Abstract
Endoplasmic reticulum (ER) stress-induced activation of ATF6, an ER membrane-bound transcription factor, requires a dissociation step from its inhibitory regulator, BiP. It has been generally postulated that dissociation of the BiP-ATF6 complex is a result of the competitive binding of misfolded proteins generated during ER stress. Here we present evidence against this model and for an active regulatory mechanism for dissociation of the complex. Contradictory to the competition model that is based on dynamic binding of BiP to ATF6, our data reveal relatively stable binding. First, the complex was easily isolated, in contrast to many chaperone complexes that require chemical cross-linking. Second, ATF6 bound at similar levels to wild-type BiP and a BiP mutant form that binds substrates stably because of a defect in its ATPase activity. Third, ER stress specifically induced the dissociation of BiP from ER stress transducers while the competition model would predict dissociation from any specific substrate. Fourth, the ATF6-BiP complex was resistant to ATP-induced dissociation in vitro when isolated without detergents, suggesting that cofactors stabilize the complex. In favor of an active dissociation model, one specific region within the ATF6 lumenal domain was identified as a specific ER stress-responsive sequence required for ER stress-triggered BiP release. Together, our results do not support a model in which competitive binding of misfolded proteins causes dissociation of the BiP-ATF6 complex in stressed cells. We propose that stable BiP binding is essential for ATF6 regulation and that ER stress dissociates BiP from ATF6 by actively restarting the BiP ATPase cycle.
The endoplasmic reticulum (ER) provides a tightly regulated environment for the folding and maturation of proteins destined to enter the secretory pathway (7, 38). The homeostasis of the ER environment can be disrupted under various stress conditions, leading to the accumulation of misfolded and unfolded proteins in the ER (termed ER stress). Cells respond to ER stress by eliciting three major cellular responses: increasing the expression of certain genes that can expand the folding capacity of the ER, limiting new protein synthesis, and accelerating the degradation of misfolded proteins. These pathways are collectively known as the ER stress response or unfolded protein response (20, 31, 33). ER stress signaling plays critical roles in nutrient sensing, glucose homeostasis, and differentiation of secretory tissues such as hepatocytes and antibody-producing plasma cells, both of which secrete large amounts of proteins (20).
ER stress induces the transcription of a large set of genes, which encode ER chaperones, protein folding enzymes, membrane trafficking factors, and components of the ER-associated degradation system (40). In the budding yeast Saccharomyces cerevisiae, where the unfolded protein response signaling mechanism is well understood, transcriptional induction is mediated by a single bZIP transcriptional factor, Hac1p (31). In metazoans, transcriptional induction is more complex, with at least three transcription factors involved in signaling: XBP1, ATF4, and ATF6. XBP1 is a bZIP transcription factor that activates the transcription of ER stress-responsive genes by binding to ER stress response elements within their promoter regions (46). Activation of XBP1 involves unconventional splicing of its mRNA by IRE1, an ER transmembrane kinase and endoribonuclease, leading to the expression of an active form of XBP1 (4, 35, 46). ER stress also induces the expression of CHOP/GADD153, a protein implicated in stress-induced apoptosis. Induction of CHOP is primarily mediated by ATF4, a bZIP transcription factor activated by another signal transducer in the ER stress pathway, PERK. PERK, also an ER transmembrane kinase, mediates ER stress-induced inhibition of translation by phosphorylating the translation factor eIF2α (14). ATF4, however, evades this inhibition and is selectively activated (13).
An essential mediator in the ER stress-induced transcriptional pathway is a third bZIP transcription factor, ATF6. ATF6 is synthesized as an ER membrane-tethered precursor with its C terminus located in the ER lumen and its N-terminal DNA-binding domain facing the cytosol (16). ATF6 translocates from the ER to the Golgi apparatus in response to ER stress, and within the Golgi apparatus it is cleaved to its active form in a two-step process by site 1 and 2 proteases (S1P and S2P) (5, 45). S1P and S2P are also involved in the proteolytic processing of sterol response element-binding proteins, transcription factors that control cellular sterol levels (32). The cytosolic fragment of ATF6 that is liberated by the proteases then moves to the nucleus to activate gene transcription. ATF6 also binds to the ER stress response elements that XBP1 binds to such that their target genes may partially overlap. Several lines of evidence support the essential role of ATF6 in the ER stress response. First, ATF6 failed to be processed to its active form and BiP induction by ER stress was completely abolished in cells deficient in S2P or in which S1P activity was inhibited (30, 45). Second, dominant negative forms of ATF6 blocked the induction of BiP reporter genes (42, 47). Third, knockdown of ATF6 levels by RNA interference reduced the induction of many ER stress-inducible genes in microarray experiments (22). Surprisingly, knockdown of ATF6 did not strongly reduce induction of BiP mRNA, but the combination of XBP1 loss by homologous recombination and ATF6 knockdown strongly reduced the induction of two tested genes, those for GRP94 and Armet (22). These results demonstrate the redundancy of the XBP1, ATF6, and additional pathways for ER stress-induced gene expression.
The central step of ATF6 activation is its translocation from the ER to the Golgi apparatus so that it can be cleaved by S1P and S2P. Trafficking of ATF6 between the ER and Golgi apparatus is controlled by the ER chaperone BiP, a member of the Hsp70 chaperone family that plays critical roles in ER protein folding and quality control (7, 11, 34). In unstressed cells, BiP binds to the lumenal domain (LD) of ATF6 and blocks its intrinsic Golgi apparatus localization signals, thereby sequestering ATF6 in the ER. In response to accumulation of misfolded proteins in the ER, ATF6 is released from BiP association, allowing its transport to the Golgi apparatus. When ER stress-induced BiP release was blocked by a mutant BiP, translocation and proteolytic processing of ATF6 were completely abolished. On the other hand, removal of BiP-binding sites from ATF6 resulted in its constitutive activation (34, 39). BiP not only regulates ATF6 activation, it also negatively regulates IRE1 and PERK, probably by sterically blocking their spontaneous oligomerization (2, 21, 23). Hence, by directly regulating all three ER stress signal transducers, BiP serves as the master regulator of the ER stress response.
ER stress-induced activation of ATF6 requires a derepression step of dissociation from the inhibitory regulator BiP; however, the molecular mechanism for this release remains unknown. Two competing models have been proposed to explain ER stress-induced BiP dissociation (2). The first, which we refer to as the competition model, assumes that the BiP-ATF6 complex is highly dynamic and that ER stress generates an excess of misfolded substrates that compete with ATF6 for BiP binding, diverting BiP from ATF6 to misfolded proteins. This competition model resembles the regulation of HSF1 by cytoplasmic chaperones (26). The second model, which can be termed an active triggering model, posits a stable BiP-ATF6 complex that is actively dissociated by a signal from misfolded proteins. In the present study we sought to distinguish between these models. We present evidence that BiP binds to ATF6 stably, arguing against the competition model. Specific sequences in ATF6 are required for ER stress-induced BiP release, supporting an active triggering model for BiP dissociation.
MATERIALS AND METHODS
Materials and constructs.
Brefeldin A (BFA), cycloheximide, protease inhibitor cocktail for mammalian cells, anti-FLAG M2 antibody, and horseradish peroxidase-conjugated goat anti-mouse, anti-rabbit, and anti-rat immunoglobulin G (IgG) sera were obtained from Sigma. Mouse monoclonal anti-T7 antibody was from Novagen. Protein A-Sepharose and ECL plus reagents were from Amersham Biosciences. Rabbit polyclonal anti-Ig heavy chain (Ig HC) serum and rat monoclonal and rabbit polyclonal anti-BiP antibodies were kindly provided by Linda Hendershot.
pBiP-myc was constructed by cloning human BiP cDNA into the vector pcDNA3.1 myc-His (Invitrogen). ATF6, LZIP, and their derivatives were cloned into the p3xFLAG-CMV7.1 vector (Sigma) to generate constructs encoding fusion proteins with three tandem copies of the FLAG epitope at their N termini. The LD of ATF6 was expressed by cloning ATF6 (amino acids [aa] 398 to 670) with a signal peptide from Ig HC at its N terminus into the pCMV-3xFLAG14 vector. This construct, pSP-ATF6(LD)-3xFLAG, contains a C-terminal 3 × FLAG epitope tag. The green fluorescent protein (GFP)-ATF6 construct was described previously (5). All of these expression constructs are under the control of a cytomegalovirus promoter. Point mutations were made with a QuikChange site-directed mutagenesis kit (Stratagene). Vectors expressing Ig HC and hamster BiP were provided by Linda Hendershot and have been previously described (3, 41). A vector encoding the temperature-sensitive variant of the vesicular stomatitis virus G (VSVG) protein was obtained from Peter Espenshade (8). The simian virus 5 (SV5) hemagglutinin-neuraminidase (HN) protein expression vector and rabbit polyclonal antiserum were obtained from Robert Lamb (Northwestern University).
Cell culture and transfection.
HeLa, COS, and NIH 3T3 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% newborn calf serum at 37°C in a 5% CO2 incubator. NIH 3T3 cells stably expressing 3 × FLAG-tagged ATF6 were prepared with a retroviral vector as previously described (5). The virus expressing ATF6 was generated by transfection of 293 cells with pBABE-puro-3xFLAG-ATF6 and a packaging site-defective Moloney murine leukemia virus construct. HeLa cells were transfected by the standard calcium phosphate DNA precipitation method as previously described (5). COS cells were transfected with Lipofectamine 2000 reagent (Invitrogen).
IP and immunoblotting.
Cells grown on 60-mm-diameter dishes were rinsed once with ice-cold phosphate-buffered saline (PBS) 24 h after transfection and then lysed in 400 μl of 1% Triton X-100 immunoprecipitation (IP) buffer (20 mM HEPES [pH 7.5], 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM EDTA, protease inhibitor cocktail). After rotating for 1 h at 4°C, the extracts were cleared by centrifugation at 13,000 × g for 10 min at 4°C. 3 × FLAG-tagged proteins were immunoprecipitated with 2 μl of anti-FLAG antibodies and 20 μl of 50% protein A-Sepharose in IP buffer. Ig HC was precipitated directly with protein A Sepharose. Immunoprecipitates were washed three times with IP buffer and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were then transferred to nitrocellulose membranes and probed with primary antibodies and horseradish peroxidase-conjugated secondary antibodies. The following dilutions of primary antibodies were used for immunoblotting: anti-BiP, 1:250; anti-FLAG M2, 1:1,000; anti-Ig HC, 1:1,000; anti-T7, 1:1,000. The blots were visualized with ECL plus reagents (Amersham Biosciences).
In vitro ATP-mediated BiP release.
For Fig. 1, Ig HC or 3 × FLAG-ATF6 was transiently expressed in HeLa cell and the cells were extracted with 1% Triton X-100 IP buffer (see above). Ig HC and 3 × FLAG-ATF6 were immunoprecipitated with either protein A-Sepharose alone or anti-FLAG antibodies plus protein A-Sepharose, respectively. The Sepharose beads were incubated with 2 mM ATP and 2 mM MgCl2 at 25°C for 30 min. The beads were then washed with IP buffer and immunoblotted.
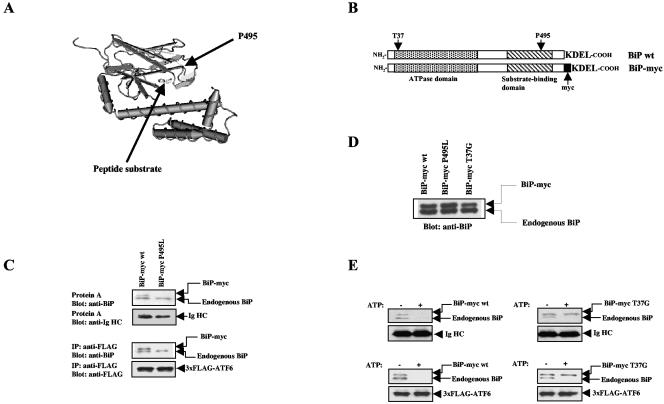
FIG. 1.
ATF6 is a chaperone substrate of BiP. (A) Structural model of the peptide-binding domain of DnaK (50). The corresponding positions of proline 495 in human BiP and the peptide substrate are highlighted with arrows. (B) Diagram of constructs encoding wt BiP and myc-tagged BiP. The positions of threonine 37 and proline 495, which were mutated in our studies, are indicated. (C) Binding of wt and mutant BiP to Ig HC and ATF6. Ig HC (top two parts) or 3 × FLAG-tagged ATF6 (bottom two parts) was transiently expressed in HeLa cells along with either wt or P495L mutant BiP-myc. Ig HC and 3 × FLAG-ATF6 were precipitated with either protein A-Sepharose or anti-FLAG antibodies plus protein A-Sepharose, respectively. The precipitated complexes were resolved by SDS-PAGE and detected by immunoblotting with the indicated antibodies. (D) Immunoblots showing the expression levels of endogenous BiP and transfected BiP-myc. wt and mutant BiP-myc were transiently expressed in HeLa cells, and cell lysates were blotted with anti-BiP antibodies. (E) Ig HC (top) or 3 × FLAG-ATF6 (bottom) was transiently expressed in HeLa cells along with either wt (left) or T37G mutant (right) BiP-myc. Ig HC and 3 × FLAG-ATF6 were immunoprecipitated, and the immunocomplexes were incubated with or without 2 mM ATP for 30 min at 25°C. The immunocomplexes were then immunoblotted for BiP, Ig HC, or 3 × FLAG-ATF6 as indicated.
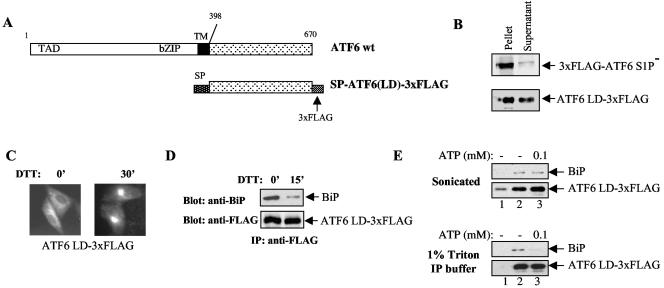
For Fig. 5, the ATF6 LD, SP-ATF6(LD)-3xFLAG, was transiently expressed in HeLa cells and extracted with either 1% Triton X-100 IP buffer or by ultrasonication in IP buffer without detergent. The extracts were treated with or without 0.1 mM ATP and 2 mM MgCl2 at 25°C for 30 min before IP with anti-FLAG antibodies plus protein A-Sepharose beads at 4°C.
FIG. 5.
(A) Diagram of constructs encoding wt ATF6 and the ATF6 LD. (B) Full-length ATF6 or the ATF6 LD was transiently expressed in HeLa cells and extracted by sonication. Cell lysates were centrifuged at 13,000 × g for 15 min before the supernatant and pellet were collected for immunoblotting analysis. (C) Localization of ATF6 LD. The ATF6 LD was transiently expressed in HeLa cells, and the cells were treated with or without 5 mM DTT as indicated. The cells were immunostained with anti-FLAG antibodies as previously described (34). (D) BiP association with the ATF6 LD. The ATF6 LD was immunoprecipitated with anti-FLAG antibodies, and BiP association was detected by immunoblotting with anti-BiP antibodies. The transfected HeLa cells were treated with 5 mM DTT as indicated to induce ER stress. (E) The ATF6 LD was extracted from HeLa cells by either 1% Triton X-100 IP buffer or sonication as indicated. The extracts were treated with or without 0.1 mM ATP at 25°C for 30 min before IP with anti-FLAG antibodies (lanes 2 and 3) and immunoblotting with either anti-BiP or anti-FLAG antibodies. For lane 1, no antibodies were added as a control for the specificity of the IP. TM, transmembrane.
Metabolic labeling.
HeLa or COS cells grown on 35-mm-diameter plates were rinsed twice with PBS 24 h after transfection and then incubated in methionine- and cysteine-free medium (ICN) for 30 min. The cells were pulse-labeled with [35S]methionine-cysteine (ICN) at 500 μCi/ml for 10 min. For lysis, the cells were rinsed with ice-cold PBS and lysed in 400 μl of 1% Triton X-100 IP buffer. The 3 × FLAG-tagged proteins were immunoprecipitated with 2 μl of anti-FLAG antibodies plus protein A-Sepharose as described above. Protein complexes were resolved by SDS-PAGE and observed by autoradiography.
Purification of BiP-ATF6 complexes from stably transfected NIH 3T3 cells.
The purification procedures were carried out essentially as previously described (44). NIH 3T3 cells stably expressing 3 × FLAG-tagged ATF6 and control uninfected NIH 3T3 cells grown on 20 14-cm-diameter dishes were washed once with ice-cold PBS and scraped into 10 ml of hypotonic buffer (50 mM HEPES-KOH [pH 7.4], 1.5 mM MgCl2, 10 mM KCl, 250 mM sucrose, 5 mM sodium EDTA, 5 mM sodium EGTA, protease inhibitor cocktail). After 30 min of incubation on ice, cells were lysed with 30 strokes of a type B Dounce homogenizer. Cell lysates were centrifuged at 1,000 × g for 5 min to remove nuclei and unlysed cells. Supernatants were then centrifuged at 10,000 × g for 15 min to purify the membrane microsome fractions, which were solubilized with 5 ml of Chapso buffer (50 mM HEPES-KOH [pH 7.4], 150 mM NaCl, 0.5% [wt/vol] Chapso, protease inhibitor cocktail). After incubation for 1 h at 4°C, the debris was removed by centrifugation at 100,000 × g for 30 min. The supernatant was incubated with 50 μl of agarose-conjugated anti-FLAG antibodies (Sigma) and rotated for 3 h. The beads were then washed five times with Chapso buffer. Protein complexes bound to beads were eluted for 12 h with 1 ml of Chapso buffer containing 0.25 mg of FLAG peptide/ml. The eluted protein complexes were separated by SDS-PAGE and visualized with a Coomassie blue staining kit (Sigma).
Live cell imaging and photobleaching experiments.
Live cells were imaged on a temperature-controlled stage of a Zeiss LSM 510 confocal microscope system with the 488-nm line of a 40-mW Ar-Kr laser for GFP and a 63× 1.4 N.A. oil objective. Qualitative fluorescence recovery after photobleaching (FRAP) experiments were performed by photobleaching a region of interest (outlined box) at full laser power and then monitoring fluorescence recovery by scanning the whole cell at low laser power. No photobleaching of the cell or adjacent cells during fluorescence recovery was observed.
Fluorescence recovery plots and diffusion measurements were obtained by photobleaching a 4-μm-wide strip as described previously (6, 27). The effective diffusion coefficient (Deff) was determined with an inhomogeneous diffusion simulation program written by Eric Siggia (37). To create the fluorescence recovery curves, the fluorescence intensities were transformed into a 0 to 100% scale in which the first postbleach time point equals 0% recovery and the recovery plateau equals 100% recovery. The plots do not represent the mobile fraction (Mf) of the GFP chimeras. The Mf was calculated by comparing the photobleach-corrected prebleach and postbleach recovery fluorescence intensity values in the photobleached region of interest as previously described (6). Image analysis was performed with NIH Image 1.62 and LSM image examiner software. Composite figures were prepared with Adobe Photoshop 5.5 and Illustrator 9.0 software (Adobe). Fluorescence recovery curves were plotted with Kaleidagraph 3.5 (Synergy Software).
The Deff is the diffusion coefficient within the cellular environment. Because of the effects of geometry, the calculated Deff may be off by as much as one-third of the actual diffusion coefficient. This is not a concern for comparisons within the same organelle. The Mf represents the percentage of the fluorescent protein available for fluorescence recovery during the time course of the experiment.
RESULTS
BiP binds to ATF6 in a way similar to that of unfolded proteins.
We first probed the molecular nature of the BiP-ATF6 interaction. Since BiP is a molecular chaperone, one possibility is that ATF6 is a chaperone substrate of BiP and that their interaction is similar to the binding of BiP to unfolded proteins. An alternative model is that BiP binds to ATF6 through a domain outside its peptide-binding pocket and that ATF6 is not a chaperone substrate. Hsp70 family chaperones interact with a number of accessory factors, including Hsp40, BAG-1, HspBP1, Hip, Hop, CHIP, and the mitochondria import receptor Tom70 through regions other than their peptide-binding domains (15, 48).
We distinguished between these two models by determining whether ATF6 binding requires a functional peptide-binding domain in BiP. Hsp70 proteins are highly conserved from bacteria to humans, and the primary sequence of human BiP can be mapped to the backbone of DnaK, the bacterial homologue of Hsp70. The crystal structure of the peptide-binding domain of DnaK was solved and has been used as a structural model to study other Hsp70 family members (50). The C-terminal peptide-binding domain of DnaK consists of a β-sheet sandwich that harbors the substrate-binding cleft. DnaK recognizes hydrophobic stretches on extended unfolded polypeptides, which are accommodated in the substrate-binding cleft (Fig. 1A). Genetic screening performed on the yeast BiP homologue Kar2p isolated several mutant forms of Kar2p defective in substrate binding (18). One of these, Kar2p-1, bears a point mutation that corresponds to proline 495 of human BiP located within loop5,6 of the β-sheet peptide-binding domain (Fig. 1A). We mutated proline 495 to leucine in human BiP and tested whether this point substitution eliminated its substrate-binding ability. Transfected BiP was tagged with a myc epitope at its COOH terminus immediately before its KDEL ER retention signal to distinguish it from endogenous BiP (Fig. 1B). Myc-tagged BiP migrates slightly slower than endogenous BiP on SDS-PAGE such that they can be distinguished (Fig. 1C). As shown below, insertion of the myc tag had no effect on the activity of BiP.
We transiently expressed the myc-tagged BiP proteins in HeLa cells and tested whether the P495L mutation affects substrate binding. Unassembled Ig HC was used as a control BiP substrate. In the absence of the Ig light chain, Ig HC is an unfolded protein and is retained in the ER by BiP binding to its CH-1 domain (41). Protein A efficiently coprecipitated Ig HC with endogenous and transfected wild-type (wt) myc-tagged BiP. As expected, the P495L mutation completely abolished BiP binding to Ig HC (Fig. 1C, top parts). Next, we determined whether the BiP P495L mutant could still associate with ATF6 by coexpressing the mutant BiP with 3 × FLAG-tagged ATF6. We found that, similar to binding to Ig HC, wt but not mutant BiP bound to ATF6 (Fig. 1C, lower parts). The BiP P495L mutant was expressed at levels similar to those of wt BiP (Fig. 1D). These results indicate that ATF6, similar to unfolded proteins, requires the intact peptide-binding pocket of BiP for binding.
Substrate binding of BiP is modulated by ATP-ADP binding to its N-terminal ATPase domain. ATP-associated BiP binds substrates loosely and rapidly dissociates, while ADP-bound BiP binds more tightly with a slower dissociation rate (43). When incubated with ATP in vitro, both transfected wt BiP-myc and endogenous BiP were released from Ig HC. In contrast, an ATPase domain mutant form, T37G mutant BiP-myc, was not released and remained bound to Ig HC (Fig. 1E, top parts). This mutant BiP is defective for an ATP-induced conformational change and therefore binds substrates tightly and does not release them in the presence of ATP (41). Similarly, in vitro ATP incubation readily dissociated ATF6 from both endogenous BiP and transfected wt BiP-myc but not from T37G mutant BiP-myc (Fig. 1E, lower parts). These data demonstrate that the binding of BiP to ATF6 is dependent on the substrate-binding domain of BiP and that this binding is sensitive to the ATP-induced conformational change in BiP. Therefore, the binding of BiP to ATF6 is identical to its binding to unassembled Ig HC, suggesting that BiP recognizes ATF6 as an unfolded protein.
The BiP-ATF6 complex is stable, and BiP does not cycle ATF6 on and off.
The finding that BiP binds to ATF6 through its peptide-binding domain supports the widely proposed competition model to explain the ER stress-induced dissociation of BiP from ER stress transducers (12, 20, 21). Since both ATF6 and misfolded proteins are chaperone substrates of BiP, they could compete for binding to free BiP molecules. An increase in misfolded proteins in the ER would reduce the free BiP available for ATF6 binding, favoring the shift of ATF6 from a BiP-bound to a free form. Free ATF6 could then be transported to the Golgi apparatus to be cleaved by S1P and S2P. Since BiP was rapidly released from ATF6 upon treatment of the cells with ER stress-inducing agents (34), for the competition model to be true the BiP-ATF6 complexes must be highly dynamic with ATF6-bound BiP in rapid equilibrium with free BiP molecules.
However, this competition model appears to be difficult to reconcile with the stability of BiP-ATF6 complexes in vitro. Whereas dynamic chaperone-substrate interactions can only be detected after stabilizing the complexes with cross-linkers (51), BiP binding to ATF6 was stable enough to endure the IP procedure without any stabilizing agents. This could be seen by the immunopurification of ATF6 complexes from NIH 3T3 cells stably expressing 3 × FLAG-tagged ATF6. The amount of BiP coprecipitated with ATF6 was 5 to 10 times more than that of ATF6 (Fig. 2A), consistent with our previous identification of multiple BiP-binding sites on ATF6 (34).
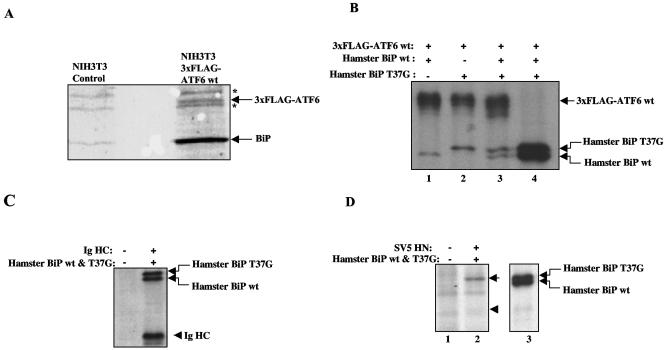
FIG. 2.
Stability of BiP-ATF6 complexes. (A) ATF6-BiP complexes were isolated by immunoaffinity purification from NIH 3T3 cells stably expressing 3× FLAG-ATF6 as described in Materials and Methods. The complexes were separated by SDS-PAGE and stained with Coomassie blue. The identities of ATF6 and BiP in the protein complexes were confirmed by matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis (data not shown). Asterisks indicate nonspecific bands. (B) COS cells transiently expressing 3× FLAG-ATF6 and hamster BiP (wt, T37G mutant, or both) were labeled with [35S]methionine-cysteine for 10 min. Cells were then lysed, and 3× FLAG-ATF6 was immunoprecipitated with anti-FLAG antibodies (lanes 1 to 3). After ATF6 was immunoprecipitated from one of the samples (lane 3), half of the remaining sample was immunoprecipitated with polyclonal antibodies specific to hamster BiP (lane 4). (C) COS cells transfected with or without Ig HC and both wt and T37G mutant hamster BiP were labeled with [35S]methionine-cysteine for 10 min. Cells were lysed, and Ig HC was precipitated with protein A-Sepharose beads. (D) HeLa cells transiently transfected with or without SV5 HN protein and hamster BiP were pulse-labeled with [35S]methionine-cysteine, and HN was immunoprecipitated with polyclonal anti-HN antiserum (lanes 1 to 2). After IP with anti-HN antisera, the supernatant of lane 2 was immunoprecipitated with polyclonal anti-BiP antiserum (lane 3). In lane 2, the position of T37G mutant BiP is indicated by an arrow and the position of SV5 HN protein is indicated by an arrowhead.
In order to test the stability of ATF6-BiP complexes in vivo, we compared the binding of ATF6 to wt and T37G mutant BiP. Substrate binding to T37G mutant BiP is stable because of defects in BiP ATPase activity and ATP-induced conformational changes (41). T37G mutant BiP failed to be released from ATF6 by either in vivo ER stress or in vitro ATP incubation (Fig. 1E) (41). We expressed wt ATF6 in COS cells along with hamster wt and T37G mutant BiP. Cells were pulse-chased with [35S]methionine for 10 min before ATF6 complexes were immunoprecipitated (Fig. 2B, lanes 1 to 3). The pulse-chase generated similar amounts of radiolabeled wt and T37G mutant BiP molecules, as detected by IP with antisera specific for hamster BiP (Fig. 2B, lane 4). We reasoned that if wt BiP constantly cycles on and off ATF6 while T37G mutant BiP binds and remains trapped on ATF6, we would observe more mutant than wt BiP bound to ATF6. However, when transfected alone or together, there was similar binding of wt and T37G mutant BiP to ATF6 (Fig. 2B, lanes 1 to 3), suggesting that wt BiP binds to ATF6 as stably as T37G mutant BiP in vivo. The level of BiP binding in this experiment appears low relative to ATF6, in contrast to the high binding detected by Coomassie blue staining (Fig. 2A). This is likely due to the difference in detection by [35S]methionine pulse-labeling in Fig. 2B, as well as possible differences due to the transient transfection of BiP and ATF6.
BiP has also been previously shown to bind stably to misfolded proteins such as unassembled Ig HC and a temperature-sensitive VSV glycoprotein (41). We also found that Ig HC bound similar levels of wt and T37G mutant BiP (Fig. 2C), consistent with previous observations (41). Next we sought to test a substrate that is known to cycle on and off BiP. We used the SV5 HN protein, which binds BiP transiently as part of the HN protein maturation process (28). As with ATF6, the cells were pulse-labeled with [35S]methionine and immunoprecipitated with either anti-HN sera or anti-hamster BiP sera. In contrast to ATF6, little binding of wt BiP was observed while T37G mutant BiP binding was still clearly detected (Fig. 2D, lane 2) even though both forms of BiP were similarly expressed (lane 3). The expression of the SV5 HN protein was weakly, but reproducibly, detected (arrowhead in lane 2) in this pulse-labeling experiment because it migrates close to a background band and perhaps because its rate of translation is lower than that of BiP. These results suggest that while SV5 HN binds strongly, as expected, to T37G mutant BiP, binding to wt BiP is weak since it likely cycles on and off during its maturation process. This contrasts with ATF6, which appears to bind stably to both wt and T37G mutant BiP, and supports the validity of this metabolic labeling approach to measure BiP-substrate dynamics.
ER stress-induced BiP dissociation is specific to ER stress transducers.
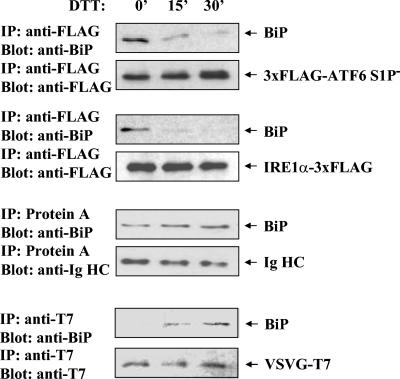
Since BiP also binds stably to unassembled Ig HC and a temperature-sensitive variant of the VSVG protein (41), we next determined whether ER stress also dissociates BiP from these two substrates. We reasoned that if BiP binding to these substrates is dynamic, as generally hypothesized for chaperone-substrate interactions, competition from misfolded proteins generated during ER stress would be expected to cause a nonselective dissociation of BiP from all of these substrates. However, we found that in dithiothreitol (DTT)-treated cells BiP remained associated with unassembled Ig HC while it was rapidly released from ATF6 and IRE1α. In fact, the amount of BiP bound to Ig HC appeared to increase during DTT treatment, probably because of the increased misfolding state of Ig HC induced by DTT (Fig. 3). We also checked the binding of BiP to the temperature-sensitive variant of VSVG. At the nonpermissive temperature (40°C), the VSVG variant is unfolded and associated with BiP (41). BiP binding to VSVG was barely detectable in unstressed cells at 40°C but increased dramatically in response to DTT treatment, possibly for the same reason as Ig HC (Fig. 3). Hence, ER stress only causes the release of BiP from ER stress transducers, but not from other BiP-binding unfolded proteins. This result is also contradictory to the competition model since increased misfolded proteins would be expected to compete with any substrate for BiP binding, including Ig HC and VSVG.
FIG. 3.
ER stress-induced BiP release is specific to ATF6 and IRE1. HeLa cells were transiently transfected with ATF6, IRE1α, Ig HC, and VSVG-T7 (tsO45) constructs as indicated to the right of each pair of panels. The S1P mutant variant of ATF6 was used to preclude any possibility of S1P digestion causing apparent dissociation of BiP. The cells were treated with 5 mM DTT for the times indicated in minutes at the top before the cells were lysed and immunoprecipitated with the indicated antibodies (or protein A-Sepharose for Ig HC). The immunocomplexes were immunoblotted for association of BiP and expression of the indicated expressed proteins.
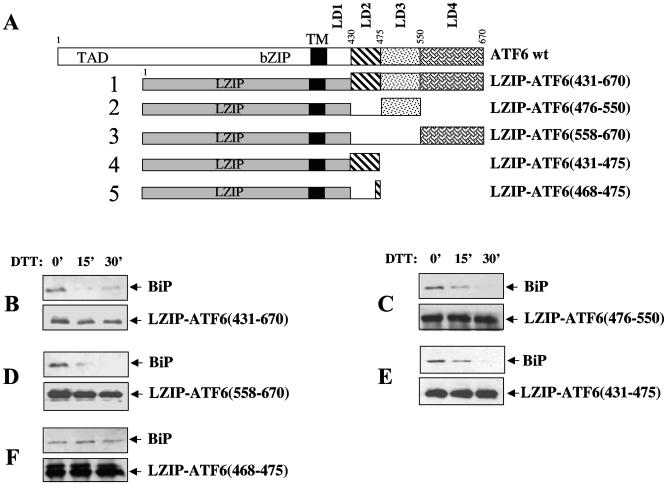
The specificity of the ER stress-induced BiP release suggests that ATF6 and IRE1 may contain an ER stress response sequence(s) that is absent in Ig HC and VSVG. To test the existence of such specific sequences in ATF6, we fused various regions of the ATF6 LD to LZIP, an ER protein that does not bind BiP (34), and determined whether BiP dissociated from these LZIP-ATF6 chimeras in stressed cells. We previously divided the LD of ATF6 into four regions (LD1 to -4) and showed that three of the four regions (LD2 to -4) could bind BiP (Fig. 4A) (34). When the cells were challenged with DTT, we found that BiP dissociated from the full-length LD (aa 431 to 670) as well as from each of the three segments (LD2 to -4) (Fig. 4B to E). This suggests that each of the three LD regions contains both BiP-binding sites and responsive sequences for ER stress-induced BiP release. The BiP-binding regions and the sequences specifying ER stress-induced BiP release may overlap or be closely spaced. We previously identified an eight-residue peptide within the LD2 region (aa 468 to 475) of ATF6 that could bind small amounts of BiP that were sufficient to retain the protein in the ER (34). We fused this eight-residue segment of ATF6 to LZIP and found that while this chimera, LZIP-ATF6(468-475), could bind BiP, BiP remained bound when the cells were treated with DTT (Fig. 4F). This contrasts with the other LZIP-ATF6 chimeras, suggesting that the eight-residue fragment (aa 468 to 475) contains a BiP-binding site but lacks an ER stress-responsive sequence. Since the binding of BiP to LZIP-ATF6(431-475) was sensitive to ER stress (Fig. 4E), these results suggest that aa 431 to 467 contain a BiP-releasing sequence. The other segments of the ATF6 LD must also contain BiP-releasing sequences since BiP independently binds to each of them and dissociates in response to ER stress. The failure of BiP to dissociate from the aa 468 to 475 region of ATF6 also provides further support for our finding that the BiP-ATF6 complex is not dynamic. For if BiP continuously cycles from ATF6, misfolded proteins should cause a rapid and nonselective dissociation of BiP from all of the chimeras, including LZIP-ATF6(468-475).
FIG. 4.
ATF6 contains responsive sequences required for ER stress-induced BiP dissociation. (A) Diagram of constructs encoding LZIP-ATF6 chimeras. These proteins were N terminally tagged with the 3× FLAG epitope. (B to F) The above constructs were transiently transfected into HeLa cells, and the cells were treated with 5 mM DTT for the times indicated in minutes at the top. The chimeras were immunoprecipitated with anti-FLAG antibodies, and the immunocomplexes were immunoblotted with either anti-FLAG (bottom) or anti-BiP (top) antibodies. Panels B to F correspond to constructs 1 to 5 in panel A. TM, transmembrane.
Detergent sensitivity of the BiP-ATF6 complex.
The apparent stability of the ATF6-BiP complex in vivo contrasts with its sensitivity to ATP-induced dissociation in vitro. Incubation of the immunoprecipitated complex in vitro with ATP resulted in complete dissociation of BiP (Fig. 1E). We reasoned that this could be due to the presence of detergents in the IP buffer that remove accessory factors required for stabilization of the complex. It has been shown for Ig HC that BiP association is assisted by a number of cofactors that can only be isolated without using detergent (25, 41). However, because ATF6 is an integral membrane protein detergents are required to free ATF6 from the membrane. To avoid this problem we expressed the LD of ATF6 as a soluble protein in the ER lumen. The signal peptide from Ig HC was fused to the N terminus of the ATF6 LD, and the C terminus of the fusion protein was tagged with a 3 × FLAG epitope to allow detection (Fig. 5A). When expressed in HeLa cells, the ATF6 LD could be efficiently extracted to a soluble fraction by ultrasonication while full-length ATF6 was predominantly found in the pellet (Fig. 5B), suggesting that the ATF6 LD was indeed expressed as a soluble ER lumenal protein. Interestingly, the ATF6 LD was localized to the ER and moved to the Golgi apparatus upon DTT treatment, similar to wt ATF6 (Fig. 5C). Moreover, the ATF6 LD bound to BiP and dissociated in response to DTT treatment, demonstrating that the ATF6 transmembrane and cytoplasmic domains are not required for these functions (Fig. 5D).
We then tested whether BiP-ATF6 LD complexes extracted from cells without using detergents could be dissociated by in vitro ATP incubation. We found that BiP-ATF6 LD complexes isolated by ultrasonication were indeed resistant to 0.1 mM ATP treatment (Fig. 5E, top parts). In contrast, when the ATF6 LD was extracted with 1% Triton X-100, the same ATP treatment effectively released BiP from the ATF6 LD (Fig. 5E, lower parts), consistent with the notion that detergents remove accessory factors involved in stabilizing BiP binding to ATF6. The resistance of BiP-ATF6 complexes to ATP-induced dissociation suggests a stable association that is maintained by detergent-sensitive cofactors. As a control for the IP, we performed the procedure without adding antibodies (Fig. 5E, lane 1). There was a low level of nonspecific precipitation of ATF6 LD in the sonicated extracts, likely because of its residual binding to protein A-Sepharose beads in the absence of detergents. Significantly higher levels of precipitation were detected when the anti-FLAG antibodies were used (lanes 2 and 3), and no BiP association was detected without the specific antibodies (lane 1). We also tried to reproduce ER stress dissociation of the ATF6-BiP complex in our sonicated extract in vitro; however, we found that DTT treatment had no effect (data not shown). DTT may not be as effective at inducing misfolded proteins in vitro as it is in vivo, or the components necessary to sense ER stress may not be active in our soluble extract.
ATF6 is mobile in the ER.
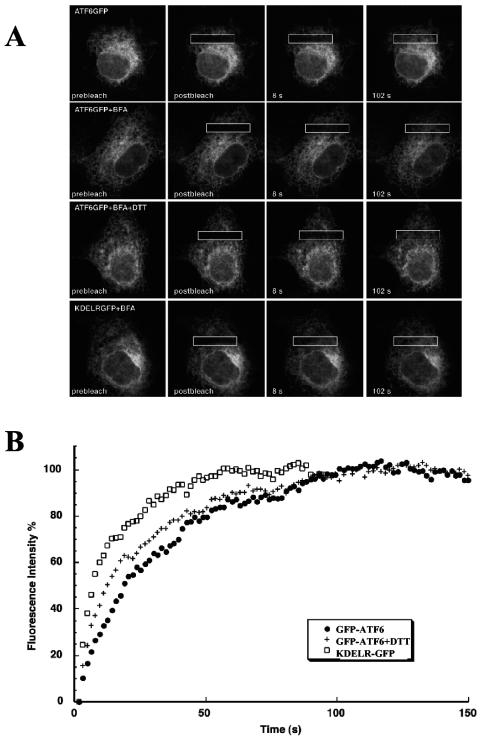
BiP binding represses ATF6 activation by preventing its export to the Golgi apparatus, and we next analyzed how stable BiP binding retains ATF6 in the ER. Export of ER proteins is mediated by anterograde trafficking vesicles that collect cargo proteins at distinct regions of the ER, the ER exit sites, and then bud and transport the cargo to the cis-Golgi apparatus (1). We previously showed that BiP retains ATF6 in the ER by blocking its intrinsic Golgi apparatus localization signals (36). It is unclear, however, how this inhibition occurs. One possibility is that the BiP-ATF6 complex cannot move to the ER exit sites because of its immobilization, aggregation, or clustering at sub-ER membrane domains. Certain nonnative proteins form aggregates in the ER and fail to be transported to the Golgi apparatus because of their immobility (7). Alternatively, the ATF6-BiP complex could be mobile and diffuse freely to the ER exit sites but fail to be recognized and packaged into the ER-to-Golgi apparatus trafficking vesicles. We distinguished between these two ER retention models by measuring the diffusional mobility of ATF6 on the ER membrane by FRAP. ATF6 was N terminally tagged with GFP, and the fusion protein was expressed in COS cells. This GFP-ATF6 chimera localized to the ER and translocated to the Golgi apparatus in response to ER stress, implying that attachment of GFP did not alter this property of ATF6 (5). The fluorescent molecules in a small region of the cell were irreversibly photobleached by a laser beam, and fluorescence recovery through the exchange of nonbleached for bleached GFP-ATF6 was measured (Fig. 6A). The Deff, which represents the lateral mobility of a protein on the ER membrane, is derived from the half time of fluorescence recovery. The Mf is the extent of fluorescence recovery in the bleached area and indicates the percentage of proteins that are free to diffuse (Table 1) (27).
FIG. 6.
ATF6 is mobile in the ER in unstressed cells. Cos-7 cells transiently expressing either GFP-ATF6 or KDELR-GFP were analyzed by FRAP. (A) Cells were untreated or incubated with BFA or BFA and DTT for at least 30 min. The indicated boxes (4-μm strips) were photobleached and analyzed for recovery as described in Materials and Methods. (B) Representative recovery curves of FRAP experiments in panel A. The recovery intensities have been transformed for comparisons of recovery rates with the recovery asymptote designated as 100%.
TABLE 1.
Mobility and diffusion of GFP fusion proteinsa
| Protein | Treatment | Deff (μm2/s) | Mf |
|---|---|---|---|
| GFP-ATF6 | None | 0.09 ± 0.03 | 74.3 ± 8.9 |
| GFP-ATF6 | BFA | 0.18 ± 0.08b | 77.4 ± 6.2 |
| GFP-ATF6 | BFA + 5 mM DTT | 0.22 ± 0.06b | 80.2 ± 4.3 |
| KDELR-GFP | BFA | 0.70 ± 0.26c | 91.3 ± 3.7c |
The data in Fig. 6 were quantitated, and the Deff and Mf were determined as described in Materials and Methods. Each value is the mean of 14 samples ± the standard deviation.
Value is statistically significantly (P < 0.001) different from that of the untreated GFP-ATF6 sample. The values for these two conditions are not statistically significantly different from one another.
Value is statistically significantly (P < 0.001) different from that of GFP-ATF6 stimulated by the two conditions.
In unstressed cells, fluorescence recovery of GFP-ATF6 was observed with a diffusion coefficient of 0.09 μm2/s and an Mf of 74% (Fig. 6 and Table 1). This suggests that in unstressed cells the majority of ATF6 molecules are mobile in the ER. We next added BFA to prevent exit of proteins from the ER, allowing the diffusional rate of ATF6 on the ER membrane to be measured with or without DTT treatment. In fact, BFA itself is an ER stress-inducing agent and caused an increase in the mobility of ATF6 (0.18 μm2/s) but no significant change in the Mf (77%). When ER stress was further induced with DTT, the diffusion coefficient of ATF6 only slightly increased to 0.22 μm2/s while the Mf remained essentially the same (80%). The KDEL receptor (KDELR), a Golgi apparatus-localized protein that shows ER localization in BFA-treated cells (27), was used as a control. KDELR had a significantly higher lateral diffusion rate (0.70 μm2/s) than ATF6 and showed a higher recovery extent of 91% (Fig. 6 and Table 1). This suggested that ATF6 was part of a higher-molecular-weight complex. Consistent with this, the Deff of ATF6 was only about two times faster than that observed for the translocon (29), a protein-synthesizing apparatus consisting of 20 different polypeptides. The LD of ATF6 binds to multiple BiP molecules, and BiP has been found in large multichaperone complexes (25, 34). The slow mobility of ATF6 may also be attributed to its transient binding to immobile components of the ER. The increase in ATF6 mobility in response to ER stress could be due to the decrease in ATF6 complex size by shedding of BiP molecules or other rearrangements of the ATF6 complex. These results indicate that in unstressed cells ATF6 is capable of freely moving throughout the ER as a large complex. Its retention in the ER, therefore, is predominantly a result of its exclusion from ER-to-Golgi apparatus trafficking intermediates.
DISCUSSION
We sought to distinguish models for ER stress-induced dissociation of the ATF6-BiP complex. Our data argue against a competition model in which increased levels of misfolded proteins compete with ATF6 for BiP binding. This model would require dynamic association between BiP and ATF6, while our evidence suggests quite stable binding. First, we easily isolated the ATF6-BiP complex from cells, in contrast to the dynamic interaction of chaperones with substrates that are often barely detectable without using chemical cross-linkers (51). Second, we compared the binding of ATF6 to wt BiP and a T37G mutant form that binds substrates stably because of a defect in its ATPase activity (41). We found that ATF6 was associated with similar amounts of wt and T37G mutant BiP, suggesting that it binds wt BiP as stably as T37G mutant BiP. This binding was similar to the stable binding of BiP previously observed with Ig HC (41) but contrasts with the BiP substrate SV5 HN protein that bound stably to T37G mutant BiP but not to wt BiP. Third, the competition model would predict that BiP binding to any substrate, not just ATF6, would be competed off by misfolded proteins accumulated during ER stress. However, we found that ER stress did not induce dissociation of BiP from Ig HC or VSVG protein substrate. Fourth, we found that the ATF6-BiP complex was resistant to ATP-induced dissociation in vitro when isolated without detergents, suggesting that the complex is maintained in a stable conformation, likely by cofactors.
In favor of an active triggering model for ER stress-induced dissociation of the ATF6-BiP complex, we found that the ER stress transducers ATF6 and IRE1α were specifically dissociated from BiP but that two other substrates, Ig HC and VSVG, were not. Mapping of regions in ATF6 required for BiP binding and ER stress-induced dissociation identified at least three regions for BiP binding. For one of these, we were able to isolate a BiP-binding region, aa 468 to 475, that was only responsive to ER stress when linked to the adjacent region of aa 431 to 467, suggesting that the latter sequence specifically senses ER stress. While we were not able to reproduce ER stress-induced BiP dissociation in vitro, our identification of an ATP-resistant ATF6-BiP complex suggests that ER stress may act on components of this complex to convert it into an ATP-sensitive form.
Stable binding of BiP to ATF6 in unstressed cells.
The finding of apparently stable ATF6-BiP is surprising since chaperone binding to substrates generally involves multiple ATP-mediated binding-and-release cycles and cycling is crucial for a polypeptide to progress along its folding pathway (15). Stable BiP binding to ATF6 was disrupted by ATP incubation in vitro when the complex was isolated with detergents; however, binding was stable in vivo or when the complex was isolated without detergents. These results suggest that the ATPase cycle of ATF6-bound BiP is stalled in vivo and that BiP is maintained in its slow-dissociating ADP-bound form. This could be achieved in several ways. The substrate-binding capacity of Hsp70 can be modulated by a number of cochaperones that regulate the Hsp70 nucleotide-binding state. For instance, DnaJ assists the loading of substrates onto Hsp70 and stimulates its ATP hydrolysis to switch it to an ADP-bound slow-release state (15). Recently several ER-localized homologues of DnaJ (ERdj) have been identified and shown to regulate the ATPase activity of BiP (36, 49). In fact, ERdj3 is part of the BiP-Ig HC complex (25). The BiP-ATF6 complex may contain one or more ERdjs that maintain BiP in its ADP-bound state and thus promote its stable binding to ATF6. Other factors like Bag1 and Hip can also stabilize the binding of Hsp70 to substrates, and their ER homologues may participate in maintaining stable BiP binding to ATF6 (15).
An active regulatory mechanism triggers the dissociation of BiP from ATF6.
Despite the stable interaction of BiP with ATF6, their dissociation during ER stress is rapid and efficient (34). The specificity of BiP dissociation from the ER stress transducers ATF6 and IRE1α suggests that an active and specific mechanism triggers their release. Our analysis of mutant BiPs suggests that BiP binds ATF6 through its peptide-binding domain and requires an ATP-dependent conformational change in BiP. In addition, the isolated complex was resistant to ATP-induced dissociation in vitro. These results suggest that misfolded proteins must reactivate the stalled BiP ATPase cycle to dissociate it from ATF6. Several cochaperones such as GrpE, Hop, and Bag1 have been shown to cause the release of substrates from Hsp70 by promoting its nucleotide exchange (15). ER homologues of these factors may sense the presence of misfolded proteins in the ER and trigger the release of BiP from ATF6 by converting BiP into its ATP-bound fast-releasing form. Alternatively, misfolded proteins may bind and dissociate cochaperones that stabilize the BiP-ATF6 complex, allowing BiP to resume its ATPase cycles. These are intriguing possibilities since many cochaperones can bind to substrates directly and they may be able to function as sensors of misfolded proteins in the ER (15). It is also possible that ER stress causes modifications of BiP, ATF6, or other components of the complex, leading to destabilization and dissociation of the complex. It has been shown that BiP can be posttranslationally modified by phosphorylation and ADP ribosylation (9, 11), although the upstream events leading to these modifications and the precise roles of the modifications of BiP activity are unknown.
The specificity of ER stress-induced BiP dissociation from ER stress transducers is particularly important since nonselective dissociation of BiP from all unfolded proteins would cause protein aggregation or exit of immature proteins from the ER, both of which would be deleterious to the cell. While we were able to identify one ER stress-responsive region within the LD of ATF6, it is unclear whether a common ER stress-releasing sequence or structural motif exists or how it is recognized. It is possible that these sequences provide docking sites for ER stress-activated BiP-releasing factors.
ATF6 has been proposed to be regulated in a manner similar to that of HSF1 since they play similar roles in two parallel stress response pathways and both of them are negatively regulated by chaperones (12, 26). Despite their similarities, our data show that the mechanism of ATF6 regulation by chaperones is markedly distinct from that of HSF1. Chaperone binding to HSF1 is highly dynamic, and dissociation during heat shock stress is caused by the competition of nonnative proteins (51). In contrast, our evidence suggests that the BiP-ATF6 complex is stable and its dissociation during ER stress is triggered by an active mechanism rather than by the passive competition of misfolded proteins generated during ER stress.
Retention of ATF6 in the ER.
It appears that ER stress exploits the quality control machinery to regulate ATF6 activation since in unstressed cells ATF6 behaves as an unfolded protein (in that it is bound by BiP through its peptide-binding domain) and is retained in the ER by BiP binding. Chaperone association is a common quality control mechanism to retain immature proteins in the ER (7, 38). Our FRAP analysis of the diffusional mobility of ATF6 suggests that the BiP-ATF6 complex is freely diffusible on the ER membrane and is not tethered to the ER matrix or included in high-molecular-weight aggregates. Therefore, retention of ATF6 in the ER is not a result of its inability to move to the ER exit sites. Rather, it is either too large to be incorporated into the anterograde trafficking vesicles or not recognized by the ER export machinery. Whereas it appears that ATF6 exists in a large protein complex, we cannot precisely estimate the size of the complex. However, the upper size limit for particles that can be transported to the Golgi apparatus is quite high. Virus particles that are ∼50 nm in diameter and procollagen fibers of ∼300 nm in length can be transported to the Golgi apparatus (7). A more likely model is that BiP binding sterically masks the ATF6 Golgi apparatus localization signals such that they cannot be recognized by the ER export machinery.
Implications of stable BiP binding to ATF6.
The stability of BiP binding to ATF6 provides a mechanism for tight control of the ER stress response. If the BiP-ATF6 complex were dynamic, transient BiP release would expose the intrinsic Golgi apparatus localization signals of ATF6, giving ATF6 an opportunity to be transported to the Golgi apparatus, where active S1P and S2P reside. This spontaneous transport of ATF6 to the Golgi apparatus, in the absence of ER stress, would result in constitutive ATF6 activation. Stable binding of BiP to unassembled Ig HC, on the other hand, appears to be critical for preventing its aggregation (41). Therefore, noncycling chaperone binding may represent a general rule in the ER for the cell to handle unfolded proteins that require partners or specific signals for their maturation. The binding of BiP to nascent polypeptides that are able to fold into their native structures, however, is transient and dynamic (11). The versatility of BiP binding to specific substrates (stable or transient) may explain its multiple roles in promoting protein folding, sensing of ER stress, and controlling the activation of ER stress transducers.
BiP also regulates the activation of the other two key ER stress transducers, IRE1 and PERK (2, 21). Binding of BiP to IRE1 also requires the functional peptide-binding domain of BiP, and the interaction of IRE1 and PERK with BiP is also dissociated in response to ER stress, suggesting that IRE1 and PERK may be bound and regulated by BiP in a manner similar to that of ATF6 (2, 21, 23). The stable rather than dynamic interaction between BiP and ER stress transducers raises the possibility that different regulatory factors may control the association and dissociation between BiP and each ER stress transducer such that BiP can be selectively dissociated from only one or two of the ER stress transducers. This would allow the ER stress transducers to be differentially activated in response to different levels or types of ER stress. In support of this notion, activation of PERK appears to precede IRE1 activation in glucose-deprived pancreatic cells (19). Studies on the differentiation of plasma cells also suggest selective ER stress activation. ATF6 cleavage preceded IRE1 activation, but there was no indication of PERK activation throughout the differentiation process (10, 17, 24). Differential activation of individual branches of the ER stress pathway can only be achieved when BiP forms stable complexes with each ER stress transducer, since dynamic binding would result in simultaneous and nonselective dissociation of BiP from all of the ER stress transducers.
Acknowledgments
We thank Linda Hendershot for valuable comments and reagents, Peter Espenshade for the VSVG expression vector, Robert Lamb for the SV5 HN vector and sera, and Kenji Kohno and Yukio Kimata for discussions of BiP mutants.
This work was supported by National Institutes of Health grant CA50329 to R.P.
REFERENCES
- 1.Antonny, B., and R. Schekman. 2001. ER export: public transportation by the COPII coach. Curr. Opin. Cell Biol. 13:438-443. [DOI] [PubMed] [Google Scholar]
- 2.Bertolotti, A., Y. Zhang, L. M. Hendershot, H. P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326-332. [DOI] [PubMed] [Google Scholar]
- 3.Bole, D. G., L. M. Hendershot, and J. F. Kearney. 1986. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J. Cell Biol. 102:1558-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calfon, M., H. Zeng, F. Urano, J. H. Till, S. R. Hubbard, H. P. Harding, S. G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92-96. [DOI] [PubMed] [Google Scholar]
- 5.Chen, X., J. Shen, and R. Prywes. 2002. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J. Biol. Chem. 277:13045-13052. [DOI] [PubMed] [Google Scholar]
- 6.Ellenberg, J., E. D. Siggia, J. E. Moreira, C. L. Smith, J. F. Presley, H. J. Worman, and J. Lippincott-Schwartz. 1997. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138:1193-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellgaard, L., and A. Helenius. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4:181-191. [DOI] [PubMed] [Google Scholar]
- 8.Espenshade, P. J., W. P. Li, and D. Yabe. 2002. Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc. Natl. Acad. Sci. USA 99:11694-11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freiden, P. J., J. R. Gaut, and L. M. Hendershot. 1992. Interconversion of three differentially modified and assembled forms of BiP. EMBO J. 11:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gass, J. N., N. M. Gifford, and J. W. Brewer. 2002. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 277:49047-49054. [DOI] [PubMed] [Google Scholar]
- 11.Gething, M. J. 1999. Role and regulation of the ER chaperone BiP. Semin. Cell Dev. Biol. 10:465-472. [DOI] [PubMed] [Google Scholar]
- 12.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575-599. [DOI] [PubMed] [Google Scholar]
- 13.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099-1108. [DOI] [PubMed] [Google Scholar]
- 14.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 15.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 16.Haze, K., H. Yoshida, H. Yanagi, T. Yura, and K. Mori. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10:3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwakoshi, N. N., A. H. Lee, P. Vallabhajosyula, K. L. Otipoby, K. Rajewsky, and L. H. Glimcher. 2003. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4:321-329. [DOI] [PubMed] [Google Scholar]
- 18.Kabani, M., S. S. Kelley, M. W. Morrow, D. L. Montgomery, R. Sivendran, M. D. Rose, L. M. Gierasch, and J. L. Brodsky. 2003. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol. Biol. Cell 14:3437-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufman, R. J. 2002. Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 110:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufman, R. J., D. Scheuner, M. Schroder, X. Shen, K. Lee, C. Y. Liu, and S. M. Arnold. 2002. The unfolded protein response in nutrient sensing and differentiation. Nat. Rev. Mol. Cell Biol. 3:411-421. [DOI] [PubMed] [Google Scholar]
- 21.Kimata, Y., Y. I. Kimata, Y. Shimizu, H. Abe, I. C. Farcasanu, M. Takeuchi, M. D. Rose, and K. Kohno. 2003. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol. Biol. Cell 14:2559-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, A.-H., N. N. Iwakoshi, and L. H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, C. Y., Z. Xu, and R. J. Kaufman. 2003. Structure and intermolecular interactions of the luminal dimerization domain of human IRE1alpha. J. Biol. Chem. 278:17680-17687. [DOI] [PubMed] [Google Scholar]
- 24.Ma, Y., and L. M. Hendershot. 2003. The stressful road to antibody secretion. Nat. Immunol. 4:310-311. [DOI] [PubMed] [Google Scholar]
- 25.Meunier, L., Y. K. Usherwood, K. T. Chung, and L. M. Hendershot. 2002. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell 13:4456-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morimoto, R. I. 2002. Dynamic remodeling of transcription complexes by molecular chaperones. Cell 110:281-284. [DOI] [PubMed] [Google Scholar]
- 27.Nehls, S., E. L. Snapp, N. B. Cole, K. J. Zaal, A. K. Kenworthy, T. H. Roberts, J. Ellenberg, J. F. Presley, E. Siggia, and J. Lippincott-Schwartz. 2000. Dynamics and retention of misfolded proteins in native ER membranes. Nat. Cell Biol. 2:288-295. [DOI] [PubMed] [Google Scholar]
- 28.Ng, D. T., S. S. Watowich, and R. A. Lamb. 1992. Analysis in vivo of GRP78-BiP/substrate interactions and their role in induction of the GRP78-BiP gene. Mol. Biol. Cell 3:143-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikonov, A. V., E. Snapp, J. Lippincott-Schwartz, and G. Kreibich. 2002. Active translocon complexes labeled with GFP-Dad1 diffuse slowly as large polysome arrays in the endoplasmic reticulum. J. Cell Biol. 158:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada, T., K. Haze, S. Nadanaka, H. Yoshida, N. G. Seidah, Y. Hirano, R. Sato, M. Negishi, and K. Mori. 2003. A serine protease inhibitor prevents endoplasmic reticulum stress-induced cleavage but not transport of the membrane-bound transcription factor ATF6. J. Biol. Chem. 278:31024-31032. [DOI] [PubMed] [Google Scholar]
- 31.Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13:349-355. [DOI] [PubMed] [Google Scholar]
- 32.Rawson, R. B. 2003. The SREBP pathway—insights from Insigs and insects. Nat. Rev. Mol. Cell Biol. 4:631-640. [DOI] [PubMed] [Google Scholar]
- 33.Ron, D. 2002. Translational control in the endoplasmic reticulum stress response. J. Clin. Investig. 110:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen, J., X. Chen, L. Hendershot, and R. Prywes. 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3:99-111. [DOI] [PubMed] [Google Scholar]
- 35.Shen, X., R. E. Ellis, K. Lee, C. Y. Liu, K. Yang, A. Solomon, H. Yoshida, R. Morimoto, D. M. Kurnit, K. Mori, and R. J. Kaufman. 2001. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107:893-903. [DOI] [PubMed] [Google Scholar]
- 36.Shen, Y., L. Meunier, and L. M. Hendershot. 2002. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 277:15947-15956. [DOI] [PubMed] [Google Scholar]
- 37.Siggia, E. D., J. Lippincott-Schwartz, and S. Bekiranov. 2000. Diffusion in inhomogeneous media: theory and simulations applied to whole cell photobleach recovery. Biophys. J. 79:1761-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sitia, R., and I. Braakman. 2003. Quality control in the endoplasmic reticulum protein factory. Nature 426:891-894. [DOI] [PubMed] [Google Scholar]
- 39.Sommer, T., and E. Jarosch. 2002. BiP binding keeps ATF6 at bay. Dev. Cell 3:1-2. [DOI] [PubMed] [Google Scholar]
- 40.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 41.Vanhove, M., Y. K. Usherwood, and L. M. Hendershot. 2001. Unassembled Ig heavy chains do not cycle from BiP in vivo but require light chains to trigger their release. Immunity 15:105-114. [DOI] [PubMed] [Google Scholar]
- 42.Wang, Y., J. Shen, N. Arenzana, W. Tirasophon, R. J. Kaufman, and R. Prywes. 2000. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275:27013-27020. [DOI] [PubMed] [Google Scholar]
- 43.Wei, J., J. R. Gaut, and L. M. Hendershot. 1995. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J. Biol. Chem. 270:26677-26682. [DOI] [PubMed] [Google Scholar]
- 44.Yang, T., P. J. Espenshade, M. E. Wright, D. Yabe, Y. Gong, R. Aebersold, J. L. Goldstein, and M. S. Brown. 2002. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110:489-500. [DOI] [PubMed] [Google Scholar]
- 45.Ye, J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355-1364. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881-891. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20:6755-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young, J. C., N. J. Hoogenraad, and F. U. Hartl. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112:41-50. [DOI] [PubMed] [Google Scholar]
- 49.Yu, M., R. H. Haslam, and D. B. Haslam. 2000. HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J. Biol. Chem. 275:24984-24992. [DOI] [PubMed] [Google Scholar]
- 50.Zhu, X., X. Zhao, W. F. Burkholder, A. Gragerov, C. M. Ogata, M. E. Gottesman, and W. A. Hendrickson. 1996. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272:1606-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou, J., Y. Guo, T. Guettouche, D. F. Smith, and R. Voellmy. 1998. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94:471-480. [DOI] [PubMed] [Google Scholar]