Abstract
The heterodimeric transcription factor PEBP2/CBF is composed of a DNA-binding subunit, called Runx1, and a non-DNA-binding subunit, called PEBP2β/CBFβ. The Runx1 protein is detected exclusively in the nuclei of most cells and tissues, whereas PEBP2β is located in the cytoplasm. We addressed the mechanism by which PEBP2β localizes to the cytoplasm and found that it is associated with filamin A, an actin-binding protein. Filamin A retains PEBP2β in the cytoplasm, thereby hindering its engagement as a Runx1 partner. The interaction with filamin A is mediated by a region within PEBP2β that includes amino acid residues 68 to 93. The deletion of this region or the repression of filamin A enables PEBP2β to translocate to the nucleus. Based on these observations, we propose that PEBP2β has two distinct domains, a newly defined regulatory domain that interacts with filamin A and the previously identified Runx1-binding domain.
The heterodimeric transcription factor PEBP2/CBF is composed of the DNA-binding protein Runx1/AML1 and the non-DNA-binding protein PEBP2β/CBFβ. The Runt domain, which is conserved among Runx family proteins, is responsible not only for the DNA-binding activity of Runx1, but also for its ability to dimerize with PEBP2β. Nuclear magnetic resonance and X-ray diffraction studies have allowed the determination of the three-dimensional structure of the Runt domain of Runx1 as well as the three-dimensional structures of PEBP2β and the heterodimer formed by both subunits (4, 9, 12, 21, 30, 36). These analyses showed that when it dimerizes with PEBP2β, the stabilized Runx1 protein can bind both the major and minor grooves of DNA (30). PEBP2β alone does not interact with DNA but enhances the DNA-binding activity of Runx1. Furthermore, Runx1 and PEBP2β homozygous knockout mice exhibit identical phenotypes, with a failure of hematopoietic stem cell development during embryogenesis. This finding provides genetic evidence that dimer formation between Runx1 and PEBP2β is vitally important for transcription factor activity (22, 24, 25, 28, 34, 35).
In humans, both Runx1 and PEBP2β are frequently targeted in leukemia-associated chromosomal abnormalities such as the t(8; 21) and inv translocations (16), which generate chimeric transcription factors that interfere with or abolish the transcriptional activity of endogenous PEBP2/CBF. For example, the inv (16)-derived PEBP2β-SMMHC protein consists of an amino-terminal fusion of the PEBP2β heterodimerization domain to the carboxy-terminal coiled-coil region of the smooth muscle myosin heavy chain.
In addition, while PEBP2/CBF was originally characterized as a transcriptional activator, recent studies have demonstrated that it can also function as a repressor, depending on the enhancer or promoter sequences it binds to and on the cofactors it interacts with. An interaction with p300/CBP or mSin3A converts Runx1 into an activator or a repressor, respectively (16, 19). Other factors such as YAP, Ear-2, ALY, Ets-1, MOZ, and Groucho/TLE also interact with Runx1 and modulate its activity (2, 5, 10, 13, 15, 17, 18, 37, 38). On the other hand, no such cofactors or modulators have been reported for PEBP2β.
Although the structure and functions of the PEBP2/CBF transcription factor have been extensively studied, little is known about how its activity is influenced by the subcellular localization of its constituent subunits. The Runx1 protein possesses nuclear localization signals and is found exclusively in the nucleus, whereas PEBP2β is located in the cytoplasm in most cells and tissues examined thus far (14, 32). The ability of Runx1 to bring PEBP2β into the nucleus has been demonstrated (1, 31). On the other hand, the mechanism that localizes PEBP2β to the cytoplasm is not known. We previously reported that cytoplasmic PEBP2β has a weak affinity for a cytoskeletal structure, namely, F-actin on stress fibers (32). We also observed that PEBP2β is located on or near the Z-line of muscle fibers, where many actin-associated proteins are abundant (7). Moreover, we found that the leukemogenic chimeric protein PEBP2β-SMMHC disorganizes cytoplasmic stress fibers and that the PEBP2β portion of this protein is necessary for interference (33). Based on these observations, we proposed that PEBP2β interacts with actin-associated proteins and that this interaction determines the cytoplasmic localization of PEBP2β (32, 33).
In the present study, we show that filamin A binds PEBP2β and retains it in the cytoplasm, thereby preventing it from acting as a partner for the Runx1 transcription factor. When filamin A is absent, PEBP2β moves into the nucleus and enhances Runx1-dependent transcription.
MATERIALS AND METHODS
Yeast two-hybrid screening.
The Matchmaker Two-Hybrid System 3 (Clontech) was used according to the instructions in the manufacturer's manual. A bait plasmid was constructed by inserting the mouse PEBP2β cDNA next to the GAL4 DNA-binding domain of the vector pGBKT7. cDNA libraries prepared from 11- or 17-day-old mouse embryos were fused to the GAL4 DNA activation domain of the vector pGAD10 and used as prey plasmids. AH109 cells were used as host cells. Plasmid DNAs were recovered from positive colonies and sequenced by use of an ABI PRISM 310 genetic analyzer (Applied Biosystems).
Plasmid construction.
A c-Myc or hemagglutinin (HA) tag was fused to the amino or carboxy terminus of filamin A-C, PEBP2β, or Runx1, as indicated in the text, by a PCR-based method. Carboxy-terminal deletion mutants of PEBP2β were constructed by PCRs using the common sense primer 5′-CGGAATTCACCATGCCGCGCGTCGTCCCGG-3′ and the following antisense primers: 5′-GGAATTCCTACTGGAGAGACAGATTGGTTC-3′ for βΔC67, 5′-GGAATTCCTACTTGCCTGCTTCTCTCTC-3′ for βΔC94, and 5′-GGAATTCCTACTGGGCTCGCTCCTCATC-3′ for βΔC133.
For preparation of an internal deletion mutant, βΔ68-93, the following primers were used in appropriate combinations for two successive rounds of PCR: 5′-AAGGTATACTTGAAGGCTCCCATG-3′, 5′-CCAATCTGTCTCTCCAGAAGGTATACT-3′, and 5′-CAAGAAGACAGCAAGACCCTAGGAATTCCG-3′. All cDNAs were subcloned into the mammalian expression vector pCAGGS-neo.
Cell culture.
HeLa cells were cultured in Dulbecco's modified Eagle's medium (Life Technologies, Inc.) supplemented with 10% (vol/vol) fetal bovine serum. M2 and A7 cells were cultured in minimal essential medium (Life Technologies, Inc.) supplemented with 8% (vol/vol) newborn calf serum and 2% (vol/vol) fetal bovine serum (8). G418 (Sigma) was added to A7 cells at 0.3 mg/ml.
siRNA-mediated repression of filamin A.
Five RNA oligonucleotides (iGENE) were synthesized by use of the following sense and antisense oligonucleotides: for filaminA-1499, 5′-CAGCUGACUUCAAGGUGUACACAAA-3′ and 5′-UUUGUGUACACCUUGAAGUCAGCUG-3′; for filaminA-4566, 5′-AGUACUGUAUGGAGAUGAAGAGGUA-3′ and 5′-UACCUCUUCAUCUCCAUACAGUACU-3′; for filaminA-5792, 5′-ACUACAGCAUUCUAGUCAAGUACAA-3′ and 5′-UUGUACUUGACUAGAAUGCUGUAGU-3′; for filaminA-6911, 5′-ACUACGAAGUCUCAGUCAAGUUCAA-3′ and 5′-UUGAACUUGACUGAGACUUCGUAGU-3′; for filaminA-7140, 5′-CACAGAAAUUGACCAAGAUAAGUAU-3′ and 5′-AUACUUAUCUUGGUCAAUUUCUGUG-3′; and for the luciferase control, 5′-ACAUCACGUACGCGGAAUACUUCGA-3′ and 5′-UCGAAGUAUUCCGCGUACGUGAUGU-3′. Small interfering RNAs (siRNAs) were introduced into HeLa cells by the use of Lipofectamine 2000 (Life Technologies, Inc.). The cells were processed for immunological detection 72 h after transfection.
Immunoprecipitation and immunoblot analysis.
Expression vectors harboring Myc-tagged filamin A-C and HA-tagged PEBP2β were cotransfected into HeLa cells by use of the Effectene reagent (Qiagen). Twenty-four hours after transfection, the cells were lysed with a buffer consisting of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% (vol/vol) Nonidet P-40, and a mixture of protease inhibitors (Complete; Roche Molecular Biochemicals). If the lysate was to be incubated with an anti-human filamin A monoclonal antibody (Chemicon), the cells were lysed with a buffer consisting of 25 mM HEPES (pH 7.5), 150 mM NaCl, 5 mM EDTA, 1% (vol/vol) Triton X-100, 10% (vol/vol) glycerol, 2 mM dithiothreitol, and a mixture of protease inhibitors. The cell lysates were incubated with the anti-c-Myc 9E10 antibody (Sigma), the anti-HA 3F10 antibody (Roche Molecular Biochemicals), or an anti-filamin A antibody, and the immunoprecipitates were adsorbed to protein G-Sepharose beads. The beads were washed five times with a buffer consisting of 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 1 mM EDTA, 1% (vol/vol) Nonidet P-40, and a mixture of protease inhibitors or, in the case of anti-filamin A precipitates, with a buffer consisting of 25 mM HEPES (pH 7.5), 150 mM NaCl, 5 mM EDTA, 1% (vol/vol) Triton X-100, 10% (vol/vol) glycerol, 2 mM dithiothreitol, and a mixture of protease inhibitors. Proteins were eluted by boiling the beads in sodium dodecyl sulfate (SDS) sample buffer, electrophoresed in an SDS-polyacrylamide gel, and transferred to a polyvinylidene difluoride membrane. Immunodetection of membrane-bound proteins was performed with the anti-HA 3F10, anti-c-Myc 9E10, or anti-PEBP2β antibody (6), and products were visualized by use of the ECL Plus reagent (Amersham Pharmacia Biotech).
Cells subjected to siRNA analysis were lysed in a urea-Triton buffer, and immunoblot analyses were performed as previously described (7). The antibodies used were an anti-human filamin A antibody, a murine anti-β-actin monoclonal antibody (Sigma), and a peroxidase-conjugated goat antibody to mouse immunoglobulin G (IgG) (Cappel Products).
Immunofluorescence analysis.
Cells grown on coverslips were transfected with expression plasmids. Twenty-four hours later, the cells were fixed with 2% (wt/vol) paraformaldehyde, permeabilized with 0.1% (vol/vol) Triton X-100, and blocked with 1% (wt/vol) bovine serum albumin in phosphate-buffered saline. Signals were detected by an indirect immunofluorescence technique using anti-HA 3F10 as a primary antibody and Cy3-conjugated goat anti-rat IgG (Chemicon) as a secondary antibody or using anti-c-Myc 9E10 or an anti-human filamin A antibody as a primary antibody and Alexa fluor 488-goat anti-mouse IgG (Molecular Probes) as a secondary antibody. Cells were viewed with a confocal laser scanning microscope (LSM410 or LSM 5 PASCAL; Zeiss).
Reporter gene assay.
Runx1 and PEBP2β expression plasmids were cotransfected with the reporter plasmid pM-CSF-R-luc (39) by use of the Effectene reagent. pRSV-β-GAL was transfected together with the reporter plasmid as an internal control, and its activity was used to normalize transfection efficiencies. Twenty-four hours after transfection, the cells were lysed in a lysis buffer (Promega), and the luciferase activities in lysates were measured by use of a luciferase assay system (Promega) and a Luminescencer-JNR AB-2100 instrument (Bio-Instrument). β-Galactosidase activity was assayed by the use of chlorophenol red-β-d-galactopyranoside (Roche Molecular Biochemicals) as a substrate. Each assay was performed at least three times. In each case, a siRNA for filamin A or a filaminA cDNA was introduced into HeLa or M2 cells, respectively. The cells were incubated for 24 h and then transfected with the expression plasmids for Runx1 and PEBP2β together with the pM-CSF-R-luc reporter and the pRL-TK vector (Promega) as an internal control.
RESULTS
Identification of filamin A as a novel PEBP2β-interacting protein.
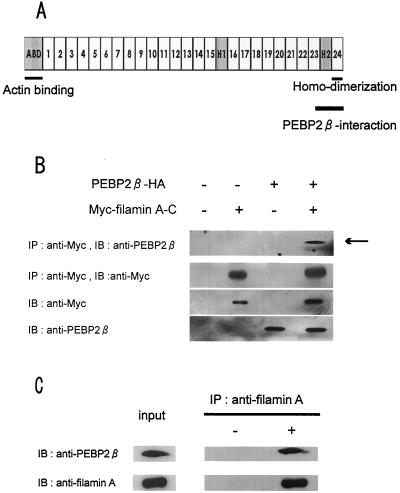
To identify molecules that interact with PEBP2β, we employed a yeast two-hybrid screening system. cDNAs derived from murine embryos were used as preys, and PEBP2β was used as a bait. We screened nine million yeast transformants by an interaction trap strategy and obtained 110 colonies that were positive for growth on both histidine and adenine. Many of the cDNAs thus isolated corresponded to Runx genes. In addition, one clone that carried a portion of the cytoskeletal gene filaminA (also known as ABP-280) was identified. The region of filaminA that was recovered as a PEBP2β-interacting sequence consisted of the extreme carboxy terminus and the adjacent hinge 2 domain (Fig. 1A). This region is hereafter referred to as filamin A-C.
FIG. 1.
Interaction of PEBP2β with filamin A. (A) Schematic diagram of structure of filamin A. The actin binding and homodimerization domains are located at the extreme amino- and carboxy-terminal ends, respectively. The repetitive domains are numbered. H represents a hinge region. A fragment of filamin A obtained in the yeast two-hybrid screening assay contained the PEBP2β-interacting region and is termed filamin A-C. (B) HeLa cells were transfected with the indicated combinations of expression plasmids, and cell lysates were immunoprecipitated (IP) with an anti-Myc antibody. Precipitates were resolved by SDS-polyacrylamide gel electrophoresis and subjected to immunoblot (IB) analysis with the indicated antibodies. An aliquot of lysate was directly immunoblotted without immunoprecipitation to evaluate the level of protein expression. (C) Lysates of untransfected HeLa cells were immunoprecipitated (IP) with or without an anti-filamin A antibody. Precipitates were resolved by SDS-polyacrylamide gel electrophoresis and subjected to immunoblot (IB) analysis with the indicated antibodies. An aliquot of lysate was directly immunoblotted without immunoprecipitation to evaluate the level of protein expression (input).
We next examined whether the PEBP2β protein interacts with filamin A in mammalian cells by transiently expressing Myc-tagged filamin A-C and HA-tagged PEBP2β in HeLa cells. As shown in Fig. 1B, PEBP2β was coimmunoprecipitated with filamin A-C from HeLa cell lysates when the two proteins were introduced by cotransfection. Similarly, filamin A-C was coimmunoprecipitated with PEBP2β (data not shown). Since HeLa cells express both the PEBP2β and filamin A proteins endogenously, their possible interaction was also examined. As shown in Fig. 1C, PEBP2β was coimmunoprecipitated with the intact filamin A molecule. These data indicate that PEBP2β forms a complex with filamin A in HeLa cells and that this is true not only for the exogenously introduced proteins, but also for those expressed endogenously.
The central region of PEBP2β is necessary for interaction with filamin A-C and for cytoplasmic localization.
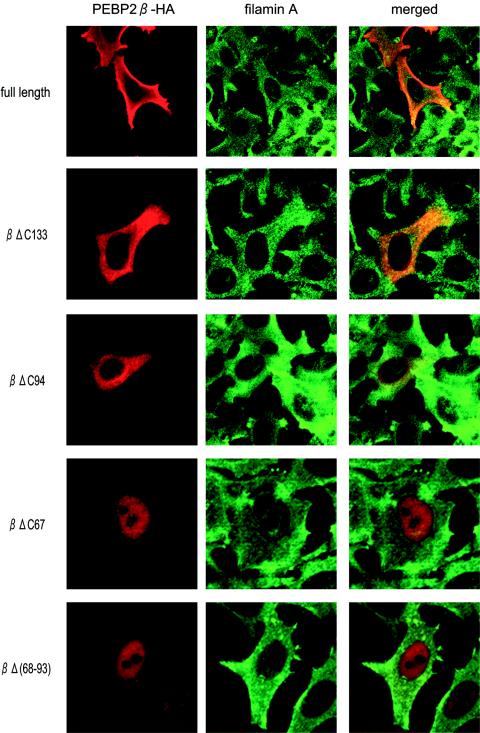
To determine which region of PEBP2β is responsible for its interaction with filamin A, we generated a series of carboxy-terminal deletion mutants. Figure 2A illustrates the relationship between each deletion derivative and the known substructures of PEBP2β. The 141-amino-acid (aa) amino-terminal region of PEBP2β is composed of four α helices, seven β strands, and six loop structures. Since three isoforms of the PEBP2β protein share the 133 amino-terminal amino acids, we constructed a derivative lacking the carboxy-terminal region (βΔC133) by truncating the protein at residue 134. The βΔC94 derivative lacked the β5-L5 region, one of the Runt-interacting domains. The βΔC67 construct possessed only the 67 amino-terminal amino acids, and the βΔ68-93 construct retained all of the Runt-interacting domains (β1-L1, L2-β3, α1, β3, and β5-L5) but lacked the L3-β4-L4 region.
FIG. 2.
Mapping of the filamin A-interacting domain of PEBP2β by yeast two-hybrid analysis. (A) Schematic diagram of domain structures found in the PEBP2β protein and description of carboxy-terminal deletion mutants of PEBP2β. α, β, and L represent α helixes, β strands, and loops, respectively. The thick lines at the top indicate areas I and II of the Runt-interacting region (30). (B) PEBP2β and its deletion mutants were tested for the ability to interact with filamin A-C. Yeast cells were transformed with the indicated combinations of plasmids and grown on nonselective (−Trp, −Leu) and selective (−Trp, −Leu, −His, 1 mM 3-amino-1,2,4-triazole) media. Lamin/large T antigen and Runx1/PEBP2β served as negative and positive controls, respectively.
By using a yeast two-hybrid system, we found that the βΔC133 and βΔC94 derivatives, as well as full-length PEBP2β, interacted with filamin A-C (Fig. 2B). We could not, however, assess the binding ability of the βΔC67 construct, since cells containing this construct alone were able to grow on selection medium. Notably, the internal deletion construct, βΔ68-93, did not interact with filamin A-C. Thus, the region of aa 68 to 93 of PEBP2β is necessary for its interaction with filamin A.
We next examined the effect of carboxy-terminal deletions on the subcellular localization of PEBP2β. Each plasmid was transfected into HeLa cells, and the expression of transduced proteins as well as of endogenous filamin A was monitored by double immunofluorescence staining (Fig. 3). While full-length PEBP2β and the βΔC133 and βΔC94 proteins were all located in the cytoplasm, the βΔC67 and βΔ68-93 proteins were both found in the nucleus. Thus, the region of aa 68 to 93 is necessary for the cytoplasmic localization of PEBP2β, since constructs that lack this region move into the nucleus.
FIG. 3.
Subcellular localization of PEBP2β protein derivatives. Each PEBP2β deletion mutant and full-length PEBP2β were transfected into HeLa cells, and cells were processed for double immunofluorescence staining. Red and green fluorescence represent PEBP2β and filamin A, respectively. Merged images are also presented.
PEBP2β is located in the nuclei of filamin A-deficient cells.
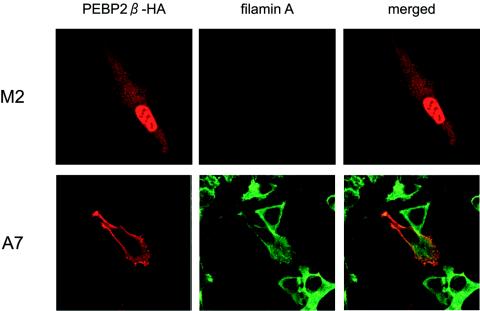
We reasoned that filamin A functions to retain PEBP2β in the cytoplasm. Therefore, we predicted that in its absence, full-length PEBP2β would not stay in the cytoplasm but would instead move into the nucleus. To test this idea, we performed experiments with a filamin A-deficient human melanoma cell line, M2, and a subline, A7, which was obtained by stably transfecting M2 cells with a human filamin A cDNA (8). A plasmid bearing full-length PEBP2β was transfected into these cells, and the expression of transduced protein as well as of endogenous filamin A was monitored by immunofluorescence (Fig. 4). PEBP2β was detected in the nuclei of filamin A-deficient M2 cells but in the cytoplasm of filamin A-expressing A7 cells. We also examined the effects of carboxy-terminal deletions on the subcellular localization of PEBP2β in M2 and A7 cells. In filamin A-expressing A7 cells, while the full-length PEBP2β, βΔC133, and βΔC94 proteins were all located in the cytoplasm, the βΔC67 and βΔ68-93 proteins were both found in the nucleus. On the other hand, in filamin A-deficient M2 cells, the full-length PEBP2β, as well as all carboxy-terminal deletion mutants, was located in the nucleus (data not shown).
FIG. 4.
Subcellular localization of PEBP2β protein in filamin A-deficient M2 cells. M2 and A7 cells were transfected with full-length PEBP2β, fixed, and processed for double immunofluorescence staining as described in the legend to Fig. 3.
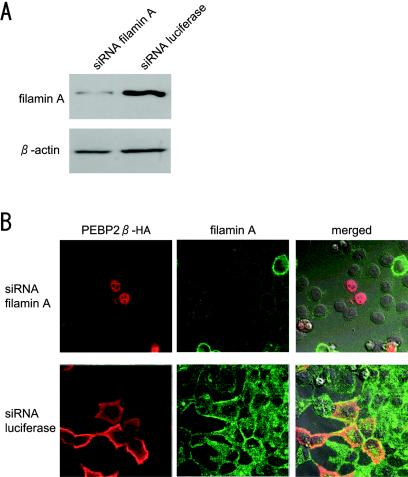
To confirm these results, we next determined whether the repression of filamin A by the siRNA method influences the subcellular localization of PEBP2β in HeLa cells. Since the same results were obtained for five different siRNAs synthesized as described in Materials and Methods, we present here representative results obtained with the siRNA filaminA-7140. A luciferase siRNA was used as a negative control. We found by immunoblot analysis (Fig. 5A) that the level of filamin A in cells treated with the filaminA-7140 siRNA was 1/10 that in luciferase siRNA-treated cells. We determined the localization of PEBP2β in these cells by immunofluorescence staining (Fig. 5B) and found the protein in the nuclei of filamin A-repressed cells but in the cytoplasm of control filamin A-expressing cells.
FIG. 5.
Effect of filamin A repression on subcellular localization of PEBP2β protein. (A) Repression of filamin A expression by the siRNA method. HeLa cells were transfected with a siRNA targeted to filamin A or luciferase. The protein level in these cells was measured by immunoblot analysis. The top panel represents filamin A, and the bottom panel shows β-actin. (B) Cells were treated with a siRNA as indicated, transfected with PEBP2β cDNA, fixed, and processed for double immunofluorescence staining as described in the legend to Fig. 3. Merged images of two-color fluorescence were captured by differential interference contrast microscopy.
These data indicate that filamin A sequesters the PEBP2β protein in the cytoplasm and that in its absence, PEBP2β moves into the nucleus.
Runx1 colocalizes with PEBP2β in the nuclei of filamin A-deficient cells.
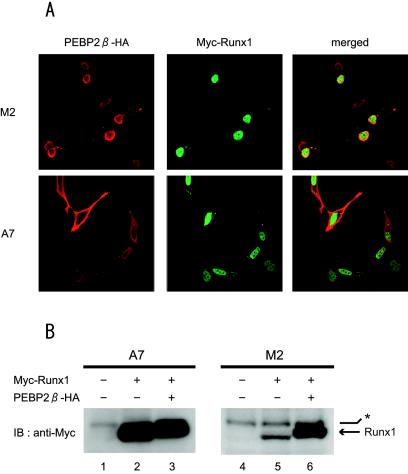
We next examined whether the reported activity of Runx1 to bring PEBP2β into the nucleus is affected by the presence of filamin A. Plasmids expressing PEBP2β and Runx1 were cotransfected into the M2 and A7 cell lines, and the transduced proteins were detected by double immunofluorescence (Fig. 6A). Runx1 colocalized with PEBP2β in the nuclei of filamin A-deficient M2 cells. In addition, PEBP2β was coimmunoprecipitated with Runx1 from cotransfected M2 cell lysates (data not shown). In contrast, although Runx1 was found in the nuclei of filamin A-expressing A7 cells, PEBP2β was found in the cytoplasm. These data suggest that the presence of filamin A prevents Runx1 from translocating PEBP2β into the nucleus. On the other hand, filamin A did not have an effect on the subcellular localization of Runx1, which was localized to the nuclei of both M2 and A7 cells.
FIG. 6.
Subcellular distribution of PEBP2β and Runx1 proteins in filamin A-deficient M2 cells and filamin A-expressing A7 cells. (A) M2 and A7 cells were cotransfected with Runx1 and PEBP2β expression plasmids, fixed, and processed for double immunofluorescence. Red and green fluorescence represent PEBP2β and Runx1, respectively. Merged images of two-color fluorescence are also presented. (B) A7 and M2 cells were transfected with the indicated combinations of Runx1 and PEBP2β expression plasmids. The protein level of Runx1 in these cells was measured by immunoblot (IB) analysis with an anti-Myc antibody. The bands indicated by the arrow represent Runx1, whereas those indicated by the asterisk represent nonspecific reactions.
PEBP2β has been reported to be capable of stabilizing the Runx1 protein, which is rather unstable by itself, by forming a heterodimer (11). We examined the protein level of Runx1 in the presence or absence of PEBP2β by using A7 and M2 cells. As shown in Fig. 6B, the band intensities of Runx1 detected by immunoblotting were roughly equal in both PEBP2β-expressing and non-PEBP2β-expressing A7 cells (compare lanes 2 and 3). In contrast, the cotransfection of PEBP2β significantly increased the band intensity of Runx1 in M2 cells (compare lanes 5 and 6). Therefore, in the nuclei of filamin A-deficient M2 cells, where the two proteins are colocalized, PEBP2β appears to increase the stability of Runx1.
The transcriptional activity of PEBP2/CBF is enhanced in filamin A-deficient cells.
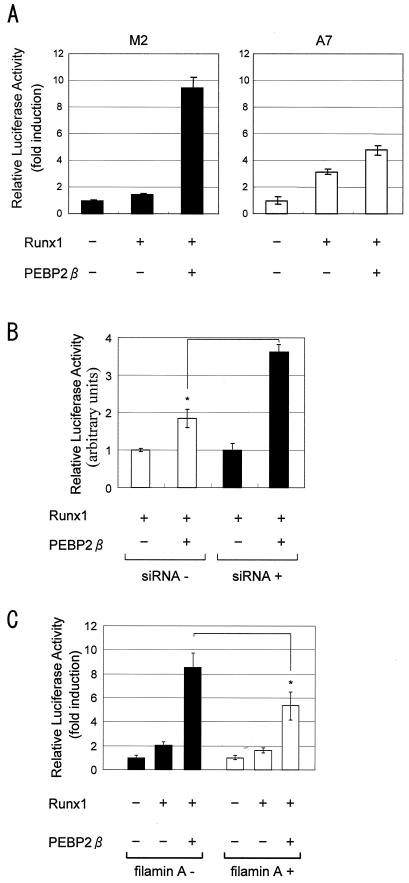
Filamin A may affect the extent of PEBP2/CBF transcriptional activity by controlling the subcellular localization of PEBP2β. In order to assess this idea, we transfected M2 and A7 cells with an M-CSF-R-luc reporter, which allowed the contribution of PEBP2β to the transcriptional activity of the PEBP2/CBF heterodimer to be measured (39). As seen in Fig. 7A, the transfection of PEBP2β in addition to Runx1 caused a sixfold increase in luciferase activity in M2 cells compared to that in cells transfected solely with Runx1. On the other hand, in A7 cells, the cotransfection of PEBP2β and Runx1 caused a 1.6-fold increase in luciferase activity compared to cells transfected only with Runx1.
FIG. 7.
PEBP2/CBF transcription activity in filamin A-deficient M2 cells and filamin A-expressing A7 cells. (A) M2 and A7 cells were cotransfected with an M-CSF-R-luc reporter construct and Runx1 and/or PEBP2β expression plasmids as indicated. Luciferase activities in cell lysates are presented as averages ± standard deviations. (B) HeLa cells pretreated with a filamin A siRNA and untreated HeLa cells were cotransfected with an M-CSF-R-luc reporter construct and Runx1 and/or PEBP2β expression plasmids as indicated. (C) M2 cells which were pretransfected with filaminA cDNA were cotransfected with an M-CSF-R-luc reporter construct and Runx1 and/or PEBP2β expression plasmids as indicated.
We also performed several control experiments in parallel. For the data shown in Fig. 7B, HeLa cells were treated or not treated with the siRNA filaminA-7140 and then were transfected with reporter and expression plasmids. The siRNA treatment increased the luciferase activity from 1.6-fold (without siRNA) to 3.6-fold (with siRNA) (the significance of the difference was valid by a t test [P < 0.002]). For the data shown in Fig. 7C, M2 cells were transfected with filaminA cDNA and then with reporter and expression plasmids. The coexpression of filamin A decreased the luciferase activity from 4.3-fold (without filamin A) to 3.4-fold (with filamin A) (the significance of the difference was valid by a t test [P < 0.03]).
Thus, the transcriptional activity of PEBP2/CBF is enhanced by an increase in the level of PEBP2β, but only in the absence of filamin A. Conversely, the presence of filamin A appears to decrease the transcriptional activity of PEBP2/CBF, probably by retaining PEBP2β in the cytoplasm. (Note that the βΔ68-93 protein did not induce transcriptional activation in a reporter assay [data not shown]. This was probably because its three-dimensional structure was unfavorably altered due to the deletion.)
DISCUSSION
Filamin A is a non-muscle-specific isoform of filamin that is ubiquitously expressed in many different cell types. As an actin binding protein, filamin A organizes a three-dimensional intracellular network of actin filaments and connects filamentous actin to plasma membrane glycoproteins. Furthermore, filamin A acts as a scaffold for intracellular proteins and is involved in various signal transduction pathways (29). In the present study, we demonstrated that filamin A binds to and retains PEBP2β in the cytoplasm, a finding which is in accord with our previous observations. For example, PEBP2β-specific staining has been observed along stress fibers and cell membrane processes in some cultured cells (32). This pattern of distribution of PEBP2β is similar to that of filamin A. Also, PEBP2β is located on or near the Z lines of muscle fibers (7). Filamin C, a muscle-specific isoform of the filamin family is present on Z lines (3) and therefore may specify the localization of PEBP2β to these structures. Indeed, we found that the cotransfected carboxy-terminal region of either filamin C or filamin B, another ubiquitously expressed isoform, was coimmunoprecipitated with PEBP2β. Moreover, the forced expression of full-length filamin B relocated PEBP2β from the nucleus to the cytoplasm of M2 cells (N. Yoshida and T. Watanabe, unpublished observations). It must be noted that since the endogenous expression of filamin B and filamin C in M2 cells was 1/10 that of filamin A or undetectable, respectively, their contribution, if any, to retaining PEBP2β in the cytoplasm may be minimal in this particular cell line.
By analyzing deletion mutants, we identified a region within the PEBP2β molecule that is important for its interaction with filamin A and for cytoplasmic localization. This region spans aa 68 through 93 and consists of loop-β strand-loop (L3-β4-L4) structures, but only 2 aa comprise the β4 strand. Therefore, this region as a whole does not appear to adopt a solid two-dimensional structure but is instead flexible. Furthermore, it contains a hydrophobic tryptophan residue (aa 73) embedded in a cluster of hydrophilic amino acid residues (30). These characteristics likely confer upon the region a tendency to interact with other molecules. In addition, an inspection of the three-dimensional structure of PEBP2β showed that the region responsible for interacting with filamin A and the Runx1-interacting region are situated on opposite sides of the molecule. The Runx1-interacting region is composed of an alpha helix and four β strands and probably adopts a rigid structure (30). Thus, based on the above observations, we propose a new model which holds that the PEBP2β protein consists of two structurally and functionally distinct domains. The first is a regulatory domain that has a loose structure and which perhaps interacts with various molecules. The binding of this domain by filamin A, for example, retains PEBP2β in the cytoplasm, thereby preventing it from being recruited as a component of a transcription factor complex. The second domain is an executive domain that has a rigid structure and which perhaps interacts only with the Runx1 protein. When it is bound to Runx1, PEBP2β can function as a transcription factor in the nucleus.
It is not clear how PEBP2β moves into the nucleus in the absence of filamin A. Previously, the Runx1 protein was thought to bring PEBP2β into the nucleus (1, 31). However, Runx1 protein expression was not detected in M2 cells by immunoblot analysis (Yoshida and Watanabe, unpublished observation), and PEBP2β was detected in the nuclei of M2 cells that were not transfected with Runx1. An unidentified mechanism appears to be involved in the nuclear localization of PEBP2β.
Runx1 and PEBP2β are known to be indispensable for the development of hematopoietic stem cells, and a precise dose of each of the Runx1 and PEBP2β proteins appears to be necessary for the proper functioning of PEBP2/CBF during this process (6, 20, 23). For example, a haploinsufficiency of Runx1 can impair the temporally and spatially regulated generation of hematopoietic stem cells in mouse embryos (6). Hematopoietic stem cells develop from hemangioblasts, a specific subset of endothelial cells. Notably, hemangioblasts, which undergo transformation from flat endothelial cells to round hematopoietic cells, are considered to accompany alterations of cytoskeletal structures, including the actin and perhaps filamin A molecules. One can imagine that the mechanism described in the present study may tune the activity of PEBP2/CBF at the site of hematopoietic stem cell generation. Efforts toward understanding this mechanism are under way.
Filamin A regulates the subcellular localization of Smad2 and of the androgen receptor (26, 27), two transcription factors that are usually found in the cytoplasm. The treatment of cells with transforming growth factor beta leads to the phosphorylation of Smad2 and to the subsequent translocation of the phosphorylated form into the nucleus. Interestingly, Smad2 is neither phosphorylated nor translocated into the nucleus after transforming growth factor beta stimulation in cells lacking filamin A. Similarly, the androgen receptor, which moves into the nucleus when bound to its ligand, remains in the cytoplasm of cells that do not express filamin A. Therefore, filamin A probably serves as a site at which a kinase and/or other ligand can also bind, and in the absence of filamin A, target molecules are not appropriately modified and thus are not translocated into the nucleus. On the other hand, PEBP2β is translocated into the nuclei of cells lacking filamin A, which enhances the transcriptional activity of PEBP2/CBF. A signal that can dissociate the interaction of PEBP2β and filamin A is not known at present. Thus, the mechanisms by which filamin A regulates the nuclear translocation and transcriptional activity of PEBP2β and of Smad2 and the androgen receptor appear to differ. Our present study has thus broadened the molecular scope of the interplay between cytoskeletal filamin A and transcription factors.
Acknowledgments
This research was supported in part by research grants from the Ministry of Education, Science, Sports, Culture and Technology of Japan. M.S. is a member of the 21st Century COE program, “Center for Innovative Therapeutic Development Towards the Conquest of Signal Transduction Diseases,” headed by K. Sugamura at Tohoku University.
We thank M. Shiina and K. Ogata for their valuable comments on the structural aspects of PEBP2β. We also thank D. Tenen for providing the p-M-CSF-R-luc reporter construct. We are grateful to M. Kuji for secretarial assistance.
REFERENCES
- 1.Adya, N., T. Stacy, N. A. Speck, and P. P. Liu. 1998. The leukemic protein core binding factor β (CBFβ)-smooth-muscle myosin heavy chain sequesters CBFα2 into cytoskeletal filaments and aggregates. Mol. Cell. Biol. 18:7432-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, M. Y., G. Huang, S. C. Bae, H. J. Wee, W. Y. Kim, and Y. Ito. 1998. Negative regulation of granulocytic differentiation in the myeloid precursor cell line 32Dcl3 by ear-2, a mammalian homolog of Drosophila seven-up, and a chimeric leukemogenic gene, AML1/ETO. Proc. Natl. Acad. Sci. USA 95:1812-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bechtel, P. J. 1979. Identification of a high molecular weight actin-binding protein in skeletal muscle. J. Biol. Chem. 254:1755-1758. [PubMed] [Google Scholar]
- 4.Berardi, M. J., C. Sun, M. Zehr, F. Abildgaard, J. Peng, H. A. Speck, and J. H. Bushweller. 1999. The Ig fold of the core binding factor α Runt domain is a member of a family of structurally and functionally related Ig-fold DNA-binding domains. Struct. Fold Des. 7:1247-1256. [DOI] [PubMed] [Google Scholar]
- 5.Bruhn, L., A. Munnerlyn, and R. Grosschedl. 1997. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCR α enhancer function. Genes Dev. 11:640-653. [DOI] [PubMed] [Google Scholar]
- 6.Cai, Z., M. de Bruijn, X. Ma, B. Dortland, T. Luteijn, R. J. Downing, and E. Dzierzak. 2000. Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity 13:423-431. [DOI] [PubMed] [Google Scholar]
- 7.Chiba, N., T. Watanabe, S. Nomura, Y. Tanaka, M. Minowa, M. Niki, R. Kanamaru, and M. Satake. 1997. Differentiation dependent expression and distinct subcellular localization of the protooncogene product, PEBP2β/CBFβ, in muscle development. Oncogene 14:2543-2552. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham, C. C., J. B. Gorlin, D. J. Kwiatkowski, J. H. Hartwig, P. A. Janmey, H. R. Byers, and T. P. Stossel. 1992. Actin-binding protein requirement for cortical stability and efficient locomotion. Science 255:325-327. [DOI] [PubMed] [Google Scholar]
- 9.Goger, M., V. Gupta, W. Y. Kim, K. Shigesada, Y. Ito, and M. H. Werner. 1999. Molecular insights into PEBP2/CBFβ-SMMHC associated acute leukemia revealed from the structure of PEBP2/CBFβ. Nat. Struct. Biol. 6:620-623. [DOI] [PubMed] [Google Scholar]
- 10.Gu, T. L., T. L. Goetz, B. J. Graves, and N. A. Speck. 2000. Auto-inhibition and partner proteins, core-binding factor β (CBFβ) and Ets-1, modulate DNA binding by CBFα2 (AML1). Mol. Cell. Biol. 20:91-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, G., K. Shigesada, K. Ito, H. J. Wee, T. Yokomizo, and Y. Ito. 2001. Dimerization with PEBP2β protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 20:723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, X., J. W. Peng, N. A. Speck, and J. H. Bushweller. 1999. Solution structure of core binding factor beta and map of the CBF α binding site. Nat. Struct. Biol. 6:624-627. [DOI] [PubMed] [Google Scholar]
- 13.Imai, Y., M. Kurokawa, K. Tanaka, A. D. Friedman, S. Ogawa, K. Mitani, Y. Yazaki, and H. Hirai. 1998. TLE, the human homolog of groucho, interacts with AML1 and acts as a repressor of AML1-induced transactivation. Biochem. Biophys. Res. Commun. 252:582-589. [DOI] [PubMed] [Google Scholar]
- 14.Kanno, T., Y. Kanno, L. F. Chen, E. Ogawa, W. Y. Kim, and Y. Ito. 1998. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol. Cell. Biol. 18:2444-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, W. Y., M. Sieweke, E. Ogawa, H. J. Wee, U. Englmeier, T. Graf, and Y. Ito. 1999. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 18:1609-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitabayashi, I., A. Yokoyama, K. Shimizu, and M. Ohki. 1998. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 17:2994-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitabayashi, I., Y. Aikawa, L. A. Nguyen, A. Yokoyama, and M. Ohki. 2001. Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein. EMBO J. 20:7184-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levanon, D., R. E. Goldstein, Y. Bernstein, H. Tang, D. Goldenberg, S. Stifani, Z. Paroush, and Y. Groner. 1998. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. USA 95:11590-11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutterbach, B., J. J. Westendorf, B. Linggi, S. Isaac, E. Seto, and S. W. Hiebert. 2000. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 275:651-656. [DOI] [PubMed] [Google Scholar]
- 20.Mukouyama, Y., N. Chiba, T. Hara, H. Okada, Y. Ito, R. Kanamaru, A. Miyajima, M. Satake, and T. Watanabe. 2000. The AML1 transcription factor functions to develop and maintain hematogenic precursor cells in the embryonic aorta-gonad-mesonephros region. Dev. Biol. 220:27-36. [DOI] [PubMed] [Google Scholar]
- 21.Nagata, T., V. Gupta, D. Sorce, W. Y. Kim, A. Sali, B. T. Chait, K. Shigesada, Y. Ito, and M. H. Werner. 1999. Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain. Nat. Struct. Biol. 6:615-619. [DOI] [PubMed] [Google Scholar]
- 22.Niki, M., H. Okada, H. Takano, J. Kuno, K. Tani, H. Hibino, S. Asano, Y. Ito, M. Satake, and T. Noda. 1997. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc. Natl. Acad. Sci. USA 94:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.North, T., T. L. Gu, T. Stacy, Q. Wang, L. Howard, M. Binder, M. Marin-Padilla, and N. A. Speck. 1999. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 126:2563-2575. [DOI] [PubMed] [Google Scholar]
- 24.Okada, H., T. Watanabe, M. Niki, H. Takano, N. Chiba, N. Yanai, K. Tani, H. Hibino, S. Asano, M. L. Mucenski, Y. Ito, T. Noda, and M. Satake. 1998. AML1(−/−) embryos do not express certain hematopoiesis-related gene transcripts including those of the PU.1 gene. Oncogene 17:2287-2293. [DOI] [PubMed] [Google Scholar]
- 25.Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld, and J. R. Downing. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321-330. [DOI] [PubMed] [Google Scholar]
- 26.Ozanne, D. M., M. E. Brady, S. Cook, L. Gaughan, D. E. Neal, and C. N. Robson. 2000. Androgen receptor nuclear translocation is facilitated by the F-actin cross-linking protein filamin. Mol. Endocrinol. 14:1618-1626. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki, A., Y. Masuda, Y. Ohta, K. Ikeda, and K. Watanabe. 2001. Filamin A associates with Smads and regulates transforming growth factor-β signaling. J. Biol. Chem. 276:17871-17877. [DOI] [PubMed] [Google Scholar]
- 28.Sasaki, K., H. Yagi, R. T. Bronson, T. K. Tominaga, T. Matsunashi, K. Deguchi, Y. Tani, T. Kishimoto, and T. Komori. 1996. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor β. Proc. Natl. Acad. Sci. USA 93:12359-12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stossel, T. P., J. Condeelis, L. Cooley, J. H. Hartwig, A. Noegel, M. Schleicher, and S. S. Shapiro. 2001. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2:138-145. [DOI] [PubMed] [Google Scholar]
- 30.Tahirov, T. H., T. Inoue-Bungo, H. Morii, A. Fujikawa, M. Sasaki, K. Kimura, M. Shiina, K. Sato, T. Kumasaka, M. Yamamoto, S. Ishii, and K. Ogata. 2001. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFβ. Cell 104:755-767. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka, K., T. Tanaka, M. Kurokawa, Y. Imai, S. Ogawa, K. Mitani, Y. Yazaki, and H. Hirai. 1998. The AML1/ETO(MTG8) and AML1/Evi-1 leukemia-associated chimeric oncoproteins accumulate PEBP2β (CBFβ) in the nucleus more efficiently than wild-type AML1. Blood 91:1688-1699. [PubMed] [Google Scholar]
- 32.Tanaka, Y., T. Watanabe, N. Chiba, M. Niki, Y. Kuroiwa, T. Nishihira, S. Satomi, Y. Ito, and M. Satake. 1997. The protooncogene product, PEBP2β/CBFβ, is mainly located in the cytoplasm and has an affinity with cytoskeletal structures. Oncogene 15:677-683. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka, Y., M. Fujii, K. Hayashi, N. Chiba, T. Akaishi, R. Shineha, T. Nishihira, S. Satomi, Y. Ito, T. Watanabe, and M. Satake. 1998. The chimeric protein, PEBP2β/CBFβ-SMMHC, disorganizes cytoplasmic stress fibers and inhibits transcriptional activation. Oncogene 17:699-708. [DOI] [PubMed] [Google Scholar]
- 34.Wang, Q., T. Stacy, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 93:3444-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, Q., T. Stacy, J. D. Miller, A. F. Lewis, T. L. Gu, X. Huang, J. H. Bushweller, J. C. Bories, F. W. Alt, G. Ryan, P. P. Liu, A. Wynshaw-Boris, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell 87:697-708. [DOI] [PubMed] [Google Scholar]
- 36.Warren, A. J., J. Bravo, R. L. Williams, and T. H. Rabbitts. 2000. Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFβ. EMBO J. 19:3004-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yagi, R., L. F. Chen, K. Shigesada, Y. Murakami, and Y. Ito. 1999. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 18:2551-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaidi, S. K., A. J. Sullivan, R. Medina, Y. Ito, Y. J. Van Wijnen, J. L. Stein, J. B. Lian and G. S. Stein. 2004. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 23:790-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, D. E., C. J. Hetherington, S. Meyers, K. L. Rhoades, C. J. Larson, H. M. Chen, S. W. Hiebert, and D. G. Tenen. 1996. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBF α2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol. Cell. Biol. 16:1231-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]