Abstract
Investigation into biological complexity, whether for a better understanding of disease or drug process, is a monumental task plaguing investigators. The lure of “omic” technologies for circumventing much of these challenges has led to widespread efforts and adoption. It is becoming clearer that a single “omic” approach (e.g., genomics) is often insufficient for completely defining the complexity in these biological systems. Hence, there is an increasing awareness that a “systems” approach will serve to increase resolution and confidence and provide a strong foundation for further hypothesis‐driven investigation. Although certain metabolites are already considered clinically important, the profiling of metabolites via metabolomics (the profiling of metabolites to fully characterize metabolic pathways) is the most recent to mature of these “omic” technologies and has been only recently adopted as compared to genomic or proteomic approaches in systems inquiries. Recent reports suggest that this “omic” may well be a key data stream in systems investigations for endeavors in personalized medicine and biomarker identification, as it seems most closely relevant to the phenotype. Clin Trans Sci 2012; Volume #: 1–4
Keywords: metabolomics, metabolome, genomics, proteomics, personalized medicine, biomarker
Introduction
To precisely target a disease state or to rationally devise new therapies, a high‐resolution understanding of the mechanistic foundation leading to disease initiation, advancement, and/or regression is required. Once appropriate targets are qualified and advanced, ideally, the deployment of these therapies will closely follow a paradigm of personalized medicine. The goal of personalized medicine is more efficient drug development, better medical outcomes, earlier interventions, and improved diagnoses. Biomarkers and the discovery of biomarkers are the foundation for personalized medicine and usually indicative of a biological state. Identification of appropriate biomarkers takes into account information about genes, proteins, and environment for the purposes of diagnosing, preventing, or treating disease. 1 Numerous strategies have been employed to identify biomarkers and the underlying mechanism and biomarkers themselves range from genes to proteins to biochemicals. “Omic” strategies, as profiling technologies, have provided a data‐rich foundation to perform unbiased investigation and have produced no shortage of data. The challenge that exists is how to mine information from these data sets and how best to integrate information across often distinctly deployed omics—genomics, transcriptomics, proteomics, and metabolomics.
Genomic analysis studies the genome and produces the vast majority of deposited data for the mouse and human. It encompasses knowledge of DNA sequence, modification, and expression. Genetic and genomic testing is used in a wide variety of diseases for presymptomatic risk assessment, diagnosis, prognosis, and treatment. To date, this approach has been most successful in oncology where some of the science of genomics has been translated into medicine. 2 For example, understanding a patient’s BRCA1/2 genes can shed light onto risk assessment in breast cancer and amenability of treatment with poly (ADP‐ribose) polymerase (PARP) inhibitors. 3 Mutation in the KRas gene is a predictor of resistance to epidermal growth factor receptor monoclonal antibodies in metastatic colorectal cancer. 4 B‐RAF specific inhibitors for a mutation in cancer have been extremely successful in the clinic. More recently because of technological advancement, large scale genome‐wide association studies (GWAS) are identifying common risk variants for various cancers in certain populations. 5 , 6 , 7 , 8 , 9 These studies take advantage of very large sample sizes which hopefully will enhance power in an attempt to manage high false positive findings. Unfortunately, many of these studies have had limited penetrance to the phenotype probably because of the polymorphisms of many diseases. Genomics research has identified numerous key biomarkers. However, given the vast amount of analyses, the successful hits have been modest. Weeding through the data, false positive results, and understanding where to focus efforts remains an enormous challenge.
Transcriptomics confers information regarding RNA and is often used to refer to messengerRNA (mRNA) expression profiling. Understanding changes within the gene is important to understand what is possible but the next step is deciphering how these changes are conveyed into differences in transcription of the gene. Relative levels of mRNA are measured and, although changes in protein expression can be inferred, mRNA levels do not necessarily correlate to the amount of expressed protein. The data sets derived from these studies are large and complex. Although some useful data has been derived from these studies, the narrowing of focus and validation of potential findings has been cumbersome therefore, this technology is prime for a method to add resolution.
Proteins undoubtedly play an important role in the underlying mechanisms of disease. 10 Protein biomarkers such as troponin I and troponin T have proven useful and are employed as diagnostic tools for myocardial injury. 11 Proteomics is also an unbiased discovery tool and discovery proteomics may provide new biomarkers and associations previously unappreciated. However, the analysis of proteins quickly becomes unwieldy because of the sheer number of proteins, huge variation in size and modifications including phosphorylation, ubiquitination, methylation, nitrosylation, and glycolation making holistic inquiry logistically challenging. Although there has been limited success using proteomics, technological advances have been lagging because of the complexity. Once a method is developed to better profile the entire proteome, this will be a very powerful unbiased tool.
Metabolomics is another “omics” tool being deployed with more frequency with the advent of methods for broad metabolic coverage. 12 , 13 In contrast to genomics and proteomics, metabolomics has trailed in adoption but this has been changing rapidly in recent years. This is likely due to a relative amount of unfamiliarity with biochemistry as well as a preoccupation with molecular biology due to seminal findings and explosive research tool development. Further, the technical challenges for measuring the diversity of chemistries represented by the metabolites of biochemistry have been burdensome. Nevertheless, the development of the appropriate technologies has recently occurred and allowed for the successful use of metabolomics for elucidation of numerous wide ranging applications such as toxicology, disease and drug mechanism of action, novel targets and biomarkers. 14 , 15 , 16 , 17 , 18 , 19 , 20 Metabolomics has also been used to successfully elucidate personalized pharmacological response to drug treatment taking into account both genetic and environmental influences. 21 These stand‐alone successes aside, examples where metabolomics has provided key insight into gene function have recently provided an argument for routine data collection of this information‐rich data stream. Common to the success achieved in these inquiries has been the ability of metabolites to readily yield functional information thus providing key insight into: a well studied genetic disorder, advancing the mechanistic understanding of the gain of function of a mutant gene associated with gliomas (discovered by whole genome sequencing), or the use in informing about wide‐scale GWAS. In contrast to a small nucleotide polymorphism (SNP), mRNA, or even an unidentified protein, the wealth of information on metabolites and metabolic pathways allows function often to be readily assigned to a metabolite and thus, easily embedded into a biological/physiological query.
Deepening Insight into Well‐Characterized Penetrant Genetic Disorders—Sickle Cell Disease
A metabolomics discovery produced an impressive potential breakthrough in a highly recognized genetic disease. Sickle cell anemia is caused by a point mutation in the β‐globin gene. People with two copies of the mutated allele suffer from formation of polymer crystals after deoxygenation which leads to red blood cell dehydration, membrane oxidation and damage and ultimately hemolysis and deformation. 22 Information about the genetic component of sickle cell disease has long been appreciated. 23 , 24 , 25 However, despite knowing the precise genetic defect, the molecular events controlling the pathophysiology are unclear. 26 Combining the genetic knowledge through use of a transgenic mouse model with a metabolomics study, adenosine and 2,3‐diphosphoglycerate levels were identified to be elevated. 26 Subsequent validation studies elucidated that elevated adenosine signaling through the adenosine A2B receptor promotes sickling by inducing 2,3‐diphosphoglycerate production. 26 Therefore, layering unbiased information allowed for resolution into a previously unappreciated key perturbation in sickle cell disease. The importance of this finding, elucidated by layering genomic information with metabolomic insight, provides a potential therapeutic breakthrough by opening a new avenue of investigation for drug development to treat this long studied, debilitating disease.
Advancing Mechanistic Understanding of Novel Hits from Large Scale Whole Genome Screens‐IDH Mutations and Cancer
A recent example of how, independently, genomics and metabolomics complemented each other to produce an impactful discovery in brain cancer is highlighted by the discovery of the mutation in the isocitrate dehydrogenase (IDH1). The mutation in the IDH1 gene was found using high‐powered genome sequencing. 27 , 28 The implications of this mutation became clear upon metabolomics profiling revealing that the mutant gene product led to the starkly elevated production of a metabolic product, 2‐hydroxyglutarate. Without this information, the impact of the mutation would remain unknown. Combining of the two “omic” strategies provided information rich data for which the results were greater than the sum of the two parts. Importantly this cross‐omics approach provided resolution and may directly improve patient care because both IDH1 and 2‐hydroxyglutarate are now novel targets for drug development. 18 , 27
Strengthening Annotation of Gene Function/Disease Association in Complex Trait Investigation via GWAS
Our classical hypothesis‐driven research is often biased by our present knowledge. 29 Because of this prejudice, “omics” on their own have strengths because of their unbiased foundation. Deploying a cross‐omic strategy could allow for higher fidelity of validation and proof of concept as well as temper false positive rates. 12 This concept has been elegantly illustrated in recent GWAS. 16 , 30 , 31 Several years ago, a smaller cohort of 284 randomly selected males between 55 and 79 were chosen to examine SNPs (genomics) combined with a targeted quantitative metabolomics platform based on 9 sugar molecules, 7 biogenic amines, 7 prostaglandins, 29 acylcarnitines, 18 amino acids, 85 sphingolipids and 208 glycerophospholipids (metabolomics). 31 One of the several SNPs resided within a gene that codes for the fatty acid delta‐5 desaturase (FADS1) and strongly correlated with a number of glycerophospholipid concentrations. In addition, the authors considered analyzing ratios of metabolites to reduce the variation in the data set. This would mean a pair of metabolites closely connected to the direct substrates and products of a given enzymatic reaction were considered together. 31 When a ratio of eicosatrienoyl‐CoA (C20:3) and arachidonyl‐CoA (C20:4), the direct substrate and product of FADS1 activity was considered, the association with the FADS1 polymorphisms p value decreased by 14 orders of magnitude. 31 This gave confidence to the finding that the FADS1 polymorphism was a true finding worthy of extensive validation. Therefore, combination of “omic” technologies provided access to functionally relevant endpoints in the framework of the polymorphisms and helped to resolve mountains of data into a feasible list of hypothesis‐driven actionable items.
More recently, another study combined the use of GWAS with metabolomics but instead of using a targeted biochemical assay approach, a global metabolic profile was assessed. 16 In addition, a much larger cohort of subjects was used. A total of 2,820 individuals from two large population‐based European cohorts were examined using GWAS and metabolomics. 16 This study was powerful from two perspectives. It confirmed the association of 14 known genomic and metabolic alterations that had only been appreciated individually. It also identified 23 previously unappreciated associations with strong correlation suggesting further validation was warranted. 16 Bradykinin and kallikrein B1 were identified to be correlated thus confirming an association of bradykinin with hypertension and identifying a potential genetic interaction with the angiotensin converting enzyme locus. 16 It was also identified that there was a polymorphism in the solute carrier SLC16A9 which had unknown function. It correlated highly with carnitine levels. Validation studies were conducted and it was confirmed that SLC16A9 is a carnitine transporter thus the cross‐omics approach was able to provide new biological and functional insight. 16 In resource limited environments, the filtering of data and the ability to prioritize investigation based on solid scientific findings is invaluable.
Summary
Although the overabundance of information and data have preoccupied “omic” technologies making it complex and difficult to decipher the most relevant of findings, their benefit of unbiased profiling makes them particularly attractive. Much of the initial efforts have centered on handling the huge amount of data using bioinformatics. With the maturing of the technologies and bioinformatics support, the ability to combine “omic” strategies is beginning to be achievable on a large scale. As illustrated by many recent examples, metabolomics may be a key data set for advancing these studies ( Figure 1 ). If these examples are reflective of a broader reality, it may be that the reason for the impact and relatively high hit rate of success for metabolomics is likely due to three major factors. First, biochemistry and the associated metabolites are well annotated in terms of function. Thus, when a metabolite is found to be associated with a particular event or trait, there is frequently a significant amount of literature to suggest how the metabolite integrates into metabolism and particular pathways but also how it may be linked to specific physiological states. Second, this function is often directly related to a gene function (e.g., via absorption, distribution, metabolism, or excretion). As illustrated in recent GWAS combined with metabolomics, when a SNP is associated with a trait or metabolite, the metabolite often leads to a gene product that is tied to the metabolic reaction, and complex trait. Third, although the technological hurdles to producing a highly precise and accurate data set were substantial given the chemical diversity of the metabolites, stable metabolomics platforms have been developed allowing for highly accurate and reproducible results. This type of data quality is likely central to assigning high significance to metabolic changes or associations in a number of studies. This is undeniably powerful and should help to provide resolution and validatable hypotheses with which to take scientific inquiry into complex diseases to a realm where huge strides can be made toward personalized medicine and understanding better underlying mechanism, diagnosis, prevention, and therapeutic strategy.
Figure 1.
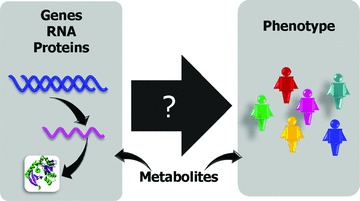
The analysis of metabolites using metabolomics is a powerful technology that can inform and provide resolution to other “omic” technologies including genomics, transcriptomics, proteomics, and underlying phenotype. The main goal of broad‐based unbiased surveying using “omics” is to yield insight into mechanism underlying phenotype. This information then provides a rich data set to mine for new therapeutic targets and biomarkers of disease progression or treatment. Metabolites are uniquely positioned to enhance this inquiry because they are well annotated and function is directly related to gene expression. Using cross‐omic approaches with the adoption of metabolomics has been successful at narrowing focus and providing actionable hypotheses therefore, expanding the use of this technology will undoubtedly provide resolution.
References
- 1. Offit K. Personalized medicine: new genomics, old lessons. Hum Genet. 2011; 130: 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Collins F. Has the revolution arrived? Nature. 2010; 464: 674–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robson M, Offit K. Clinical practice. Management of an inherited predisposition to breast cancer. N Engl J Med. 2007; 357: 154–162. [DOI] [PubMed] [Google Scholar]
- 4. De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. Kras, braf, pik3ca, and pten mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011; 12: 594–603. [DOI] [PubMed] [Google Scholar]
- 5. Haiman CA, Chen GK, Vachon CM, Canzian F, Dunning A, Millikan RC, Wang X, Ademuyiwa F, Ahmed S, Ambrosone CB, et al. A common variant at the tert‐clptm1l locus is associated with estrogen receptor‐negative breast cancer. Nat Genet. 2011; 43: 1210–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu X, Scelo G, Purdue MP, Rothman N, Johansson M, Ye Y, Wang Z, Zelenika D, Moore LE, Wood CG, et al. A genome‐wide association study identifies a novel susceptibility locus for renal cell carcinoma on 12p11.23. Hum Mol Genet. 2011; 21: 456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garcia‐Closas M, Ye Y, Rothman N, Figueroa JD, Malats N, Dinney CP, Chatterjee N, Prokunina‐Olsson L, Wang Z, Lin J, et al. A genome‐wide association study of bladder cancer identifies a new susceptibility locus within slc14a1, a urea transporter gene on chromosome 18q12.3. Hum Mol Genet. 2011; 20: 4282–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schumacher FR, Berndt SI, Siddiq A, Jacobs KB, Wang Z, Lindstrom S, Stevens VL, Chen C, Mondul AM, Travis RC, et al. Genome‐wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011; 20: 3867–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smedby KE, Foo JN, Skibola CF, Darabi H, Conde L, Hjalgrim H, Kumar V, Chang ET, Rothman N, Cerhan JR, et al. GWAS of follicular lymphoma reveals allelic heterogeneity at 6p21.32 and suggests shared genetic susceptibility with diffuse large b‐cell lymphoma. PLoS Genet. 2011; 7: e1001378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thomas A, Lenglet S, Chaurand P, Deglon J, Mangin P, Mach F, Steffens S, Wolfender JL, Staub C. Mass spectrometry for the evaluation of cardiovascular diseases based on proteomics and lipidomics. Thromb Haemost. 2011; 106: 20–33. [DOI] [PubMed] [Google Scholar]
- 11. Gerszten RE, Asnani A, Carr SA. Status and prospects for discovery and verification of new biomarkers of cardiovascular disease by proteomics. Circ Res. 2011; 109: 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ala‐Korpela M, Kangas AJ, Inouye M. Genome‐wide association studies and systems biology: together at last. Trends Genet. 2011; 27: 493–498. [DOI] [PubMed] [Google Scholar]
- 13. Baraldi E, Carraro S, Giordano G, Reniero F, Perilongo G, Zacchello F. Metabolomics: moving towards personalized medicine. Ital J Pediatr. 2009; 35:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takei M, Ando Y, Saitoh W, Tanimoto T, Kiyosawa N, Manabe S, Sanbuissho A, Okazaki O, Iwabuchi H, Yamoto T, et al. Ethylene glycol monomethyl ether‐induced toxicity is mediated through the inhibition of flavoprotein dehydrogenase enzyme family. Toxicol Sci. 2010; 118: 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Watson M, Roulston A, Belec L, Billot X, Marcellus R, Bedard D, Bernier C, Branchaud S, Chan H, Dairi K, et al. The small molecule gmx1778 is a potent inhibitor of nad+ biosynthesis: strategy for enhanced therapy in nicotinic acid phosphoribosyltransferase 1‐deficient tumors. Mol Cell Biol. 2009; 29: 5872–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wagele B, Altmaier E, Deloukas P, Erdmann J, Grundberg E, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011; 477: 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zgoda‐Pols JR, Chowdhury S, Wirth M, Milburn MV, Alexander DC, Alton KB. Metabolomics analysis reveals elevation of 3‐indoxyl sulfate in plasma and brain during chemically‐induced acute kidney injury in mice: investigation of nicotinic acid receptor agonists. Toxicol Appl Pharmacol. 2011; 255: 48–56. [DOI] [PubMed] [Google Scholar]
- 18. Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer‐associated idh1 mutations produce 2‐hydroxyglutarate. Nature. 2009; 462: 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abate‐Shen C, Shen MM. Diagnostics: the prostate‐cancer metabolome. Nature. 2009; 457: 799–800. [DOI] [PubMed] [Google Scholar]
- 20. Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009; 457: 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 21. Clayton TA, Lindon JC, Cloarec O, Antti H, Charuel C, Hanton G, Provost JP, Le Net JL, Baker D, Walley RJ, et al. Pharmaco‐metabonomic phenotyping and personalized drug treatment. Nature. 2006; 440: 1073–1077. [DOI] [PubMed] [Google Scholar]
- 22. Gladwin MT. Adenosine receptor crossroads in sickle cell disease. Nat Med. 2011; 17: 38–40. [DOI] [PubMed] [Google Scholar]
- 23. Ingram VM. A specific chemical difference between the globins of normal human and sickle‐cell anaemia haemoglobin. Nature. 1956; 178: 792–794. [DOI] [PubMed] [Google Scholar]
- 24. Madigan C, Malik P. Pathophysiology and therapy for haemoglobinopathies. Part I: sickle cell disease. Expert Rev Mol Med. 2006; 8: 1–23. [DOI] [PubMed] [Google Scholar]
- 25. Urbinati F, Madigan C, Malik P. Pathophysiology and therapy for haemoglobinopathies. Part II: thalassaemias. Expert Rev Mol Med. 2006; 8: 1–26. [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y, Dai Y, Wen J, Zhang W, Grenz A, Sun H, Tao L, Lu G, Alexander DC, Milburn MV, et al. Detrimental effects of adenosine signaling in sickle cell disease. Nat Med. 2011; 17: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ledford H. Big science: the cancer genome challenge. Nature. 2010; 464: 972–974. [DOI] [PubMed] [Google Scholar]
- 28. Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006; 314: 268–274. [DOI] [PubMed] [Google Scholar]
- 29. Pfeiffer T, Hoffmann R. Large‐scale assessment of the effect of popularity on the reliability of research. PLoS One. 2009; 4: e5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, Holm H, Sanna S, Kavousi M, Baumeister SE, et al. Genome‐wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011; 43: 1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gieger C, Geistlinger L, Altmaier E, Hrabe de Angelis M, Kronenberg F, Meitinger T, Mewes HW, Wichmann HE, Weinberger KM, Adamski J, et al. Genetics meets metabolomics: a genome‐wide association study of metabolite profiles in human serum. PLoS Genet. 2008; 4: e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]


