguishable from cysts of the ‘G. duodenalis species complex’ and G. microti (Fig. 1, Fig. 2). G. peramelis cysts had an average length of 12.68 μm (standard deviation 0.72 μm; range 11.07–14.15 μm). Cyst width averaged 7.88 μm (standard deviation 0.47 μm; range 6.78–9.03 μm).
Fig. 1.
Cyst of Giardia peramelis-light microscopy.
Fig. 2.
Cysts of Giardia peramelis-immunofluorescence microscopy.
3.2. Giardia peramelis molecular characterisation
Of the 111 individual quenda faecal samples found positive for Giardia spp. by immunofluorescence microscopy or PCR, 75 (67.6%) were successfully amplified and sequenced at one or more loci. Sixty-four samples were sequenced at the 18s rRNA locus, 50 samples sequenced at ITS1-5.8s-ITS2 and two samples sequenced at the gdh locus (Table 1). Two of the 75 samples sequenced at all three loci, 37 samples sequenced at both 18S and ITS1-5.8s-ITS2, and the remaining 36 samples sequenced at only one locus (Table 2).
Table 1.
Molecular characterization results obtained at three loci for each quenda sample positive for Giardia spp. via immunofluorescence microscopy or PCR and sequencing (n = 111).
| Number of Giardia spp. positive quenda samples successfully amplified and sequenced at tested loci: |
|||
|---|---|---|---|
| Sample source (n) | 18S rRNA | ITS1-5.8s-ITS2 | gdh |
| trapped, IMFa + ve quenda (n = 99) | 54 | 42 | 2 |
| trapped, IMFa –ve quenda (n = 1) | 0 | 1 | 0 |
| quenda carcasses (n = 11) | 10 | 7 | 0 |
| Total no. successfully characterised at locus/ total Giardia spp. positives |
64/111 (57.7%) |
50/111 (45.0%) |
2/111 (1.8%) |
IMF = immunofluorescence microscopy.
Table 2.
Combined results of loci amplification for quenda confirmed to be infected with Giardia peramelis on PCR and sequencing (18S, ITS1-5.8s-ITS2 and gdh) (n = 75).
| Loci at which samples were successfully amplified and sequenced: |
|||||
|---|---|---|---|---|---|
| Sample source (n) | 18S only | ITS1-5.8s-ITS2 only | gdh only | 18S & ITS1-5.8s-ITS2 only | 18S & ITS1-5.8s-ITS2 & gdh |
| trapped, IMFa + ve quenda (n = 63) | 21 | 9 | 0 | 31 | 2 |
| trapped, IMFa –ve quenda (n = 1) | 0 | 1 | 0 | 0 | 0 |
| quenda carcasses (n = 11) | 4 | 1 | 0 | 6 | 0 |
| Total no. Characterised at locus combination/total G. peramelis positive quenda | 25/75 (33.3%) | 11/75 (14.7%) | 0/75 (0%) | 37/75 (49.3%) | 2/75 (2.7%) |
IMF = immunofluorescence microscopy.
Of the 116 sequences obtained, 114 were the novel G. peramelis; two sequences obtained at the 18S rRNA locus were suspected to be mixed infections of Giardia duodenalis/G. peramelis and Giardia canis/G. peramelis. These ‘mixed infection’ samples were also amplified at the ITS1-5.8s-ITS2 region locus-the resultant sequences were clearly identified as G. peramelis, with no ambiguous nucleotides evident.
Utilizing the BLAST tool within NCBI, the 18S rRNA sequences were highly similar to the published sequence AY309064, previously reported as the ‘quenda genotype’. Alignment in Sequencher™ revealed one single nucleotide polymorphism (SNP) between sequences obtained in this study (represented by QBY95 and QM22) and AY309064. In addition, one SNP was identified within a small subset of sequences obtained in this study, represented by sequence QBN13. A large region of extreme variability was identified between all sequences and another reported ‘quenda genotype’. Of the 292 bp published for this sequence (accession number HQ398319), a region of approximately 150 bp showed extreme mismatch.
Sequences obtained at ITS1-5.8S-ITS2 required the BLAST program selection of ‘somewhat similar sequences’ in order to achieve a result. The ‘G. duodenalis species complex’ was the primary match, but achieved low coverage and identities with all comparisons. Alignment of these sequences in Sequencher™ revealed one SNP, represented by samples QBY95/QM22 and QBN13. Similar BLAST results were obtained with the two gdh sequences.
3.2.1. Phylogenetic analysis
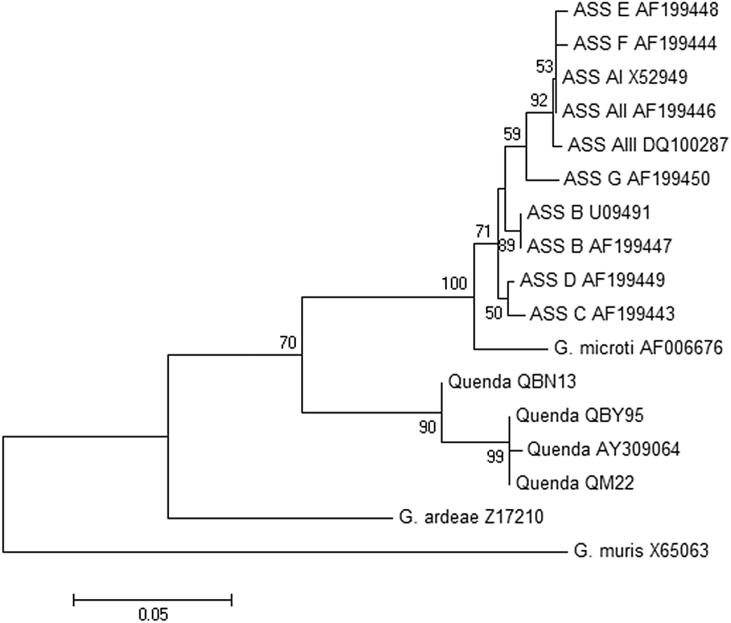
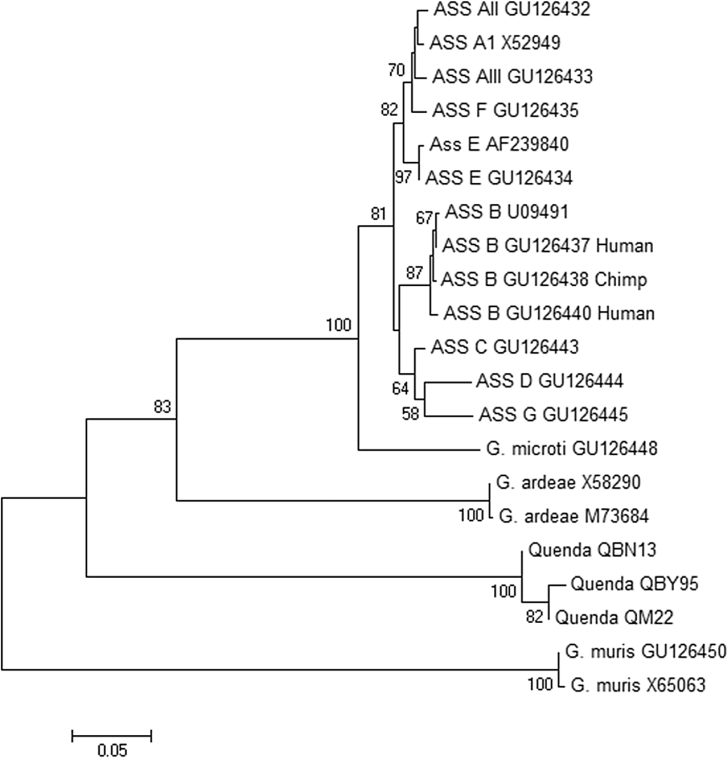
Very similar trees were obtained by neighbour-joining, maximum likelihood and maximum parsimony methods; only the neighbour-joining trees are presented here. Phylogenetic analysis of sequence data obtained at 18s rRNA confirmed that the Giardia genotype obtained from quenda in this study (G. peramelis) formed a separate clade with the ‘quenda genotype’ reference AY309064, and was distinct from all assemblages within the ‘G. duodenalis species complex’ and G. microti (Fig. 3). A similar topology was observed based on genetic data obtained at ITS1-5.8S-ITS2, with the exception that G. peramelis was also placed external to G. ardeae (Fig. 4). Phylogenetic analysis of gdh also placed the G. peramelis external to all assemblages within the ‘G. duodenalis species complex’ (results not shown).
Fig. 3.
Phylogenetic relationships of Giardia peramelis isolates obtained in this study (quenda QBN13, QM22, QBY95) with published reference material available at the 18s rRNA locus. Evolutionary history inferred using the neighbour-joining method supported with bootstrap test of 1000 replicates (values > 50% shown). G. muris is used as the out group.
Fig. 4.
Phylogenetic relationships of Giardia peramelis isolates obtained in this study (quenda QBN13, QM22, QBY95) with published reference material available at the ITS1-5.8S-ITS2 locus. Evolutionary history inferred using the neighbour-joining method supported with bootstrap test of 1000 replicates (values > 50% shown). G. muris is used as the out group.
4. Discussion
4.1. Giardia peramelis taxonomic summary
Type host: I. obesulus-quenda (syn. southern brown bandicoot).
Mammalian additional hosts: unknown.
Type locality: Perth, Western Australia.
Additional locations: other locations in Western Australia (Adams et al., 2004, Thompson et al., 2010).
Site of Infection: unknown; presumably small intestine, based on the known Giardia spp. site of infection in other mammalian hosts (Monis et al., 2009).
Prepatent and patent periods: unknown.
Material deposited: A sample of formalin-preserved and ethanol-preserved G. peramelis cysts, and photomicrographs of G. peramelis cysts, were deposited at the Western Australian Museum (specimen registration no. WAM Z68785).
Etymology: The specific epithet peramelis is derived from the subfamily Peramelinae/family Peramelidae/order Peramelemorphia-taxonomic classifications of the quenda, the only confirmed host of G. peramelis. This is in line with current taxonomic nomenclature for Giardia spp. (Monis et al., 2009).
This study confirms that G. peramelis (previously the ‘quenda genotype’ of Giardia) is a unique species. Based on genotyping at 18s rRNA, all G. peramelis isolates in this study strongly aligned with the original ‘quenda genotype’ data, generated by Adams et al. (2004). We have further confirmed the genetic novelty of G. peramelis with genetic data obtained at ITS1-5.8S-ITS2, which identified G. peramelis as belonging to the genus Giardia but did not match any published sequences of Giardia spp. Phylogenetic analyses of both genes place G. peramelis external to the ‘G. duodenalis species complex’ and external to G. microti. This supports the proposals of both Adams et al. (2004) and Thompson and Monis, 2004, Thompson and Monis, 2012 that G. peramelis is a novel lineage, distinct from described species.
The extreme mismatch observed between the G. peramelis 18S rRNA sequences reported here and in Adams et al. (2004), and the previously reported ‘quenda genotype’ (HQ398319) obtained from a calf (Ng et al., 2011a), suggests that a partial ‘quenda genotype’ may have been sequenced from the calf, along with other genetic material.
As G. peramelis cysts are morphologically indistinguishable from several other species of Giardia, molecular characterisation is required to differentiate G. peramelis from other Giardia spp. Our results suggest that the 18S rRNA and ITS1-5.8s-ITS2 loci are the most successful target regions for this purpose.
The substantial number of quenda observed to be infected with G. peramelis, sampled across a large number of locations in this study, suggests that quenda are a natural host for G. peramelis.
5. Conclusions
This study confirms that G. peramelis (formerly the ‘quenda genotype’ of Giardia) is a unique species of Giardia. It expands on known genetic and morphological data of this species, and describes techniques used in the genetic characterisation of G. peramelis.
Acknowledgements
The authors gratefully acknowledge the expert laboratory advice provided by Louise Pallant throughout this project; the Department of Parks and Wildlife; Whiteman Park, Perth Airport, the Botanic Gardens and Parks Authority, John Forrest National Park and the Cities of Joondalup, Cockburn and Armadale; WWF- Australia; participating property owners, managers and staff; expert advisors and volunteer field assistants; and Kanyana Wildlife Rehabilitation Centre. This project was supported by The Holsworth Wildlife Research Endowment- Equity Trustees Charitable Foundation, and the Australian Research Council. A.H was supported by an Australian Postgraduate Award.
References
- Adams P.J., Monis P.T., Elliot A.D., Thompson R.C.A. Cyst morphology and sequence analysis of the small subunit rDNA and ef1α identifies a novel Giardia genotype in a quenda (Isoodon obesulus) from Western Australia. Infect. Genet. Evol. 2004;4:365–370. doi: 10.1016/j.meegid.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Brooks D.R., Hoberg E.P. Triage for the biosphere: the need and rationale for taxonomic inventories and phylogenetic studies of parasites. Comp. Parasitol. 2000;67:1–25. [Google Scholar]
- Caccio S.M., Beck R., Almeida A., Bajer A., Pozio E. Identification of Giardia species and Giardia duodenalis assemblages by sequence analysis of the 5.8 S rDNA gene and internal transcribed spacers. Parasitology. 2010;137:919–925. doi: 10.1017/S003118200999179X. [DOI] [PubMed] [Google Scholar]
- Caccio S.M., Ryan U. Molecular epidemiology of giardiasis. Mol. Biochem. Parasitol. 2008;160:75–80. doi: 10.1016/j.molbiopara.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Hopkins R.M., Meloni B.P., Groth D.M., Wetherall J.D., Reynoldson J.A., Thompson R.C.A. Ribosomal RNA sequencing reveals differences between theC. genotypes of Giardia isolates recovered from humans and dogs living in the same locality. J. Parasitol. 1997;83:44–51. [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- McCarthy S., Ng J., Gordon C., Miller R., Wyber A., Ryan U.M. Prevalence of cryptosporidium and Giardia species in animals in irrigation catchments in the southwest of Australia. Exp. Parasitol. 2008;118:596–599. doi: 10.1016/j.exppara.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Monis P.T., Caccio S.M., Thompson R.C.A. Variation in Giardia: towards a taxonomic revision of the genus. Trends Parasitol. 2009;25:93–100. doi: 10.1016/j.pt.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Ng J., Yang R., McCarthy S., Gordon C., Hijjawi N., Ryan U. Molecular characterization of cryptosporidium and Giardia in pre-weaned calves in Western Australia and New South Wales. Vet. Parasitol. 2011;176:145–150. doi: 10.1016/j.vetpar.2010.10.056. [DOI] [PubMed] [Google Scholar]
- Ng J., Yang R., Whiffin V., Cox P., Ryan U. Identification of zoonotic cryptosporidium and Giardia genotypes infecting animals in Sydney's water catchments. Exp. Parasitol. 2011;128:138–144. doi: 10.1016/j.exppara.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Read C., Walters J., Robertson I.D., Thompson R.C.A. Correlation between genotype of Giardia duodenalis and diarrhoea. Int. J. Parasitol. 2002;32:229–231. doi: 10.1016/s0020-7519(01)00340-x. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasz A. Murdoch University; Perth: 2014. Parasites of the carnivorous marsupial Dasyurus geoffroii and the influence of translocation. [Google Scholar]
- Thompson J., Yang R., Power M., Hufschmid J., Beveridge I., Reid S., Ng J., Armson A., Ryan U. Identification of zoonotic Giardia genotypes in marsupials in Australia. Exp. Parasitol. 2008;120:88–93. doi: 10.1016/j.exppara.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Thompson R.C.A., Monis P. Variation in Giardia: implications for taxonomy and epidemiology. Adv. Parasit. 2004;58:69–137. doi: 10.1016/S0065-308X(04)58002-8. [DOI] [PubMed] [Google Scholar]
- Thompson R.C.A., Monis P. Giardia- from genome to proteome. Adv. Parasitol. 2012;78:57–95. doi: 10.1016/B978-0-12-394303-3.00003-7. [DOI] [PubMed] [Google Scholar]
- Thompson R.C.A., Smith A., Lymbery A.J., Averis S., Morris K.D., Wayne A.F. Giardia in western australian wildlife. Vet. Parasitol. 2010;170:207–211. doi: 10.1016/j.vetpar.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Vermeulen E.T., Ashworth D.L., Eldridge M.D., Power M.L. Investigation into potential transmission sources of Giardia duodenalis in a threatened marsupial (Petrogale penicillata) Infect. Genet. Evol. 2015;33:277–280. doi: 10.1016/j.meegid.2015.05.015. [DOI] [PubMed] [Google Scholar]