Abstract
Background
Bio-aerosols originate from different sources and their potentially pathogenic nature may form a hazard to healthcare workers and patients. So far no extensive review on existing evidence regarding bio-aerosols is available.
Objectives
This study aimed to review evidence on bio-aerosols in healthcare and the dental setting. The objectives were 1) What are the sources that generate bio-aerosols?; 2) What is the microbial load and composition of bio-aerosols and how were they measured?; and 3) What is the hazard posed by pathogenic micro-organisms transported via the aerosol route of transmission?
Methods
Systematic scoping review design. Searched in PubMed and EMBASE from inception to 09-03-2016. References were screened and selected based on abstract and full text according to eligibility criteria. Full text articles were assessed for inclusion and summarized. The results are presented in three separate objectives and summarized for an overview of evidence.
Results
The search yielded 5,823 studies, of which 62 were included. Dental hand pieces were found to generate aerosols in the dental settings. Another 30 sources from human activities, interventions and daily cleaning performances in the hospital also generate aerosols. Fifty-five bacterial species, 45 fungi genera and ten viruses were identified in a hospital setting and 16 bacterial and 23 fungal species in the dental environment. Patients with certain risk factors had a higher chance to acquire Legionella in hospitals. Such infections can lead to irreversible septic shock and death. Only a few studies found that bio-aerosol generating procedures resulted in transmission of infectious diseases or allergic reactions.
Conclusion
Bio-aerosols are generated via multiple sources such as different interventions, instruments and human activity. Bio-aerosols compositions reported are heterogeneous in their microbiological composition dependent on the setting and methodology. Legionella species were found to be a bio-aerosol dependent hazard to elderly and patients with respiratory complaints. But all aerosols can be can be hazardous to both patients and healthcare workers.
Introduction
Aerosols are defined as liquid or solid particles suspended in the air by humans, animals, instruments, or machines. Bio-aerosols are aerosols consisting of particles of any kind of organism [1, 2]. The characteristics of bio-aerosols differ depending on environmental influences such as humidity, air flow, and temperature. Aerosols, which are responsible for the transmission of airborne micro-organisms by air, consist of small particles named droplet nuclei (1–5μm) or droplets (>5μm). Droplet nuclei can stay airborne for hours, transport over long distances and contaminate surfaces by falling down [1]. It has been proven that droplets can contaminate surfaces in a range of 1 meter (3ft) [2]. The droplets are capable of penetrating deep into the alveoli, offering a potential route of infection [3]. The susceptibility of acquiring an infectious agent is determined by factors such as: virulence; dose; and pathogenicity of the micro-organism; and the host’s immune response [3–5]. Humans generate bio-aerosols by talking, breathing, sneezing or coughing [1]. Based on the infectious status of a person, the bio-aerosols are proven to contain influenza or rhinoviruses [6, 7], Mycobacterium tuberculosis [3], Staphylococcus aureus, Varicella Zoster Virus, Streptococcus spp. or Aspergillus spp. [8]. Moreover, bio-aerosols can be generated by devices such as ventilation systems, showers and high energetic instruments running on tap water. Showers and instruments cooled with tap water are able to spread environmental microbes such as Legionella spp. or other bacteria originating from water sources or water derived biofilms from tubing [4, 5, 9].
Due to the nature of their profession, healthcare workers (HCWs) are at higher risk to acquire pathogenic micro-organisms. Their risk of exposure is in line with the infectious nature of their patients, interventions or instruments that produce bio-aerosols. HCWs working in wards with patients suffering from pneumonia, who produce high virulence bio-aerosols, or HCWs exposed to bio-aerosol sources in dental practices, are at higher risk for developing disease or allergies [10, 11]. According to a risk assessment study, conducted in a hospital with HCWs exposed to high risk procedures, a risk ratio (RR) of 2.5 was found for acquiring viral or bacterial infection [12]. Multiple studies have found that HCWs were at higher risk to acquire an infectious disease, observing a high serological status of Legionella spp. and high rates of asymptomatic tuberculosis in dental practitioners and hospital staff [10, 13–15]. It is plausible that other diseases could also be acquired via bio-aerosols. Finally, evidence shows that patients with cystic fibrosis, who are immunosuppressed, are highly susceptible to airborne agents like Pseudomonas spp. [16]. Knowing this, we can assume that bio-aerosols with a high load of micro-organisms are a threat to immunocompromised patients suffering from leukemia, psoriasis, aplastic anemia and others [17]. Thus, the risk of acquiring pathogenic agents by bio-aerosols may be a hazard to both healthy and immunosuppressed patients as well as to HCWs.
To our knowledge, no detailed summary of the evidence regarding bio-aerosols in dental and hospital settings is available. Therefore, we chose to perform a scoping review on the present body of evidence regarding bio-aerosols. This results in an up-to-date summary of the literature, allowing us to make recommendations for future research by identifying gaps in current knowledge, and to underline the risks for HCW and immunocompromised. Since this is a scoping review, our objectives are broad and cover three areas concerning bio-aerosols in hospital and dental settings [18, 19]:
What are the sources that generate bio-aerosols?
What is the microbial load and composition of bio-aerosols and how were they measured?
What is the hazard posed by pathogenic micro-organisms transported via the aerosol route of transmission?
Methods
Design and search strategy
A scoping review was performed systematically according to the PRISMA statement for transparent reporting of systematic reviews and meta-analysis [20] and JBI Briggs Reviewers Manual [21] (see S1 PRISMA checklist). Three search strings were run in PubMed and EMBASE from inception to 09-03-2016. In PubMed the following strings were combined: Hospitals [Mesh] OR hospital OR hospitals OR "health care category" [Mesh] OR "health care" OR "Cross infection" [Mesh] OR "cross infection" OR cross-infection OR nosocomial OR "health facilities"[Mesh] OR "health facility" OR "health facilities" AND aerosols [Mesh] OR aerosol OR aerosols OR bioaerosol OR bio-aerosol OR "bio aerosol" OR bio-aerosols OR "bio aerosols" AND bacteria [Mesh] OR bacteria OR bacterial OR bacteremia OR bacteraemia OR sepsis OR septicaemia OR septicemia OR virus OR viruses OR viral OR viridae OR viral OR viruses [Mesh] OR Amoebozoa [Mesh] OR amoebozoa OR amoebe OR amoebas OR amoebic OR fungi [Mesh] OR fungus OR fungal OR fungi OR fungating OR parasites [Mesh] OR parasitic OR parasite OR parasites OR parasitemia OR parasitemias OR “micro organism” OR “micro organisms” microorganism OR microorganisms OR micro-organism OR micro-organisms OR “health care associated infections” OR infections OR infection OR infectious. For EMBASE we used the following strings combined: ‘hospitals/exp OR hospitals OR (health AND care AND category) OR healthcare OR ‘cross infection’OR ‘health facility’ OR ‘health facilities’ AND Infection/exp OR microorganism/exp OR fungi/exp OR virus/exp OR sepsis/exp OR bacteria/exp AND Aerosol OR aerosols/exp OR bioaerosol OR bio-aerosol OR bioaerosols OR aerosols.
Screening process and inclusion criteria
References yielded from the search strategy were imported in Covidence, an online web application for screening systematic reviews, and duplicates were removed. C.Z. and A.L. screened and scored the relevance of the hits independently, based on their title and abstract. The full text manuscripts were retrieved via Endnote, Google, Research Gate or by addressing the corresponding and/or first author. Subsequently, the studies were assessed on their eligibility for inclusion based on the full text. A study was included for final data extraction and summary when it met one of the following criteria: bio-aerosol composition; pathogenicity; sources; conducted in healthcare or the dental setting; published in English, German, French, Spanish or Dutch. Discussion papers, letters to the editor, animal studies, protocols, prevention of bio-aerosols, technical studies, reviews without pooled data, narrative reviews, development of drug therapy, or studies conducted in other settings besides healthcare were excluded. Additionally, a reference check and search through grey literature was conducted and included in the flowchart termed ‘snowballing’.
Data extraction and summary
Data on the origin of bio-aerosols was categorized based on sources. Studies on the microbial composition of the bio-aerosols were summarized based on the colony forming units (CFU). References that reported sampling time were recalculated for a sampling time of 10 minutes and finally Log-transformed to make comparison possible between studies. These studies are presented in figures. References not reporting sampling time were not summarized and are presented in the study of characteristics table. The micro-organisms reported in individual studies were summarized per type of organism and setting. Potential hazard for patients and HCWs were summarized narratively.
Results
A total of 5823 studies were retrieved, of which 678 duplicates and 4797 irrelevant studies were removed. After reading 311 abstracts, 201 full text studies were assessed for eligibility. This eventually resulted in 62 studies including references from snowballing (see Fig 1. PRISMA flowchart).
Fig 1. PRISMA flowchart.
Generation of bio-aerosols
One study reported solely on the generation of bio-aerosols [22]. Therefore, we extracted data on the generation of bio-aerosols from papers selected for the other objectives [23–44]. The sources of bio-aerosols in dental clinics were: ultrasonic scalers, high speed hand pieces, air turbines, three in one syringes, and air water syringes. Studies conducted in hospitals reported 30 different bio-aerosol generating sources. Humans produced aerosols by coughing, and sneezing. Patients with cystic fibrosis positive for Burkoderia cepacia were also capable of producing pathogenic aerosols. Interventions conducted by HCWs that produced aerosols were: colonoscopy, tracheal intubation, suction before and after intubation, manipulation oxygen mask, bronchoscopy, non-invasive ventilation, insertion of nasogastric tube, defibrillation, chest physiotherapy, and washing the patient. Bed making, ward rounds, tea trolley round, activity at bed, floor mopping, moving furniture, lunch time, drugs round, evening meal, vacuum cleaner, toilet use, cold-mist humidifier, shower, cleaning patients room and the nebulizer were found to be other activities in a hospital to produce aerosols [22].
Hospital environment
Thirty-one studies analyzed the microbial composition of bio-aerosols in the hospital environment [11, 30, 35–37, 39, 41–65]. The studies combined identified a total of 111 organisms by using culture techniques (see Table 1 for overview of micro-organisms identified and Table 2 study characteristics hospital setting). Fifty-six bacterial species (23 Gram-negative and 32 Gram-positive; 1 mycobacteria), 45 fungal genera and ten viral species were identified[11, 30, 35–37, 39, 41–52, 54, 56–66] Most bacteria originated from human skin or the human gut, the environment or water. The identified viruses originated from the human respiratory tract. The methods for collecting air samples from the bio-aerosols and the methods for culturing micro-organisms were heterogeneous. The method most frequently used to actively collect micro-organisms was the Andersen air sampler (N = 9). Four studies used passive collection of micro-organisms by placing Petri dishes with agar. In all studies, 21 different culture methods were used, wherefrom Tryptic soy agar (N = 7) was most frequently used.
Table 1. Overview of micro-organisms identified in hospital setting.
| Bacteria N = 56 | ||||
|---|---|---|---|---|
| Gram negative | Gram positive | |||
| Achromobacter xylosoxidans | Moraxella | Bacillus cereus | Rodococcus spp | Staphylococcus saprophyticus |
| Acinetobacter baumannii | Neisseria spp | Bacillus spp | Staphylococcus aureus | Staphylococcus schleiferi |
| Acinetobacter calcoaceticus | Ochrobactrum anthropic | Bacillus sutilis | Staphylococcus auriculans | Staphylococcus sciuri |
| Branhamella catarrhalis | Pantoea agglomerans | Brevibacterium casei | Staphylococcus capitis | Staphylococcus werneri |
| Burkholderia cepacia | Proteus | Clostridium difficile | Staphylococcus caprae | Staphylococcus xylosus |
| Enterobacter spp | Pseudomonas aeruginosa | Corynebacterium | Staphylococcus chromogenes | Sterptococcus pyogens |
| Escherichia coli | Pseudomonas fluorescens | Diphtheroid spp | Staphylococcus cohnii | Streptococcus spp. |
| Flavobacterium spp | Pseudomonas putida | Kocuria kristinae | Staphylococcus epidermidis | Viridans Streptococci |
| Klebsiella oxytoca | Rahnella aquatilis | Kocuria varians | Staphylococcus haemolyticus | Other strain:Mycobacterium tuberculosis |
| Klebsiella pneumonia | Shigella | Micrococcus luteus | Staphylococcus hominis | |
| Legionella pneumophila | Strenotrophomonas maltohilia | Micrococcus lylae | Staphylococcus lentus | |
| Neisseria flavescens | Micrococcus spp | Staphylococcus saprophyticus | ||
| Viruses N = 10 | ||||
| Human metapneumovirus | Human adenovirus | Influenza A H1N1 | Influenza B virus | Influenza virus |
| Parainfluenza virus 1–3 | Picornavirus | Respiratory syncytial virus | Rhinovirus | Toque teno virus |
| Parasites N = 0 | ||||
| None reported | ||||
| Fungi N = 45 | ||||
| Absidia spp | Candida spp | Epicoccum spp | Nigrospora spp | Scopulariopsos spp |
| Acremonium spp | Chaetomium spp | Exserohilum spp | Paecilomyces spp | Sepedonium spp |
| Alternaria spp | Chrysonilia spp | Fusarium spp | Penicillium spp | Sporotrichum |
| Aspergillosis fumigatus | Chrysoporium spp | Geotrichum candidum | Phoma spp | Stemphylium spp |
| Aspergillus flavus | Cladosporium spp | Geotrichum spp | Pithomyces spp | Syncephalastrum spp |
| Aspergillus niger | Conidiobulus spp | Gliocladium spp | Rhinocladiella spp | Trichoderma album |
| Aspergillus spp | Curvalaria spp | Monilia spp | Rhizopus nigricans | Trichosporon |
| Aureobasidium spp | Dactylaria spp | Mucor spp | Scedosporium spp | Ulocladium spp |
| Bipolaris spp | Emonsia spp | Myclia sterilia | Scopulariopsis brevicaulis | Verticillium spp |
Table 2. Study characteristics hospital setting.
| Study | Set up | Findings |
|---|---|---|
| Anderson 1996 | Setting: Pediatric hematology and oncology ward | Fungi:Aspergillosis fumigatus |
| Sampling method: Air dust sampler L100; 100L/min; 10 min before and after vacuum cleaning; 0.5m above cleaner. Fungal isolates by morphology. Sampling: Before and after vacuuming | Before: 24 CFU/m3 After: 62 CFU/m3 | |
| Augustowska 2006 | Setting: Pneumological ward, hospital | Mean monthly CFU/m3 range of airborne bacteria:257.1–436.3Mean monthly CFU/m3 range of fungi:9.9–96.1 |
| Sampling method Air sampled with a custom-designed particle-sizing slit sampler; 20L/min; daily at 9:00 and 13:00h. Blood and Sabouraud agar. | Bacteria: | |
| Gram positive cocci: 34.4–46.4% à Staphylococcus epidermidis; Micrococcus; Streptococcus. | ||
| Gram negative: 11.8–27.5%à Flavobacterium spp; Acinetobacter calcoaceticus; Pantoea agglomerans; Escherichia coli; Enterobacter spp; Klebsiella oxytoca; Pseudomonas auruginosa; Branhamella catarrhalis; Neisseria flavescens; Corynebacterium; Rodococcus spp; Bacillus spp.Fungi: 7.6–42.5%Aspergillus fumigatus: 77%Aspergillus niger; Aspergillus flavus; Aspergillus spp; Penicillium spp; Geotrichum candidum; Trichoderma album; Mucor spp; Rhizopus nigricans. | ||
| Best, 2012 | Setting: Toilet, hospital | Mean CFU: |
| Sampling method: Air sampled using AirTrace Environmental portable sampler placed at toilet seat height; 250-500L, 10cm above seat and 25 cm at handle height; 28.3L/min. Selective agar plate placed around the toilet; placed before flushing and remained for 90 min. | After flushing: 36Recovery of C. difficile mean CFU: | |
| Toilet lid closed vs open 0–30 min: 4 vs 1630–60 min: 4 vs 360–90 min: 0 vs 1 | ||
| Bacteria:C. difficile | ||
| Cabo Verde, 2015 | Setting: OT, SW, ES hospital | CFU/m3 range bacteria:ES: 240-736SW: 99-495OT: 12–170 |
| Sampling method: Air sampler MAS-100 100L/min; 1m above floor when possible.Trypic soy agar; malt extract agar; gram staining. | CFU/m3 range fungiES: 27–933 SW: 1–32 OT: 0–1 | |
| Total % bacteriaStaphylococcus aureus, capitis, hominis, epidermis, werneri: 51%Micrococcus luteus, lylae: 37%Neisseria: 4%Brevibacterium casei: 1%Shigella: 0.3%Proteus: 2% | ||
| Total % fungi | ||
| Penicillium spp: 41%Aspergillus spp: 24%Cladosporium spp: 14%Chrysonilia spp: 5%Chrysoporium spp: 3%Scopulariopsis brevicaulis: 3% | ||
| Chen, 2008 | Setting: TB and non TB area; hospital | Average concentration: |
| Sampling method: Air sampled using Nucleipore filter; 20L/min; 2x4h a day; 1.2–1.5m height. DNA isolation qPCR | TB area:3.8 x 103 ± 1.7 x 103 copy/m3 | |
| Non-TB area:3.9 x 10 ± 2.1 x 10 copy/ m3 | ||
| Emergency department:4.0 x 103 ± 2.6 x 103 copy/m3Ward area medical department:102 ± 5.5 x 101 copy/m3 | ||
| Chen, 2007 | Setting: TB and Non TB area; Hospital | Range concentration:1.43 x 10 copy/m3 to 2.06 x 105 copy/m3 |
| Sampling method: Air sampled using Nucleipore filter; 22L/min; 8h sampling; 1m from patients bed at 1.2–1.5m height. DNA isolation qPCR | ||
| Fekadu, 2015 | Setting: MW, SW hospital | Mean CFU/m3 (range): |
| Sampling method: Air samples were taken by passive method. Petri dishes placed 1m above the floor in room’s center. Samples collected in the morning and evening time of exposure: 30, 60, 90 min. Nutrient and Sabouraud agar plates. | Bacteria: 5583 (3106–9733) | |
| Fungi: 3008 (2123–4168) | ||
| Fennelly, 2012 | Setting: TB and Non TB area; Hospital | TB median CFU (range):16 (1–710) |
| Sampling method: Cough bio-aerosol sampling system method used to collect bio-aerosols in TB positive patients. Andersen six-stage sampler with 7H11 agar collection for 5 minutes. | ||
| Humphreys, 1994 | Setting: Hospital | Bacteria:B. cepacia |
| Sampling method: CF patients colonized with B. cepacia. Air sampled with Surface Air System air sampler, 900 l for 5 min; 100 cm from patient. Samples taken 15 min intervals for one hour after patient had left the room and 18 h after vacating. Selective B. cepacia agar medium. | During stay CFU/m3:Range: 1-158Mean: 32 | |
| After stay CFU/m3:Range: 3-53Mean: NR | ||
| Huynh, 2008 | Setting: Laboratory | Virus:Influenza virus; para influenza virus; Rhinovirus; Influenza A virus. |
| Sampling method: Reading aloud (20 min), quiet breathing (20 min), 20 voluntary coughs over a 3–5 min period in a separate sampling mask. PCR | ||
| Heimbuch, 2016 | Setting: Hospital | Mean CFU/cm2Mask 1 external layer: 24.15Mask 2 external layer: 3.33 |
| Sampling method: No direct bio-aerosol sampling. Viable bacteria found on mask was measured. Tryptic soy agar; Gram staining. | Bacteria:Kocuria kristinae, varians; Micrococcus spp.; Staphylococcus aureus, S. auriculans, S. capitis, S. caprae, S. chromogenes, S. cohnii, S. epidermidis, S. haemolyticus, S. hominis, S. lentus, S. saprophyticus, S. schleiferi, S. sciuri, S. warneri, S. xylosus; Acinetobacter baumannii; Ochrobactrum anthropic; Pesudomonas fluorescens /putida; Rahnella aquatilis; Stenotrophomonas matophilia. | |
| Knibbs, 2014 | Setting: Children’s hospital | Total mean CFU/m3 (range) per distance:1m: 52.6 (40.9–67.6)2m: 37.3 (28.9–48.0)4m: 24.8 (19.2–32.0) |
| Sampling method: CF patients colonized with B. cepacia. Cough bio-aerosols collected by Andersen Impactor (28.3L/min); chocolate bacitracin agar for B. cepacia and isolates by RT-PCR. | Total mean CFU/m3 (range) per duration:5 min: 14.6 (11.0–19.2)15 min: 11.9 (8.9–15.7)45 min: 7.7 (5.4–11.0) | |
| % of subjects positive for bacteria:P. aeruginosa: 100Mucoid P. aeruginosa: 78.9Non-mucoid P. aeruginosa: 94.7S. aureus: 26.3Strenotrophomonas maltohilia: 10.5 | ||
| Subjects positive for fungi (%):Trichosporon: 15.8Aspergillus spp: 15.8Scedosporium spp: 10.5Candida spp.: 5.7 | ||
| Kulkarni, 2016 | Setting: Infant nursing bay, hospital | Mean PFU of RSV:315.189; range: 82.600–1.120.000 |
| Sampling method: Air sampled with Westech six-stage Microbial sampler; 28.3L/min for 30 min; 1m distance from infected infant. PCR | 2h after discharge:6.175. | |
| Virus:RSV | ||
| Kumar, 2010 | Setting: GW, PW, OT, ICU, hospital | Bacteria %:S. aureus: 43.8Escherichia coli: 17.9Klebsiella pneumonia: 16Sterptococcus pyogens: 13.2Pseudomonas aeruginosa: 8.9 |
| Sampling method: Air sampled by passive settling plate technique. Nutrient, blood, and MacConkey’s agar 5 min exposure. | ||
| Lindsley, 2015 | Setting: Laboratory | Range 5 to 538 PFU, mean (SD): 142 (125). |
| Sampling method: Bio-aerosols collected using SKC BioSampler with 5ml vessel containing viral transport media: Hank Balanced Salt Solution. qPCR | Virus: Influenza A | |
| Li, 2003 | Setting: ICU, hospital | Range of mean CFU/m3 Fungi: 0–3115 |
| Sampling method: Air was sampled using Andersen six stage sampler; 28.3L/min; 1m height. Sampling time 20–30 min. Tryptic soya and malt extract agar. | Bacteria: 0–423 | |
| Martins-Diniz, 2005 | Setting: SW, ICU, hospital | Mean total fungi CFU/m3 |
| Sampling method: Air samples taken by Andersen sampler. 80L of air sampled on Sabouraud agar and chloramphenicol culture medium. Samples taking during the morning and immediately after cleaning, late afternoon and end of regular shift, monthly collections. | SC morning: 13,362 | |
| SC afternoon: 20,939 | ||
| ICU morning: 16.925 | ||
| ICU afternoon: 16,392 | ||
| Total CFU/m3 SW & ICU morning/afternoon: | ||
| Cladosporium spp.: 6,338/16,587 11,587/11,192 | ||
| Fusarium spp: 2,350/900 514/612 | ||
| Pencillium spp: 912/813 1,425/950 | ||
| Chrysosporium spp.: 401/562 637/950 | ||
| Aspergillus spp.: 362/289 775/413 | ||
| Aureobasidium spp.: 562/200 238/476 | ||
| Myclia sterilia; 350/300 64/237 | ||
| Monilia spp.: 325/100 62/250 | ||
| Paecilomyces spp.: 89/275 162/175 | ||
| Curvalaria spp.: 262/200 26/75 | ||
| Chaetomium spp.: 275/12 75/212 | ||
| Stemphylium spp.: 162/100 38/63 | ||
| Rhinocladiella spp.: 75/38 137/0 | ||
| Exserohilum spp.: 25/0 87/38 | ||
| Epicoccum spp: 0/75 88/150 | ||
| Phoma spp.: 100/25 201/125 | ||
| Alternaria spp.: 26/26 137/88 | ||
| Nigrospora spp.: 162/25 13/100 | ||
| Syncephalastrum spp.: 51/87 137/25 | ||
| Bipolaris spp.: 25/25 0 | ||
| Dactylaria spp.: 12/0 37/37 | ||
| Acremonium spp: 25/12 87/26 | ||
| Conidiobulus spp.: 0 12/112 | ||
| Verticillium spp.: 12/0 87/0 | ||
| Gliocladium spp.: 62/37 87/0 | ||
| Pithomyces spp.: 0/25 100/0 | ||
| Sepedonium spp.: 0 0 | ||
| Scopulariopsos spp.: 0/25 25/12 | ||
| Sporotrichum: 0/12 50/0 | ||
| Ulocladium spp.: 0/12 0/25 | ||
| Scedosporium spp.: 25/0 0 | ||
| Emonsia spp.: 0 0/12 | ||
| Geotrichum spp.: 12/0 0 | ||
| Menzies, 2003 | Setting: Screening center for TB | Average airborne viable CFU/m3 at induction: |
| Sampling method: Sputum induction by ultrasonic nebulizer for 15 min. Samples taken with Andersen six stage sampler 15L/min, at the height of the therapists breathing zone (1.5m height). Sheep blood agar. | Before: 233 | |
| During: 351 | ||
| After: 106 | ||
| Mirhoseini, 2015 | Setting: OT, ICU, SW, IM, hospital | Total mean CFU/m3: |
| Sampling method: Air sampled with AGI 12.5L/min 180–240 min. Tryptic soy agar culture. | OT: 396 | |
| ICU: 222 | ||
| SW: 537 | ||
| IM: 524 | ||
| Total: 420 | ||
| Mirzai, 2014 | Setting: OR, ED, Hospital | Total mean CFU/m3 ED & OR: |
| Sampling method: Air samples taken by eight step Andersen 28.1L/min. Once a month for a year every morning before start of the shift. Plates at 1m height and 1m from obstacles exposed for 10 min. Blood, BHI, and McConkey agar. | Total ER: 103.88 (33.84) | |
| Total OR: 63.32 (32.94) | ||
| Micrococcus: 14.85/16.09 | ||
| Streptococcus A: 1.26/2.19 | ||
| Viridans Streptococci: 2.92/1.72 | ||
| Pneumococcus: 7.81/3.81 | ||
| Escherichia coli: 6.91/2.0 | ||
| Bacillus sutilis: 6.64/1.63 | ||
| S. aereus: 14.17/10.92 | ||
| S.epidermis: 10.95/5.72 | ||
| S. saprophyticus: 11.35/5.45 | ||
| Bacilluscereus: 7.14/1.73 | ||
| Diphtheroid spp: 5.28/2.27 | ||
| Pseudomonas spp: 4/3.47 | ||
| Klebsiella: 4.19/1.09 | ||
| Enterobacter: 1.17/0.27 | ||
| Citrobacter: 0.77/1.62 | ||
| Branhamla: 0.19/0.36 | ||
| Bacterial, 20°C CFU/m3 mean (range): | ||
| Conv. Flow morning: 141 (42–325) | ||
| Conv. Flow evening: 82 (49–141) | ||
| Lam. Flow morning: 25 (0–92) | ||
| Lam. Flow evening: 82 (7–233)Bacterial, 30°C CFU/m3 mean (range): | ||
| Conv. Flow morning: 49 (14–92) | ||
| Conv. Flow evening:77 (35–170)Lam. Flow morning: 110 (14–572) | ||
| Lam. Flow evening: 11 (0–42)Gram negative CFU/m3 mean (range): | ||
| Conv. Flow morning:2 (0–14) | ||
| Conv. Flow evening: 0 | ||
| Lam. Flow morning: 0 | ||
| Lam. Flow evening: 0Fungi CFU/m3 mean (range): | ||
| Conv. Flow morning: 22 (0–71) | ||
| Conv. Flow evening: 38 (0–99) | ||
| Lam. Flow morning: 5 (0–21) | ||
| Lam. Flow evening: 80 (0–455) | ||
| Ortiz, 2009 | Setting: General hospital | Average 2 years TAC: |
| Sampling method: Air sampler MAS-100 100L/min; petri dish. Total aerobic count; rose-bengal agar, microscopy and staining. | OT: 25.6 CFU/m3 | |
| Sampling: Before and after intervention. 2 years. Plate count agar; Rose Bengal agar | MW: 67 CFU/m3 | |
| HR: 124.4 CFU/m3 Average 2 years FL: | ||
| OT: 0.05 CFU/m3 | ||
| MW: 6.9 CFU/m3 | ||
| HR: 10.6 CFU/m3 Fungi: | ||
| Aspergillosis fumigatus, flavus: 89%, 11%. | ||
| Bacteria: | ||
| not specified. | ||
| Sudharsanam, 2012 | Setting: OW, Hospital | Total range of bacteria: |
| 45–150 CFU/plate. | ||
| Sampling method: Passive air sampling and active by active using filter and impinger. Petri dishes at 60–70 cm height, exposed for 30 min. 3.5L/min of air for 20 min. Samples taken in multiple months. Blood, Sabouraud’s Dextroser, and MacConkey agar. | Total range of fungi: 0–13 CFU/plate | |
| Bacteria: | ||
| Coagulase negative staphylococci; micrococci; Enterobacter; Pseudomonas. | ||
| Fungi: | ||
| Aspergillus fumigatus; flavus; niger.; Absidia spp. | ||
| Stelzer-Braid, 2009 | Setting: Hospital | Virus: |
| Sampling method: Bio-aerosols collected with face mask. RT-PCR | Influenza A; Influenza B; parainfluenza 1, 2, 3, respiratory syncytial virus, human metapneumovirus, picornavirus. | |
| Thompson, 2013 | Setting: Hospital | Median RNA copy no/L (IQR): Airway suction: 1.852 (1.543–2.7521) |
| Sampling method: Air sampled with Glass May-3stage impingers at 55L/min at 1m height; 1m from patients head; 40 min collection. RT-PCR. | Baseline: 7.913 (2.436–11.613) | |
| Bronchoscopy: 148.805 (12.735–284.875) | ||
| Intubation: 2.838 (2.838–2.838)Virus: | ||
| Influenza A H1N1 | ||
| Vavricka, 2010 | Setting: Endoscopy unit, hospital | Mean CFU/m3 (range) with air suction: |
| Sampling method: Air sampled with MAS-100; 100L/min; before first colonoscopy and end of evening; 30 sec; 1.2m height; 0.3m distance. Colombia agar. | Before: 4 (1–7) | |
| End: 16 (9–22) | ||
| Mean CFU/m3 (range) without air suction: | ||
| Before: 14 (0–142) | ||
| End: 7 (1–88)Bacteria: | ||
| Staphylococci spp. | ||
| Vélez-Pereira, 2014 | Setting: ICU, hospital | Range mean (SD) CFU/m3: |
| Sampling method: Air sampled with cascade impactor; 28.3L/min; 1.5m height; mannitol salt; pseudomonas agar. | Staphylococcus spp 67.3 (1.7)– 277 (59.2) | |
| Pseudomonas spp 6.4 (1.7)– 204 (9.2) | ||
| Bacteria %: | ||
| Staphylococcus spp.: 71.5 | ||
| Pseudomonas aeruginosa: 64.6 | ||
| Verani, 2014 | Setting: Nephrology, hospital | Mean concentration/cm2 (SD): |
| Sampling method: Air sampled with microflow impactor sampler; 1,000L for virus and 180L for bacteria sampled; tryptone soy agar; samples before and after disinfection toilet. RT-PCR | Human adenovirus | |
| Before disinfection: 349 (51) | ||
| After disinfection: 1,371 (49) | ||
| Total viruses %: | ||
| Human adenovirus: 77 | ||
| Toque teno virus: 15 | ||
| Norovirus: 0 | ||
| Combination: 7 | ||
| Total bacteria %: 41 | ||
| Before disinfection: 1.57 | ||
| After disinfection: 1,371 (49) | ||
| Wainwright, 2009 | Setting: Children’s Hospital | Voluntary cough range CFU: 0–13,485 |
| Sampling method: Air sampled with Cough bio-aerosol sampling system and Anderson six stage impactors for 5 min in children with CF positive for Pseudomonas aeruginosa. Chocolate agar. | Mean CFU settle plate: 6 (95%CI 3–14) | |
| Bacteria: | ||
| Pseudomonas aeruginosa; Strenotrophomonas maltophilia; Achromobacter xylosoxidans. | ||
| Wan, 2011 | Setting: OR, hospital | Median CFU/m3 (range): |
| Sampling method: Air sampled with Andersen one-stage viable impactor at 28.3L/min volume for 3 min; 1.2–1.5m height; 1.5m from operation tables. Tryptic soy agar. | Transplantation room: 76.0 (9–477) | |
| Trauma room: 121.5 (13–756) | ||
| Cardiovascular surgery: 13 (0–163) | ||
| Colon surgery: 73.5 (0–567) | ||
| Orthopedic surgery: 89 (4–243) | ||
| Bacteria % in transplantation, trauma, cardiovascular surgery, rectal and orthopedic surgery room: | ||
| Bacillus spp: 32; 24; 16; 16; 34 | ||
| Micrococcus spp: 35; 25; 26; 39; 9 | ||
| Staphylococcus spp: 16; 20; 33; 26; 23 | ||
| Acinetobacter spp: 0; 8; 3; 0; 6 | ||
| Moraxella spp: 1; 0; 6; 0; 0; | ||
| Pseudomonas spp: 1; 0; 0; 0; 0 | ||
| Woo, 1986 | Setting: Hospital | Distance (cm) and colonies/plate: |
| Sampling method: Air sampled by passive technique placing plates in different places within a hospital shower. Buffered charcoal yeast extract agar. | Adaptor / adaptor + extension: | |
| 0: 3 / 3 | ||
| 1: 10 / 5 | ||
| 4: 7 / 5 | ||
| 10: 4 / 2 | ||
| 20: 2 / 1 | ||
| Bacteria: | ||
| Legionella pneumophila |
Abbreviation: AMHB = aerobic mesophilic heterotrophic bacteria; BHI = blood heart infusion; CFU = Colony forming units; CF = cystic fibrosis; cm = centimeters; ED = emergency department; ER = emergency room; ES = emergency service; FL = fungal load; GW = general ward; HR = hospital room; IM = internal medicine; IQR = inter quartile range; L = liters; min = minutes; m = meters; MW = maternity ward; NR = not reported; NTM = nontuberculous mycobacteria; OR = operation room; OT = operation theatre; OW = operation ward; PFU = plaque forming units; PW = pulmonary ward; RSV = respiratory syntical virus; SC = surgical center; SD = standard deviation; SW = Surgical Ward; TAC = total aerobic count; TB = tuberculosis; qPCR = quantitative polymerase chain reaction.
Fourteen studies analyzed the bacterial load of the bio-aerosols [11, 39, 41, 42, 45–47, 50, 55, 58, 60, 61, 64, 65]. The mean Log-10 of CFU/m3 ranged from 0.8 to 3.8 (see Fig 2). Additionally, five studies analyzed the bio-aerosol contamination before and/or after treatment, intervention or of a room when a patient with an infectious disease was present. The measured bacterial or fungal load ranged from Log 0.6–4.2 at baseline to Log 1.2–4.3 after the second measurement (see Fig 3) [30, 35, 43, 56, 57]. Seven studies reported on the fungal load in bio-aerosols during the day when patients were present in a hospital room. Fungal loads ranged from Log 0.8–3.5 CFU/m3 in various hospital wards [45, 47, 50, 56, 59, 61, 66]. Multiple studies quantified the air in patient specific areas or via specific methods.
Fig 2. Bacterial or fungal loads in mean Log-10 CFU/m3 in hospitals.
* = passive sampling method; # active sampling method.
Fig 3. Bacterial or fungal loads in mean Log-10 CFU/m3 in hospitals, measured twice.
* = passive sampling method; # active sampling method.
Two studies identified multiple viruses in bio-aerosols after patients with symptoms of a cold coughed, however both studies did not report on the viral load [51, 62]. Viral loads in the bio-aerosol ranged between Log 2.2 plaque forming units /m3 in the air of an infant nursery positive for RSV and Log 5.5 PFU/m3 in the air contaminated by patients positive for Influenza A virus [52, 54]. Another study reported the RNA copy/L and found Log 3.3–5.2 in aerosols produced by patients positive for Influenza A virus [33].
Dental environment
Seventeen studies analyzed the microbial composition of dental clinics [23–29, 67–76]. The studies cumulatively identified 38 types of micro-organisms by using culture techniques (see Table 3 for complete overview of micro-organisms identified and Table 4 for study characteristics in dental setting). Wherefrom nineteen bacteria (7 Gram-negative and 12 Gram-positive) and 23 fungal genera were detected. The bacteria originated from water, human skin and the oral cavity. None of the included studies looked for viruses or parasites. Similar to the hospital setting, the active Andersen air sampler (N = 4) and the passive culturing method by placing Petri dishes with agar (N = 6) were the most frequent used air sampling techniques. Thirteen different culture methods were used to identify the collected micro-organisms, of which Tryptic soy agar (N = 3) and blood agar (N = 3) were used most often.
Table 3. Complete overview micro-organisms identified in the dental setting.
| Bacteria N = 19 | |||||
|---|---|---|---|---|---|
| Gram negative | Gram positive | ||||
| Acinetobacter wolffii | Staphylococcus capitis | Staphylococcus chromogenes | Micrococcus luteus | Diphteroids | |
| Legionella spp. | Staphylococcus lentus | Staphylococcus haemolyticus | Micrococcus spp. | Corynebacteria | |
| Pseudomonas aureus | Staphylococcus xylosus | Staphylococcus epidermidis | Micrococcus lylae | Bacillus spp. | |
| Staphylococcus aureus | Staphylococcus fominis | Bacillus pumilus | Actinomycetes | ||
| Viruses N = 0 | |||||
| None reported | |||||
| Parasites N = 0 | |||||
| None reported | |||||
| Fungi N = 23 | |||||
| Alternaria alternata | Aspergillus flavus | Cladosporium cucumerinum | Geotrichum spp | Stemphylium spp | |
| Alternaria brassicicola | Aspergillus fumigatus | Cladosporium ramotenellum | Monocillim indicum | Stemphylium spp | |
| Alternaria citri | Aspergillus niger | Cladosporium sphaerospermum | Monodictys glauca | Ulocladium alternariae | |
| Arthrinium phaesospermum | Botrytis spp | Cladosporium spp | Pencillium spp | ||
| Aspergillus | Cladosporium cladosporiodias | Cladosporium spongiosum | Penicillium chrysogenum | ||
Table 4. Study characteristics in the dental setting.
| Study | Set up | Findings |
|---|---|---|
| Almaghlouth, 2007 | Setting: Dental clinic | Before: Diphteroids spp; Micrococcus spp |
| Sampling method: Passive air sample: Blood agar and Brain Heart agar culture plates. 15–20 min exposure during; 30 min in advance and 2h after treatment. | During: Diphteroids spp; Micrococcus spp; Staphylococcus epidermidis. | |
| After: Diphteroids spp; Micrococcus spp; Staphylococcus epidermis, aureus. | ||
| Bennett, 2000 | Setting: Dental clinic | Range max peak CFU/m3 |
| Sampling method: Air sampled with The Casella slit sampler; 30L/min; 2 min during treatment; 1m from patients mouth at bench height. Andersen sampler 28L/min 5 min; during treatment; 2m from patient; at foot of dental chair. Tryptone yeast extract Cystine agar; Columbia blood agar. | Winter: 6.3 x 103–8.7 x 103 | |
| Summer: 3.0 x 103–6.2 x 103 | ||
| Bacteria: | ||
| EPS producing streptococci: 50% | ||
| Barlean, 2010 | Setting: Dental clinic | Mean (range) CFU/m3 mesophilic bacteria: |
| Sampling method: Passive air sample: Tryptone soy agar, sheep blood; Sabouraud agar; exposed 15 min; 30cm and 2.5m distance from dental unit. | Before: 129 (42–273) | |
| After: 429.6 (105–1018) | ||
| Range CFU/m3 fungi: | ||
| Before: 21–29 | ||
| After: 52–808 | ||
| Bacteria: | ||
| Mesophilic bacteria: Staphylococcus aureus: 6.6.% | ||
| Fungi: NR | ||
| Cellini, 2000 | Setting: Dental clinic | CFU/m3 range: |
| Sampling method: Air sampled with Air Microbial index, plate method; placed prior to exposure; 1h exposure at 1m height and 1m from wall; before and after | 4–18 | |
| Dutil, 2007 | Setting: Dental clinic | Mean endotoxin in air CFU/m3: |
| Sampling method: Andersen six-stage air sampler; 28.3L.min during 20 min; 2h before; during and 2h after dental treatment. 30cm from patients mouth. R2A and blood agar. | Before: 0.34 | |
| During: 0.54 | ||
| After: 0.33 | ||
| Duell, 1970 | Setting: Dental clinic | TB (not found) |
| Sampling method: Andersen six stage air sampler; petri dishes; 30 cm from patient; air sampling 15min. | ||
| Grenier, 1995 | Setting: Dental clinic, closed and multichair | Mean CFU/m3 (SD) 1: |
| Sampling method: Air sampled with Slit-to-agar biological air sampler, 20L/min; 30 min before, during, after treatment duration of 30 min; 122cm from patient at 91 cm height | Before: 12 (4) | |
| Three settings: | During: 216 (75) | |
| 1. Closed dental operatory using ultrasonic scaler. | After: 44 (14) | |
| 2. Operative treatments in closed dental operatory. | 2h after: 10 (1) | |
| 3. Multichair | 4h after: 6 (2) | |
| Mean CFU/m3 (SD) 2: | ||
| Before: 14 (4) | ||
| During: 75 (22) | ||
| After: 51 (22) | ||
| 2h after: 12 (4) | ||
| 4h after: 9 (4) | ||
| Mean CFU/m3 (SD NR!)3: | ||
| Before: 13.5 | ||
| After: 97.75 | ||
| 3h after initiation: 82.75 | ||
| 7h after: 9 | ||
| Guida, 2012 | Setting: Dental clinic within a hospital | Active sampling CFU/m3 (SD) |
| Sampling method: Air sampled using Surface Air System; 180L/min 500L volume; 1m height; 1m from unit. Passive sampling with petri dish exposed for 1h. Before; during and after treatment. Tryptone soya agar. | Before: 86.6 (50.6) | |
| During: 82.3 (48.3) | ||
| After: 86.5 (64.8)\ | ||
| Passive sampling CFU/m3 (SD): | ||
| Before: 38.3 (21) | ||
| During: 39.6 (28.2) | ||
| After: 20 (11.5) | ||
| Kadaifciler, 2013 | Setting: Dental clinic | AMHB range mean CFU/m3 (SD): |
| Sampling method: Air sampled with HiAirflow, 100L/min, 1.4 height, near to dental unit, before; during and after treatment. Trypton soya agar for AMHB; Sabouraud and RB-agar for fungi. | Before treatment: 10 (0)– 453 (21) | |
| After treatment: 10 (0)– 2.477 (57) | ||
| Fungi range mean CFU/m3 (SD): | ||
| Before treatment: 0–42 (23) | ||
| After treatment: 0–94 (95) | ||
| Yeast range mean CFU/m3 (SD): | ||
| Overall: 3–25 | ||
| Bacteria: | ||
| Micrococcus spp.; Staphylococcus xylosus; Staphylococcus lentus; Staphylococcus chromogenes. | ||
| Fungi %: | ||
| NSF: 27.30 P. chrysogenum: 21.27; Pencillium: 11.11; C. Cucumerinum: 5.55; Alternaria brassicicola: 4.96; Cladosporium spp: 4.49; Aspergillus flavus: 3.86; Alternaria alternata: 2.83; Alternaria citri: 2.60; C. Cladosporiodias: 2.48; C. spongiosum: 2.48; Aspergillus: 1.93 Aspergillus niger: 1.30; Ulocladium alternariae: 0.82; Arthrinium phaesospermum: 0.82; Stemphylium spp.: 0.82; Acremonium zonatum: 0.82; Botrytis spp.: 0.82; Cladosporium sphaerospermum: 0.82; Monocillim indicum: 0.82; Cladosporium ramotenellum: 0.82; Monodictys glauca: 0.82. | ||
| Yeast %: | ||
| Geotrichum spp: 12.62 | ||
| Kimmerle, 2012 | Setting: Dental clinic, multi chair | Bacteria average CFU/m3: |
| Sampling method: Air sampled with Andersen biological air sampler total of 100L at different time points. Colombia blood agar plates and enterococci-selective bile esculin agar; 16S RNA gene sequencing; Gram staining. | Micrococcus luteus: 188.8; Staphylococcus epidermidis & haemolyticus: 114.5; Micrococcus lylae: 16.6; Pseudomonas: 10.6; Chrysemona luteda: 0.5; Staphylococcus hominis: 9.0; Acinetobacter wolffii: 5.1; Pseudomonas stutzeri: 0; Staphylococcus capitis: 3.7; Bacillus pumilus: 6.8Fungi %: | |
| Aspergillus fumigatus: 4.8; Aspergillus niger: 0.9; Aspergillus flavus: 0.2 | ||
| Kedjarune, 2000 | Setting: Multi chair dental clinic | Total bacteria |
| Sampling method: Air sampled with Slit-to-agar air sampler 75cm height and 30-35cm from dentists. | Total mean (SD) CFU/m3: | |
| Before: 177.41 (211.14); During: 232.49 (163.35) | ||
| After: 44.58 (46.82) | ||
| Total mean (SD) before/ during CFU/m3: | ||
| Endodontic: 264.16 (143.53) / 270.29 (158.33) | ||
| Operative: 188.28 (114.74) / 186.23 (69.26) | ||
| Scaling: 245.10 (134.55) / 182.57 (63.99) | ||
| Bacillus: | ||
| Total mean (SD) CFU/m3: | ||
| Before work: 10.89 (9.9); during work: 9.84 (20.14); after work: 3.34 (7.41) | ||
| Total mean before and during (SD) CFU/m3: | ||
| Endodontic: 11.15 (9.39) / 6.32 (4.98) | ||
| Operative: 16.20 (32.39) / 6.17 (7.58) | ||
| Scaling: 17.05 (11.74) / 8.69 (4.78) | ||
| Pankhurst, 1995 | Setting: Dental clinic | Bacteria: |
| Sampling method: CF patients colonized with B. cepacia. Air sampled before, during and after treatment by Surface Air System sampler, 540L per patient, 0.25m from patient. Selective culture for B. cepacia. | B. cepacia (not found) | |
| Pasquarella, 2012 | Setting: Dental clinic | Total mean bacteria CFU/m3before and after treatment: |
| Sampling method: Passive air sample placing Tryptone Soya Agar for 1 hour, 1 m above the floor. Active air sampling using SAS sampler, 180L/min volume 500 L. | Passive sampling: 78/110Active sampling: 12/14 | |
| Rautemaa, 2006 | Setting: Dental clinic | High speed: |
| Sampling method: Passive air sample: Horse blood chocolate agar; 6 locations; 0.5m – 2m from patient. Gram staining. Sampling 1.5 and 3 hours exposure at start of treatment | <1m: mean CFU 823/m2 | |
| >1.5m: mean CFU 1120/m2Periodontal: mean CFU 598/m2 | ||
| Bacteria: | ||
| Gram positive cocci; Gram negative cocci; Gram positive rods; Gram negative rods. | ||
| Szymanska, 2008 | Setting: Dental clinic | Total bacteria % before disinfection: |
| Sampling method: Air samples collected with portable Air Sampler RCS plus, placed 25 cm distance from patient, 100L air per sample. Tryptic soy agar and GP2 microplate. Sampling before and after disinfection (H2O2) of DUWL. | Gram negative: 1.31; Staphylococci and other catalase positive cocci: 15.70; Streptococci and other catalase negative cocci: 79.23; Endospore forming bacilli: 1.45%; Corynebacteria and related organisms: 2.30; Actinomycetes: 0.01% | |
| Total bacteria % after disinfection: | ||
| Gram negative: 1.08; Staphylococci and other catalase positive cocci: 61.19; Streptococci and other catalase negative cocci: 24.28; Endospore forming bacilli: 7.92; Corynebacteria and related organisms: 4.18; Actinomycetes: 1.35 | ||
| Mean CFU/m3 before disinfection: | ||
| Gram negative: 52; Staphylococci and other catalase positive cocci: 624; Streptococci and other catalase negative cocci: 480; Endospore forming bacilli: 50; Corynebacteria and related organisms: 30; Actinomycetes: 0 | ||
| Mean CFU/m3 after disinfection: | ||
| Gram negative: 0; Staphylococci and other catalase positive cocci: 640; Streptococci and other catalase negative cocci: 90; Endospore forming bacilli: 50; Corynebacteria and related organisms: 30; Actinomycetes: 0 | ||
| Timmerman 2003 | Setting: Dental clinic | Total CFU during high volume evacuation: |
| Sampling method: Passive air sampling by placing Petri dishes during ultrasonic scaling. Baseline measure for 10min exposed dishes in middle of operatory. 2 plates on tray table over patient’s chest 40cm away from mouth. 2 plates on cart 150cm from patient’s mouth next to wall, behind patient and dentist at 100cm height exposed for 20min. BHI agar. | Before: 0.2 (0.4) | |
| 0–5 min 40cm: 0.4 (0.7) | ||
| 20–25 min 40cm: 1.6 (1.3) | ||
| 0–20 min 150cm: 5.4 (8.3) | ||
| 20–40 min 150cm: 2.7 (3.2) | ||
| 0–40 min 150cm: 8.1 (11.3) | ||
| Total CFU during conventional dental suction: | ||
| Before: 0.6 (0.7) | ||
| 0–5 min 40cm: 2.5 (2.9) | ||
| 20–25 min 40cm: 1.8 (1.6) | ||
| 0–20 min 150cm: 4.3 (3.5) | ||
| 20–40 min 150cm: 6.3 (5.9) | ||
| 0–40 min 150cm: 4.0 (4.1) |
Abbreviations: BHI = brain heart infusion; TB = tuberculosis; CFU = colony forming units; cm = centimeters; DUWL = dental unit waterline; h = hours; m = meters; min = minutes; SD = standard deviation.
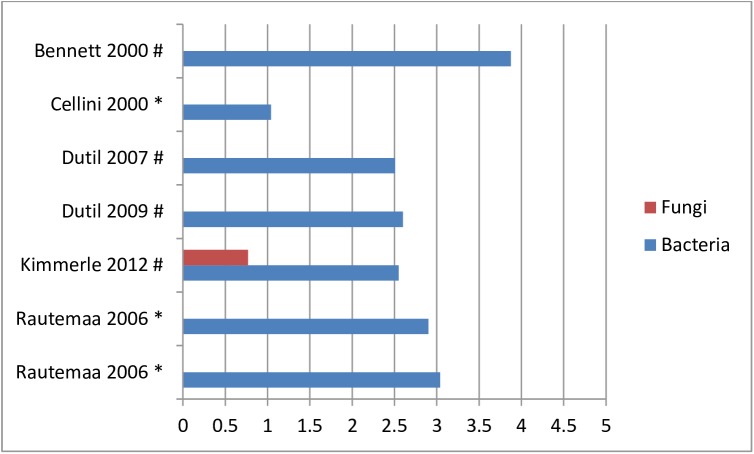
The mean bacterial load in the bio-aerosols ranged from Log 1–3.9 CFU/m3 (see Fig 4). Furthermore, six studies analyzed the bio-aerosol contamination before and after treatment. The bacterial or fungal load ranging from Log -0.7–2.4 CFU/m3 at baseline and from Log 1–3.1 CFU/m3 after treatment (see Fig 5) [25, 68, 71, 72]. Only one study reported on the relation between the distance from the bio-aerosol generating source and the bacterial load. They found a higher bacterial load in the bio-aerosols at 1.5 meter from the oral cavity of the patient than in the bio-aerosols within 1 meter from the patient [26]. One study screened for B. cepacia and one screened for M. tuberculosis, however both studies could not retrieve these organisms after regular patient treatment [24, 28].
Fig 4. Bacterial or fungal loads in mean Log-10 CFU/m3 in dental.
* = passive sampling method; # active sampling method.
Fig 5. Bacterial or fungal loads in mean Log-10 CFU/m3 in dental, measured twice.
* = passive sampling method; # active sampling method.
Hazard of a bio-aerosol
Seven studies reported on the hazard of micro-organisms to HCWs and/or patients, see Table 5 study characteristics hazard in healthcare and the dental setting [12, 31, 34, 45, 77–79] Three studies looked into the risks for patients when exposed to Legionella pneumophila containing sources that may produce bio-aerosols [34, 38, 78]. They reported that cooling towers, air conditioning or mechanical ventilation systems could be a source of L. pneumophila. Kool et al. concluded that patients had an increased risk to acquire L. pneumophila in hospitals when they used corticosteroids (OR = 13; 95CI% 1.6–102) and when intubated (OR = 10; 95%CI 1.3–73) [34]. Another study identified smoking, drinking alcohol, having chronic lung disease and cancer as risk factors for getting an infection with L. pneumophila [78]. For the dental clinic there is one case study reported that reported of irreversible septic shock and died after two days in a patient that was infected with L. pneumophila [79].
Table 5. Study characteristics hazard in healthcare and the dental setting.
| Study | Set up | Findings |
|---|---|---|
| Augustowska 2006 | Study the variability of airborne microflora of a hospital ward in pneumonological department.Organism: | Decrease of lung function: |
| Mesophilic bacteria; fungi | - Decrease spirographic indices in asthmatic patients at increase of bacteria/fungi in air: 37.5% | |
| Study design: | - Decrease of vital capacity. | |
| Cross-sectional study with microbiological examination of the air and the lung function in asthmatic patients. | - Decrease of forced expiratory volume in 1 second. | |
| Browning 2012 | Awareness of the risks involved in treating active herpes labialis in a dental clinic | Case 4:Dental hygienist got both eyes infected with HSV-1. Three possibilities: 1) high exposure to active herpes patients; 2) ultrasonic scaling producing HSV-1 bio-aerosol; 3) rubbing eyes while working. |
| Organism: HSV-1 | ||
| Study design: Case series, N = 4 | ||
| Kool 1998 | Investigate a clustered of cases of legionnaires disease among patients at a hospital. | Exposure factors OR univariate: |
| Organism: L. pneumophila | - Corticosteroids: 5.1 (1–50) SIG—Intubation: 4.6 (1.1–27) SIG | |
| Study design: Case-control study N = 74 | - Showering: 0.1 (0–1.3) NS—Medication nebulizer: 1.5 (0.3–6.8) NS | |
| - Drinking tap water: 1.3 (0.3–5.4) NS | ||
| Exposure factors OR multivariate: | ||
| - Walking in hallway 1–3 days: 0.07 (0.01–0.6) NS- Corticosteroids: 13 (1.6–102) SIG. | ||
| - Walking in hallway >3 days: 0.3 (0.03–2.6) NS—Intubation: 10 (1.3–73) SIG. | ||
| MacIntyre 2014 | Description of the range of exposure to HRP in HCWs. | RR infection high risk procedure performed vs not performed: |
| Organism: Adenovirus; human metapneumovirus; corona virus; parainfluenza virus; influenza virus A, B; rhinovirus A,B; streptococcus pneumonia; mycoplasma pneumonia; B. pertussis; Legionella spp.; chlamydophilia; Haemophilus influenza type B. | Clinical respiratory infection: 2.5 (1.3–6.5) P<0.01 | |
| Confirmed virus: 2.8 (0.9–8.7) P = 0.07 | ||
| Study design: | Virus or bacteria: 2.6 (1.4–5) P<0.01 | |
| Prospective study, participants recording following procedures on a daily basis: Provision of nebulizer medications; suction; intubation; bio-aerosol-generating procedures; chest physiotherapy. N = 481 | Influenza: 1.5 (0.2–13) NS | |
| RR bacteria of virus in HCWs: | ||
| Influenza vaccine: 1.3 (0.56–2.8) NS | ||
| Hand washing: 0.7 (0.3–1.63) NS | ||
| HRP performed: 2.9 (1.37–6.22) P = 0.01 | ||
| Palusinska-Szysz 2009 | Description of the pathogenicity of legionella | Symptoms: |
| Organism: L. pneumophila | Early: mild cold; low fever; malaise; anorexia; muscles aches; puss forming sputum; blood streaked sputum; cough blood. | |
| Later: high fever; bronchiolitis; alveolitis; lung damage with infiltrated regions. | ||
| Study design: Review | Risk factors instruments: | |
| Air conditioning; cooling towers; whirlpool spas; water delivery systems; contaminated birth bathtub; intubation; mechanical ventilation; aspiration; respiratory equipment. | ||
| Risk factors in human: | ||
| neonates; elderly; diabetes; chronic lung disease; chronic severe renal failure; hematologic malignancies; lung cancer; male gender; alcohol; gluco-corticosteroids; immunosuppression; high iron; smokers. | ||
| Ricci, 2012 | Description infection source of legionella | An 82-year-old woman admitted to intensive care unit with fever and respiratory distress. The woman was positive for L. pneumophila. The woman has acquired the infection from the contaminated DUWL biofilm and died 2 days after diagnosis. |
| Organism: L. pneumophila | OR per procedure: | |
| - Tracheal intubation: 6.6 (2.3–18.9)–SIG | ||
| Study design: Case control | - Non-invasive ventilation: 3.1 (1.4–6.8) SIG | |
| - Suction before intubation: 3.5 (0.5–24.6) NS | ||
| Tran 2012 | Risk of transmission of acute respiratory infections to HCWs exposed to bio-aerosol generating procedures | - Suction after intubation: 1.3 (0.5–3.4) NS |
| Organism: SARS | - Nebulizer: 0.9 (0.1–13.6) NS | |
| Study design: Systematic review and meta-analysis | - Manipulation oxygen mask: 4.6 (0.6–32.5) NS | |
| - Bronchoscopy: 1.9 (0.2–14.2) NS | ||
| - Insertion of nasogastric tube: 1.2 (0.4–4.0) NS | ||
| - Chest compression: 1.4 (0.2–11.2) NS | ||
| - Defibrillation: 2.5 (0.1–43.9) NS | ||
| - Chest physiotherapy: 0.8 (0.2–3.2) NS |
Abbreviation: HCWs = healthcare workers; HSV-1: herpes simplex virus-1; HRP = high risk procedures; NS = not significant; OR = odds ratio; RR = risk ratio; SARS = severe acute respiratory syndrome; SIG = significant; N = number of subjects.
One systematic review reported on the pooled odds ratio (OR) for the transmission and exposure to Severe Acute Respiratory Syndrome (SARS) in HCWs during bio-aerosol generating procedures. Tracheal intubation (OR = 6.6; 95%CI 2.3–18.9) and noninvasive ventilation (OR = 3.1; 95%CI 1.4–6.8) were risk factors for acquiring SARS. Other bio-aerosol generating interventions such as manipulation of an oxygen mask were not significant risk factors [31]. Another study calculated that the risk ratio for acquiring clinical respiratory infections was 2.5 (95%CI 1.3–6.5) for HCWs performing a high risk procedure [12]. Augustowska et al. studied the effect of bacteria and fungi on asthmatic patients. They reported a decrease in maximum breathing capacity due to the increase of bacterial or fungal load in the air [45].
A case-study in a dental clinic described the risk of acquiring Herpes Simplex Virus (HSV)-1 for the dentist and the dental hygienists when they treated a patient with active HSV-1. One member of the treatment team became infected with HSV-1, probably by the bio-aerosol containing HSV-1, induced by ultrasonic scaling or by rubbing her eyes while working. The infected HCWs manifested recurrent HSV-1 infections [77].
Discussion
By conducting a scoping review we were able to summarize existing evidence on the generation, composition, load and hazards of bio-aerosols in hospital and dental environment. We found that bio-aerosols are generated via multiple sources such as machines, different types of interventions; instruments; and human activity. The composition of bio-aerosols depended on the method of sampling (active versus passive), microbiological techniques (culture based versus DNA-based, different culture plates used) and the setting of the study (specific clinics versus general dental clinics). Bio-aerosols can be hazardous to both patients and HCWs. Multiple studies described the threat of Legionella species to elderly and patients with respiratory complaints.
The composition of bio-aerosols was extensively studied in hospital environments (N = 31) compared to dental environments (N = 16). Regarding the micro-organism composition of bio-aerosols, we conclude that bio-aerosols contain a high variety of bacterial and fungal strains from different sources such as the human skin and intestine; and the environment such as soil and water. Based on the sampling and culturing techniques, fungi and Gram-positive bacteria were found most often. Pathogens such as Legionella and Pseudomonas species were found in bio-aerosols that were distributed by instruments using tap water. Few studies looked for viruses, and in total only ten different viruses were identified, because no open ended detection or identification methods are available for viruses. Therefore only specific targeted techniques were used. Moreover, none of the studies conducted in dental practice have used methods to identify the presence of viruses in the generated aerosols. Therefore, we must keep in mind that the yielded results were dependent on the methodology of the individual study. The results of the individual studies, and the heterogeneity we found in this review, are dependent on the methods leading to an over- or under estimation of the complete bio-aerosol profile. The same inconsistency is discussed in previous studies in which the researchers compared two main sampling methods [80]. The methodological variety between studies, e.g. differences in method of sampling and culturing or sequencing, differences in sampling time and sampling area; and differences in distance to the bio-aerosol generating source caused difficulties in comparing results. When a study used selective medium or agar it results in an overview of selected micro-organisms. This leaves out other micro-organisms that were present at that moment. The same accounts for duration of sampling or passive versus active sampling. In passive sampling, the researcher waits for a certain amount of time for micro-organisms to fall on a Petri dish, while other micro-organisms were still floating in the air and take more time to fall on surfaces. The spread in a bio-aerosol is heterogeneous, so whatever is ‘catched’ on that moment may vary from the second, third or even fourth sampling attempt. So, the method chosen (active or passive) should be dependent on the aim of the air quantification [80]. Furthermore, in many studies variables in the experimental setup were not described, like sampling time, distance and sampling location. Also, no standard deviations of the microbiological loads were reported consistently. Besides, the data might be an underestimation of reality since studies looked for specific micro-organisms, in specific settings by selective sampling and culture dependent techniques, thereby missing other micro-organisms present in the bio-aerosols. Also, there was very little data available on the persistence of micro-organisms in the air over time and the spatial distribution. None of the included studies looked for parasites, although it has been reported that these are present in many tap water dependent bio-aerosol producing systems with plastic tubing [81, 82].
We found little evidence to state the presence or absence of direct threats or health risks for patients or HCWs with regards to bio-aerosols. In the hospital setting, two studies reported on the hazard for the staff [12, 31], and four on the hazard for patients [34, 45, 78, 79]. The search yielded one study for this objective assessing the hazard of an infectious disease to dental staff [77]. However, it is known that on average, dental practitioners carry elevated levels of Legionella antibodies [83], but the hazard to non-healthy HCWs and patients remains, based on our findings, unknown. An estimation of the hazard of bio-aerosols is usually made based on the microbial content and load of the bio-aerosols. We conclude that bio-aerosols can be hazardous to certain populations that are extensively exposed to bio-aerosol generating procedures or immunocompromised people.
Limitations
The search yielded 40 references that were to be screen based on full text. However, we could not recover these 40 full text manuscripts to assess the their eligibility for inclusion, even though we tried to contact the first and/or corresponding author, by retrieving his/her email via the abstract or Google. We assume that the body of evidence for each objective would have been larger if all 40 studies, or at least a part, would have been available and included. Another limitation was that the outcomes and methods were inconsistent throughout all included studies. Also, there was little data on the hazard of bio-aerosols, thus no strong conclusions could be drawn.
Recommendation for future research
We recommend that future research on bio-aerosols should create an extensive and complete methodology for the quantification of air contamination. Time and frequency of air sampling, distance from sources, location of sampling in both passive and active air sampling techniques should be described thoroughly. Furthermore, the identification of micro-organisms should be done by both selective and non-selective methods and cover organisms that find their origin in water, human, and environment. Also, we believe that infections due to a bio-aerosol should be structurally reported so that the risk for HCWs and patient can be analyzed. Finally, the risks for HCWs, especially dentists, working in an environment in which they are continuously exposed to bio-aerosols, and to their patients remain unclear and therefore need further research. This is needed in order to comprehend the risks of bio-aerosols generated in clinical settings to attention to staff and patients to improve awareness, hygienic standards, risks, and prevention methods.
Conclusion
Bio-aerosols are generated via multiple sources such as different interventions, instruments and human activity. Bio-aerosols have different microbiological profiles depending on the setting and the used methodology. Bio-aerosols can be hazardous to both patients and healthcare workers. Legionella species were found to be a bio-aerosol dependent hazard to elderly and patients with respiratory complaints.
Supporting information
(DOC)
Acknowledgments
The authors would like to thank Prof. dr. Geert van der Heijden for his consulting role during the methodology process.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.ASHRAE. ASHRAE position document on airbone infectious diseases. 2014.
- 2.Barker J, Jones MV. The potential spread of infection caused by aerosol contamination of surfaces after flushing a domestic toilet. J Appl Microbiol. 2005;99(2):339–47. doi: 10.1111/j.1365-2672.2005.02610.x [DOI] [PubMed] [Google Scholar]
- 3.WHO. Infection prevention and control measures for acute respiratory infections in healthcare settings: An update. 2007:39–47. [PubMed]
- 4.Szymanska J. Dental bioaerosol as an occupational hazard in a dentist's workplace. 2007:203–7. [PubMed]
- 5.Laheij AM, Kistler JO, Belibasakis GN, Valimaa H, de Soet JJ, European Oral Microbiology W. Healthcare-associated viral and bacterial infections in dentistry. J Oral Microbiol. 2012;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brankston G, Gitterman L, Hirji Z, Lemieux C, Gardam M. Transmission of influenza A in human beings. The Lancet Infectious Diseases. 2007;7(4):257–65. doi: 10.1016/S1473-3099(07)70029-4 [DOI] [PubMed] [Google Scholar]
- 7.Tellier R. Aerosol transmission of influenza A virus: a review of new studies. J R Soc Interface. 2009;6 Suppl 6:S783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gralton J, Tovey E, McLaws ML, Rawlinson WD. The role of particle size in aerosolised pathogen transmission: a review. J Infect. 2011;62(1):1–13. doi: 10.1016/j.jinf.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuttlebee CM, O'Donnell MJ, Keane CT, Russell RJ, Sullivan DJ, Falkiner F, et al. Effective control of dental chair unit waterline biofilm and marked reduction of bacterial contamination of output water using two peroxide-based disinfectants. J Hosp Infect. 2002;52(3):192–205. [DOI] [PubMed] [Google Scholar]
- 10.Pankhurst CL, Coulter WA. Do contaminated dental unit waterlines pose a risk of infection? J Dent. 2007;35(9):712–20. doi: 10.1016/j.jdent.2007.06.002 [DOI] [PubMed] [Google Scholar]
- 11.Mirhoseini SH, Nikaeen M, Khanahmd H, Hatamzadeh M, Hassanzadeh A. Monitoring of airborne bacteria and aerosols in different wards of hospitals—Particle counting usefulness in investigation of airborne bacteria. Ann Agric Environ Med. 2015;22(4):670–3. doi: 10.5604/12321966.1185772 [DOI] [PubMed] [Google Scholar]
- 12.MacIntyre R, Dwyer D, Seale H, Quanyi W, Yi Z, Yang P, et al. High risk procedures and respiratory infections in hospital health care workers—Quantifying the risk. 2014:e379.
- 13.Reinthaler FF, Mascher F, Stunzner D. Serological examinations for antibodies against Legionella species in dental personnel. J Dent Res. 1988;67(6):942–3. doi: 10.1177/00220345880670061001 [DOI] [PubMed] [Google Scholar]
- 14.Borella P, Bargellini A, Marchesi I, Rovesti S, Stancanelli G, Scaltriti S, et al. Prevalence of anti-legionella antibodies among Italian hospital workers. J Hosp Infect. 2008;69(2):148–55. doi: 10.1016/j.jhin.2008.03.004 [DOI] [PubMed] [Google Scholar]
- 15.Cadmus SI, Okoje VN, Taiwo BO, van Soolingen D. Exposure of dentists to Mycobacterium tuberculosis, Ibadan, Nigeria. Emerg Infect Dis. 2010;16(9):1479–81. doi: 10.3201/eid1609.100447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaffer K. Epidemiology of infection and current guidelines for infection prevention in cystic fibrosis patients. J Hosp Infect. 2015;89(4):309–13. doi: 10.1016/j.jhin.2015.02.005 [DOI] [PubMed] [Google Scholar]
- 17.Aerosol safety in the OR: help staff breathe easier. Mater Manag Health Care. 1995;4(8):32, 4. [PubMed] [Google Scholar]
- 18.Institute TJB. The Joanna Briggs Institute Reviewers' Manual 2015: 2015 edition / Supplement. The Joanna briggs Institute; 2015. [Google Scholar]
- 19.Peters MD, Godfrey CM, Khalil H, McInerney P, Parker D, Soares CB. Guidance for conducting systematic scoping reviews. Int J Evid Based Healthc. 2015;13(3):141–6. doi: 10.1097/XEB.0000000000000050 [DOI] [PubMed] [Google Scholar]
- 20.Moher Liberati, Tetzlaff Altman, group tP. Preferred Reporting items for systematic reviews and meta analyses: THE PRISMA statement. Plos Medicine. 2009;6(7):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.JoannaBriggsInstitute T. The Joanna Briggs Institute Reviewers Manual 2015 methodology for JBI scoping reviews. 2015.
- 22.Roberts K, Hathway A, Fletcher LA, Beggs CB, Elliott MW, Sleigh PA. Bioaerosol production on a respiratory ward. 2006:35–40.
- 23.Al Maghlouth A, Al Yousef Y, Al Bagieh N. Qualitative and quantitative analysis of bacterial aerosols. 2007:91–100. [PubMed]
- 24.Duell RC, Madden RM. Droplet nuclei produced during dental treatment of tubercular patients. A preliminary study. 1970:711–6. [DOI] [PubMed]
- 25.Grenier D. Quantitative analysis of bacterial aerosols in two different dental clinic environments. 1995:3165–8. [DOI] [PMC free article] [PubMed]
- 26.Rautemaa R, Nordberg A, Wuolijoki-Saaristo K, Meurman JH. Bacterial aerosols in dental practice—a potential hospital infection problem? 2006:76–81. [DOI] [PMC free article] [PubMed]
- 27.Szymańska J, Dutkiewicz J. Concentration and species composition of aerobic and facultatively anaerobic bacteria released to the air of a dental operation area before and after disinfection of dental unit waterlines. 2008:301–7. [PubMed]
- 28.Pankhurst CL, Harrison VE, Philpott-Howard J. Evaluation of contamination of the dentist and dental surgery environment with Burkholderia (Pseudomonas) cepacia during treatment of children with cystic fibrosis. 1995:243–7. [DOI] [PubMed]
- 29.Dutil S, Veillette M, Mériaux A, Lazure L, Barbeau J, Duchaine C. Aerosolization of mycobacteria and legionellae during dental treatment: low exposure despite dental unit contamination. 2007:2836–43. [DOI] [PubMed]
- 30.Vavricka SR, Tutuian R, Imhof A, Wildi S, Gubler C, Fruehauf H, et al. Air suctioning during colon biopsy forceps removal reduces bacterial air contamination in the endoscopy suite. 2010:736–41. [DOI] [PubMed]
- 31.Tran K, Cimon K, Severn M, Pessoa-Silva CL, Conly J. Aerosol generating procedures and risk of transmission of acute respiratory infections to healthcare workers: a systematic review. 2012:e35797. [DOI] [PMC free article] [PubMed]
- 32.Macintyre CR, Seale H, Yang P, Zhang Y, Shi W, Almatroudi A, et al. Quantifying the risk of respiratory infection in healthcare workers performing high-risk procedures. 2013:1802–8. [DOI] [PMC free article] [PubMed]
- 33.Thompson KA, Pappachan JV, Bennett AM, Mittal H, Macken S, Dove BK, et al. Influenza aerosols in UK hospitals during the H1N1 (2009) pandemic—the risk of aerosol generation during medical procedures. 2013:e56278. [DOI] [PMC free article] [PubMed]
- 34.Kool JL, Fiore AE, Kioski CM, Brown EW, Benson RF, Pruckler JM, et al. More than 10 years of unrecognized nosocomial transmission of legionnaires' disease among transplant patients. 1998:898–904. [DOI] [PubMed]
- 35.Anderson K, Morris G, Kennedy H, Croall J, Michie J, Richradson MD, et al. Aspergillosis in immunocompromised paediatric patients: Associations with building hygiene, design, and indoor air. 1996:256–61. [DOI] [PMC free article] [PubMed]
- 36.Verani M, Bigazzi R, Carducci A. Viral contamination of aerosol and surfaces through toilet use in health care and other settings. 2014:758–62. [DOI] [PMC free article] [PubMed]
- 37.Woo AH, Yu VL, Goetz A. Potential in-hospital modes of transmission of Legionella pneumophila. Demonstration experiments for dissemination by showers, humidifiers, and rinsing of ventilation bag apparatus. 1986:567–73. [DOI] [PubMed]
- 38.Yiallouros PK, Papadouri T, Karaoli C, Papamichael E, Zeniou M, Pieridou-Bagatzouni D, et al. First outbreak of nosocomial Legionella infection in term neonates caused by a cold mist ultrasonic humidifier. 2013:48–56. [DOI] [PubMed]
- 39.Heimbuch BK, Wallace WH, Balzli CL, Laning ML, Harnish DA, Wander JD. Bioaerosol Exposure to Personnel in a Clinical Environment Absent Patients. 2016:0. [DOI] [PubMed]
- 40.Bischoff WE, McNall RJ, Blevins MW, Turner J, Lopareva EN, Rota PA, et al. Detection of Measles Virus RNA in Air and Surface Specimens in a Hospital Setting. [DOI] [PubMed]
- 41.Fennelly KP, Jones-López EC, Ayakaka I, Kim S, Menyha H, Kirenga B, et al. Variability of Infectious Aerosols Produced during Coughing by Patients with Pulmonary Tuberculosis. 2012:450–7. [DOI] [PMC free article] [PubMed]
- 42.Wainwright CE, France MW, O'Rourke P, Anuj S, Kidd TJ, Nissen MD, et al. Cough-generated aerosols of Pseudomonas aeruginosa and other Gram-negative bacteria from patients with cystic fibrosis. 2009:926–31. [DOI] [PMC free article] [PubMed]
- 43.Humphreys H, Peckham D, Patel P, Knox A. Airborne dissemination of Burkholderia (Pseudomonas) cepacia from adult patients with cystic fibrosis. 1994:1157–9. [DOI] [PMC free article] [PubMed]
- 44.Knibbs LD, Johnson GR, Kidd TJ, Cheney J, Grimwood K, Kattenbelt JA, et al. Viability of Pseudomonas aeruginosa in cough aerosols generated by persons with cystic fibrosis. 2014:740–5. [DOI] [PMC free article] [PubMed]
- 45.Augustowska M, Dutkiewicz J. Variability of airborne microflora in a hospital ward within a period of one year. 2006:99–106. [PubMed]
- 46.Best EL, Sandoe JAT, Wilcox MH. Potential for aerosolization of Clostridium difficile after flushing toilets: The role of toilet lids in reducing environmental contamination risk. 2012:1–5. [DOI] [PubMed]
- 47.Cabo Verde S, Almeida SM, Matos J, Guerreiro D, Meneses M, Faria T, et al. Microbiological assessment of indoor air quality at different hospital sites. 2015:557–63. [DOI] [PubMed]
- 48.Chen PS, Li CS. Concentration profiles of airborne Mycobacterium tuberculosis in a hospital. 2007:194–200.
- 49.Chen PS, Li CS. Quantification of airborne Mycobacterium tuberculosis in health care setting using real-time qPCR coupled to an air-sampling filter method. 2008:371–6.
- 50.Fekadu S, Getachewu B. Microbiological Assessment of Indoor Air of Teaching Hospital Wards: A case of Jimma University Specialized Hospital. 2015:117–22. [DOI] [PMC free article] [PubMed]
- 51.Huynh KN, Oliver BG, Stelzer S, Rawlinson WD, Tovey ER. A new method for sampling and detection of exhaled respiratory virus aerosols. 2008:93–5. [DOI] [PubMed]
- 52.Kulkarni H, Smith C, Hirst R, Baker N, Easton A, O'Callaghan C. Airborne transmission of respiratory syncytial virus (RSV) infection. 2016.
- 53.Kumar S, Atray D, Paiwal D, Balasubramanyam G, Duraiswamy P, Kulkarni S. Dental unit waterlines: source of contamination and cross-infection. 2010:99–111. [DOI] [PubMed]
- 54.Lindsley WG, Noti JD, Blachere FM, Thewlis RE, Martin SB, Othumpangat S, et al. Viable influenza A virus in airborne particles from human coughs. 2015:107–13. [DOI] [PMC free article] [PubMed]
- 55.Li CS, Hou PA. Bioaerosol characteristics in hospital clean rooms. 2003:169–76. [DOI] [PubMed]
- 56.Martins-Diniz JN, da Silva RAM, Miranda ET, Mendes-Giannini MJS. Monitoring of airborne fungus and yeast species in a hospital unit. 2005:398–405. [DOI] [PubMed]
- 57.Menzies D, Adhikari N, Arietta M, Loo V. Efficacy of environmental measures in reducing potentially infectious bioaerosols during sputum induction. 2003:483–9. [DOI] [PubMed]
- 58.Mirzaei R, Shahriary E, Qureshi MI, Rakhshkhorshid A, Khammary A, Mohammadi M. Quantitative and qualitative evaluation of bio-aerosols in surgery rooms and emergency department of an educational hospital. 2014:e11688. [DOI] [PMC free article] [PubMed]
- 59.Nasir ZA, Mula V, Stokoe J, Colbeck I, Loeffler M. Evaluation of total concentration and size distribution of bacterial and fungal aerosol in healthcare built environments. 2013:269–79.
- 60.Ortiz G, Yagüe G, Segovia M, Catalán V. A study of air microbe levels in different areas of a hospital. 2009:53–8. [DOI] [PubMed]
- 61.Sudharsanam S, Swaminathan S, Ramalingam A, Thangavel G, Annamalai R, Steinberg R, et al. Characterization of indoor bioaerosols from a hospital ward in a tropical setting. 2012:217–25. [DOI] [PMC free article] [PubMed]
- 62.Stelzer-Braid S, Oliver BG, Blazey AJ, Argent E, Newsome TP, Rawlinson WD, et al. Exhalation of respiratory viruses by breathing, coughing, and talking. 2009:1674–9. [DOI] [PubMed]
- 63.Thompson K, Thomson G, Mittal H, Parks S, Dove B, Speight S, et al. Transmission of influenza to health-care workers in intensive care units—Could Aerosol generating procedures play a role? 2013:S5.
- 64.Vélez-Pereira A, Camargo DY. Staphylococcus sp. airborne in the intensive care unit of an University Hospital. 2014:183–90.
- 65.Wan GH, Chung FF, Tang CS. Long-term surveillance of air quality in medical center operating rooms. 2011:302–8. [DOI] [PubMed]
- 66.Tynelius-Bratthall G, Ellen RP. Fluctuations in crevicular and salivary anti-A. viscosus antibody levels in response to treatment of gingivitis. J Clin Periodontol. 1985;12(9):762–73. [DOI] [PubMed] [Google Scholar]
- 67.Bennett AM, Fulford MR, Walker JT, Bradshaw DJ, Martin MV, Marsh PD. Microbial aerosols in general dental practice. 2000:664–7. [DOI] [PubMed]
- 68.Barlean., Iancu., Minea., Danila., Baciu. Airborne microbial contamination in dental practice in iasi, Romania. OHDMBSC. 2010;9(1). [Google Scholar]
- 69.Cellini L, Di Campli E, Di Candia M, Chiavaroli G. Quantitative microbial monitoring in a dental office. 2000:301–5. [DOI] [PubMed]
- 70.Dutil S, Meriaux A, de Latremoille MC, Lazure L, Barbeau J, Duchaine C. Measurement of airborne bacteria and endotoxin generated during dental cleaning. J Occup Environ Hyg. 2009;6(2):121–30. doi: 10.1080/15459620802633957 [DOI] [PubMed] [Google Scholar]
- 71.Guida M, Gallé F, Di Onofrio V, Nastro RA, Battista M, Liguori R, et al. Environmental microbial contamination in dental setting: A local experience. 2012:207–12. [PubMed]
- 72.Kadaifciler DG, Cotuk A. Microbial contamination of dental unit waterlines and effect on quality of indoor air. 2013:3431–44. [DOI] [PubMed]
- 73.Kimmerle H, Wiedmann-Al-Ahmad M, Pelz K, Wittmer A, Hellwig E, Al-Ahmad A. Airborne microbes in different dental environments in comparison to a public area. 2012:689–96. [DOI] [PubMed]
- 74.Kedjarune U, Kukiattrakoon B, Yapong B, Chowanadisai S, Leggat P. Bacterial aerosols in the dental clinic: effect of time, position and type of treatment. Int Dent J. 2000;50(2):103–7. [DOI] [PubMed] [Google Scholar]
- 75.Timmerman MF, Menso L, Steinfort J, van Winkelhoff AJ, van der Weijden GA. Atmospheric contamination during ultrasonic scaling. J Clin Periodontol. 2004;31(6):458–62. doi: 10.1111/j.1600-051X.2004.00511.x [DOI] [PubMed] [Google Scholar]
- 76.Pasquarella C, Veronesi L, Napoli C, Castiglia P, Liguori G, Rizzetto R, et al. Microbial environmental contamination in Italian dental clinics: A multicenter study yielding recommendations for standardized sampling methods and threshold values. Sci Total Environ. 2012;420:289–99. doi: 10.1016/j.scitotenv.2012.01.030 [DOI] [PubMed] [Google Scholar]
- 77.Browning WD, McCarthy JP. A case series: Herpes simplex virus as an occupational hazard. 2012:61–6. [DOI] [PMC free article] [PubMed]
- 78.Palusinska-Szysz M, Cendrowska-Pinkosz M. Pathogenicity of the family Legionellaceae. 2009:279–90. [DOI] [PubMed]
- 79.Ricci ML, Fontana S, Pinci F, Fiumana E, Pedna MF, Farolfi P, et al. Pneumonia associated with a dental unit waterline. The Lancet. 2012;379(9816):684. [DOI] [PubMed] [Google Scholar]
- 80.Napoli C, Marcotrigiano V, Montagna MT. Air sampling procedures to evaluate microbial contamination: a comparison between active and passive methods in operating theatres. BMC Public Health. 2012;12:594 doi: 10.1186/1471-2458-12-594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singh T, Coogan MM. Isolation of pathogenic Legionella species and legionella-laden amoebae in dental unit waterlines. J Hosp Infect. 2005;61(3):257–62. doi: 10.1016/j.jhin.2005.05.001 [DOI] [PubMed] [Google Scholar]
- 82.Trabelsi H, Sellami A, Dendena F, Sellami H, Cheikh-Rouhou F, Makni F, et al. Free-living Amoebae (FLA): morphological and molecular identification of Acanthamoeba in dental unit water. Parasite. 2010;17(1):67–70. doi: 10.1051/parasite/2010171067 [DOI] [PubMed] [Google Scholar]
- 83.Pankhurst CL, Coulter W, Philpott-Howard JJ, Harrison T, Warburton F, Platt S, et al. Prevalence of legionella waterline contamination and Legionella pneumophila antibodies in general dental practitioners in London and rural Northern Ireland. Br Dent J. 2003;195(10):591–4; discussion 81. doi: 10.1038/sj.bdj.4810735 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.