Abstract
Animal cells counteract oxidative stress and electrophilic attack through coordinated expression of a set of detoxifying and antioxidant enzyme genes mediated by transcription factor Nrf2. In unstressed cells, Nrf2 appears to be sequestered in the cytoplasm via association with an inhibitor protein, Keap1. Here, by using the yeast two-hybrid screen, human Keap1 has been identified as a partner of the nuclear protein prothymosin α. The in vivo and in vitro data indicated that the prothymosin α-Keap1 interaction is direct, highly specific, and functionally relevant. Furthermore, we showed that Keap1 is a nuclear-cytoplasmic shuttling protein equipped with a nuclear export signal that is important for its inhibitory action. Prothymosin α was able to liberate Nrf2 from the Nrf2-Keap1 inhibitory complex in vitro through competition with Nrf2 for binding to the same domain of Keap1. In vivo, the level of Nrf2-dependent transcription was correlated with the intracellular level of prothymosin α by using prothymosin α overproduction and mRNA interference approaches. Our data attribute to prothymosin α the role of intranuclear dissociator of the Nrf2-Keap1 complex, thus revealing a novel function for prothymosin α and adding a new dimension to the molecular mechanisms underlying expression of oxidative stress-protecting genes.
The defense against oxidative stress and electrophilic attack is mediated in animal cells by activation of a battery of genes encoding detoxification enzymes [such as glutathione S-transferase (GST) and NAD(P)H-quinone oxidoreductase-1 (NQO1)] and antioxidant proteins [such as γ-glutamylcysteine synthetase and heme oxygenase-1 (HO-1)] (for a review, see reference 15). Coordinated up-regulation of expression of these genes is attained through the presence of cis-acting antioxidant response elements (AREs), which are recognized by the Nrf2 transcription factor in their regulatory regions (35). Heterodimerization of Nrf2 with small Maf proteins may be required for efficient DNA binding and transcription activation (18, 29). Nrf2 has been demonstrated to regulate both the basal and induced expression of many ARE-responsive genes (27), as the response to electrophilic and reactive oxygen species-producing agents is profoundly impaired in Nrf2-deficient cells (17). Accordingly, nrf2-deficient mice displayed elevated sensitivity to chemical carcinogenesis and oxidative stress (31).
The ability of Nrf2 to direct transcription of the target genes is subject to regulation through an interaction with an ubiquitously expressed inhibitor protein, Keap1 (19). Because Keap1 displays similarity to a Drosophila Kelch protein, which is an actin-binding protein (44), Keap1 was proposed to bridge Nrf2 to the cytoskeleton in the cytoplasm of nonstressed cells, thus mediating its inhibitory action (8, 19, 22). Recently, Keap1 was also reported to target Nrf2 for cytoplasmic ubiquitination and degradation by the proteosome (28, 45). Induction of oxidative stress and treatment of cells with chemopreventive agents enable Nrf2 to escape Keap1-dependent cytoplasmic sequestration and degradation, leading to stabilization of Nrf2, increased nuclear accumulation of Nrf2, and activation of Nrf2-dependent cytoprotective genes (19, 28, 45). The importance of Keap1-mediated regulation of Nrf2 activity is emphasized by the observations that in cells lacking Keap1, Nrf2 is constitutively accumulated in the nucleus (20) and that mice with constitutively activated Nrf2 due to the absence of Keap1 died postnatally, with the phenotype being reversed in keap1 nrf2 double mutants (40).
Attempts to identify how the stress signals are transduced to the target genes point to the constituents of the Nrf2-Keap1 complex as well. In particular, Nrf2 has been demonstrated to be a protein kinase C and a PERK substrate, and in both cases Nrf2 phosphorylation led to destabilization of the Nrf2-Keap1 complex in stress-induced cells and promoted Nrf2 nuclear accumulation and transcriptional activity (3, 7, 16). On the other hand, Keap1 was also reported to be a sensor of oxidative and electrophilic stress due to the presence of several reactive Cys residues. Keap1 Cys273 and Cys288 mutants were impaired in their ability to repress Nrf2-dependent transcriptional activation under basal conditions and in Keap1-dependent ubiquitination and degradation of Nrf2 (24, 41, 45). In vitro, exposure to electrophiles disrupted the interaction of Keap1 with the Neh2 domain of Nrf2 (9).
Thus, the cytoplasmic Nrf2-Keap1 complex emerged as the critical regulator of ARE-dependent transcription. Here, we report the identification of a novel Keap1 partner which, quite unexpectedly, turned out to be the nuclear protein prothymosin α (ProTα). We demonstrate that ProTα releases Nrf2 from the Nrf2-Keap1 complex in vitro and contributes to Nrf2-dependent gene expression in vivo. To lend mechanistic support for the involvement of ProTα in the regulation of ARE-dependent transcription, we provide evidence that Keap1 is a nuclear-cytoplasmic shuttling protein equipped with a functional nuclear export signal (NES), which apparently confers nuclear-cytoplasmic shuttling to the Nrf2-Keap1 complex.
ProTα is a ubiquitously and abundantly expressed small nuclear protein (6, 11, 25) that is involved in proliferation of mammalian cells (14) and in their protection against apoptosis (13, 21). Overexpression of ProTα in NIH 3T3 and HL-60 cells was shown to accelerate proliferation (32, 43), whereas inhibition of ProTα synthesis prevented cell division (36) and induced apoptosis in HL-60 cells (33). Overexpression of ProTα in a rat fibroblast cell line resulted in loss of contact inhibition, anchorage-independent growth, and decreased serum dependence (30). Consistent with its properties, ProTα is particularly abundant in tumor cells (10, 38). The antiapoptotic activity of ProTα likely arises from its ability to inhibit apoptosome formation (21), whereas the mechanism of ProTα action in stimulating proliferation is not clearly established. Mounting evidence suggests ProTα involvement in transcription regulation (23, 26, 39). The newly identified ProTα-Keap1 interaction may have important implications for the mechanism(s) of Nrf2-dependent gene expression.
MATERIALS AND METHODS
Yeast two-hybrid screen.
Manipulations with yeast cells were performed essentially as described previously (Yeast protocols handbook, Clontech, Palo Alto, Calif., 2000). Saccharomyces cerevisiae strain L40ccU containing pBTM117c-ProTα was transformed with GAL4 cDNA human brain (HB) and human bone marrow (HBM) libraries (Matchmaker; Clontech). Totals of 6 × 106 and 4 × 106 independent transformants, respectively, were plated on a minimal medium lacking tryptophan, leucine, histidine, and uracil. After incubation at 30°C for 4 to 8 days, a total of 47 colonies formed were picked to SD plates lacking Trp and Leu. After incubation at 30°C for 2 days, β-galactosidase (β-Gal) activity was tested. β-Gal-positive clones were picked to SD plates lacking Leu and containing canavanine to eliminate the bait, and β-Gal activity was retested after incubation at 30°C for 2 days. Total DNA was prepared from β-Gal-negative clones and transformed into Escherichia coli JM109. To check for true positives, isolated plasmids were transformed into L40ccU harboring pBTM117c, pBTM117c-ProTα, pBTM117c-Sim1, or pBTM117c-Rev and tested for selective growth on SD plates lacking Trp, Leu, His, and Ura and for β-Gal activity as described above. The inserts of library plasmids of positive clones, pACT2-HBM and pACT2-HB, were sequenced.
Human cells and fluorescence microscopy.
HeLa and HepG2 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (ICN). Transfections were done with Lipofectamine 2000 (Invitrogen), using 2 to 6 μg of plasmid DNA, according to the manufacturer's protocol. At 24 h after transfection, cells were fixed and processed for microscopy as described previously (13). When indicated, cells were treated with 2 nM leptomycin B (LMB) (Sigma) for 4 h prior to fixation. Protein fusions with enhanced green fluorescent protein (EGFP) were intracellularly localized as described previously (12). For immunolocalization, anti-Keap1 2H5 and 2E4 monoclonal antibodies and rabbit anti-Nrf2 polyclonal antibodies (C-20; Santa Cruz Biotechnology) at a dilution of 1:300 were used as primary antibodies. For Keap1 competition assay, the 2H5 antibody (0.64 μg) was preincubated with 12.8 μg of recombinant GST-Keap1(308-624) expressed in and purified from E. coli or with 6.4 μg of GST at 4°C for 1 h prior to its addition to fixed cells. Fluorescein isothiocyanate-conjugated goat anti-mouse antibodies and rhodamine-conjugated goat anti-rabbit antibodies (Santa Cruz Biotechnology) were used as secondary antibodies at a dilution of 1:400. Images were acquired through an Axiovert 200 M fluorescence microscope equipped with an LSM510 confocal laser scanning module or with an AxioCam digital camera (Carl Zeiss).
Plasmids.
pBTM117c-ProTα and its derivatives encoding wild-type human ProTα and various deletion mutants thereof fused to LexA were constructed by in-frame ligation of the ProTα-encoding DNA fragments derived from pHT15A (12) downstream from the LexA-encoding sequence in pBTM117c (42). pHT15A(44,50) was obtained by replacing the 110-bp StyI-AccI DNA fragment of pHT15A with that containing the E44G E50G double mutation (34). The MProTα(44,50)-encoding sequence was then used to replace the wild-type ProTα-encoding sequence in pHP12A (37), pKTL1 (5), pcDNA4-enh-wt ProTα (13), and pBTM117c-ProTα.
To obtain the complete ORF of human Keap1, the missing N-terminal Keap1-encoding DNA fragment was amplified by PCR from a SW480 cDNA library by using primers 5′-GGAACCGCATGCAGCCAGAT-3′ and 5′-GTGTAGGCGAATTCAATGAG-3′. The SphI-EcoRI-digested PCR product and the EcoRI-XhoI DNA fragment from pACT2-HBM were then ligated in frame into the SphI-SalI-digested pQE32 (Qiagen) to produce pQE32-Keap1. pACT2-Keap1 was generated by inserting the BamHI-EcoRI DNA fragment from pQE32-Keap1 into pACT2-HBM. The complete Keap1 ORF in pACT2-Keap1 was then replaced with DNA fragments encoding different domains of Keap1. pQE-Keap1(1-139) was constructed by deleting the Keap1-encoding region downstream from the EcoRI site in pQE32-Keap1. pGEX-Keap1(308-624) was obtained by inserting the EcoRI-XhoI DNA fragment from pACT2-HB into pGEX-4T-2 (Amersham Pharmacia Biotech).
To produce Keap1 and its mutants as EGFP fusions in human cells, the Keap1-encoding DNA fragments were excised from pACT2-Keap1 and ligated in frame into the polylinker regions of the pEGFP-C series of vectors (Clontech). The L(308,310)A double mutation was introduced in Keap1 by PCR with a 5′-TCGAGGAGgcCACCgcGCACAAGC-3′ mutagenic primer (lowercase letters indicate the nucleotide substitutions) and the pUC19 direct and reverse sequencing primers. The structure of the cloned PCR product was verified by sequencing. The Keap1mNES-encoding sequence was inserted into pQE32 and then transferred, as the BamHI-HindIII and BamHI-XhoI DNA fragments, into pEGFP-C2 and pcDNA4/HisMaxC (Invitrogen) vectors, respectively.
pcDNA4/HisMax-Keap1 was constructed by transferring the BamHI-XhoI fragment from pACT2-Keap1 into pcDNA4/HisMaxA (Invitrogen). For overproduction of zz-Keap1 in yeast, the zz-encoding DNA fragment from pQEzz59 (5) and the Keap1-encoding DNA fragment from pACT2-Keap1 were inserted in frame between the EcoRI and BamHI sites of the pYeDP1/8-2 yeast shuttle vector (34).
The cDNA fragment encoding human Nrf2 was amplified by PCR from a HeLa cDNA library (Clontech) with primers 5′-CGGGATCCCATGGATTTGATTGACATA-3′ and 5′-CCGCTCGAGTCTAGTTTTTCTTAACATCT-3′. The PCR product was digested with BamHI and XhoI, inserted into pcDNA4/HisMaxB (Invitrogen), and then transferred into the BamHI-SalI-digested pQE31 (Qiagen).
pARE/luc was constructed by inserting a synthetic ARE of the human NQO1 gene (5′-GTACCGCAGTCACAGTGACTCAGCAGAATCGCTAG-3′) between the NheI and KpnI sites of pGL3-Promoter (Promega).
pSuper-RNAi was constructed by ligating annealed double-stranded oligonucleotide 5′-CCCGAGACGCCCCTGCTAACGGTTCAAGAGACCGTTAGCAGGGGCGTCTCTTTTTGGAAA-3′ with 5′ overhangs GATC (sense strand) and AGCT (antisense strand) into the BglII-HindIII-digested pSuper (4) (kindly provided by R. Agami, The Netherlands Cancer Institute, Amsterdam, The Netherlands). The structure of the resulting construct was confirmed by sequencing.
Antibodies and Western blotting analysis.
Anti-ProTα monoclonal antibodies 2F11 and 4F4 and anti-GFP monoclonal antibody 3A9 were described previously (37). Anti-Nrf2 (C-20), anti-IκBα (C-21), anti-histone H1 (AE-4), antiactin (C-2), and anti-LexA (N-19) antibodies were from Santa Cruz Biotechnology. Anti-caspase-3 antibodies were kindly provided by Y. Lazebnik (Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.). Mouse Keap1-specific monoclonal antibodies 2H5 and 2E4 and rabbit anti-Keap1(1-139) and anti-Keap1(308-624) polyclonal antibodies were generated essentially as described previously (37), using recombinant GST-Keap1(308-624) [for 2H5], His6-Keap1(1-139) [for 2E4 and anti-Keap1(1-139)] and His6-Keap1(308-624), respectively, as immunogens. Antibodies were affinity purified from ascitic fluids and sera by protein A-Sepharose (Amersham Pharmacia Biotech) chromatography, and polyclonal antibodies were additionally purified with membrane-immobilized Keap1 fragments. Specificity of the antibodies was confirmed by enzyme-linked immunosorbent assay, immunoblotting, and immunoprecipitation techniques. Notably, anti-Keap1 polyclonal antibodies and the 2E4 monoclonal antibody could bind both native and denatured Keap1, whereas the 2H5 monoclonal antibody could recognize nondenatured Keap1 only.
Immunoprecipitations of ProTα and Western blot analyses were performed as described previously (37). Detection of protein bands was performed by using sheep anti-mouse immunoglobulin (Ig) or goat anti-rabbit Ig conjugated with horseradish peroxidase as secondary antibodies and ECL Western blotting detection reagents (Amersham Pharmacia Biotech) or Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer Life Sciences). For coimmunoprecipitation of endogenous Keap1, 3 × 106 HeLa cells, either nonstressed or treated with 100 μM diethyl maleate (DEM) (Sigma) for 4 h, or HepG2 cells were lysed for 40 min on ice in 200 μl of EBC buffer (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 0.5% NP-40, 1 mM EDTA) supplemented with 1 mM phenylmethylsulfonyl fluoride. The cleared lysates were preincubated with 20 μg of control mouse IgG and protein A-Sepharose at 4°C for 2 h. After centrifugation for 5 min, the supernatants were incubated with 20 μg of 2F11 anti-ProTα antibody or with control mouse IgG at 4°C overnight. Protein A-Sepharose was added, and incubation was continued for 1 h. The beads were collected by brief centrifugation, washed two times with EBC buffer and two times with EBC buffer containing 0.5 M NaCl and 0.1% sodium dodecyl sulfate (SDS) at 0°C, and boiled in Laemmli sample buffer for 3 min. The samples were subjected to Western blot analysis with affinity-purified rabbit anti-Keap1(1-139) or anti-Keap1(308-624) polyclonal antibodies.
Protein purification and in vitro protein binding assays.
Recombinant human ProTα and its derivatives were isolated from E. coli BL21(DE3) cells transformed with appropriate plasmids by a phenol extraction procedure and further purified by DEAE-chromatography (12). 32P labeling of the ProTα derivatives containing a protein kinase recognition site was as described by Chichkova et al. (5). The specific radioactivity of 32P-ProTα thus obtained was 107 cpm/μg. Recombinant His6-Nrf2, His6-Keap1(1-139), His6-Keap(308-624), and GST-Keap1(308-624) were overproduced in E. coli JM109 carrying an appropriate plasmid and isolated with Ni-nitrilotriacetic acid agarose (Qiagen) and glutathione-Sepharose 4B (Amersham Pharmacia Biotech) chromatography, respectively. zz-Keap1 was isolated from galactose-induced S. cerevisiae 2805 cells transformed with pYeDP1/8-2/zzKeap1 by cell disruption with glass beads at 0°C for 10 min and absorption of the recombinant protein onto IgG-Sepharose (Amersham Pharmacia Biotech).
Interaction of 32P-ProTα (30,000 cpm per sample) with IgG-Sepharose-immobilized zz-Keap1 (150 ng of fusion protein per sample) or zz alone (30 ng) was assessed in binding buffer (20 mM HEPES [pH 6.8], 100 mM NaCl, 0.1% Tween 20). When indicated, the binding buffer was supplemented with 100 μM ZnSO4, CaCl2, or MgCl2. After 1 h of incubation at 0°C with shaking, the resin was washed with the same buffer, and the amounts of 32P-ProTα in pulled-down fractions and in supernatants were determined.
For competition assays, aliquots of IgG-Sepharose with immobilized zz-Keap1 (50 ng of fusion protein per sample) were incubated in binding buffer containing 100 μM ZnSO4 with either 32P-ProTα (30,000 cpm) or Nrf2 (250 ng) at 0°C for 1 h with shaking. The resin was washed with binding buffer and then incubated in the same buffer containing 250 ng of Nrf2 (for the Keap1-ProTα complex) and with the indicated amounts of wild-type ProTα or MProTα(44,50) mutant (for the Keap1-Nrf2 complex) at 0°C. Incubations with buffer alone served as controls. At the indicated time intervals, the amounts of 32P-ProTα and Nrf2 in pulled-down fractions and in supernatants were measured by radioactivity counting and by immunoblotting with anti-Nrf2 antibodies, respectively
To assess binding of endogenous Keap1 in the lysates of HeLa and HeG2 cells to matrix-immobilized ProTα, wild-type ProTα and its MProTα(44,50) mutant (1 mg each) were coupled in parallel to 1 ml of CNBr-activated Sepharose 4B (Amersham Pharmacia Biotech) according to the manufacturer's protocol. The beads were then preincubated with 5 mg of bovine serum albumin per ml in buffer A (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 1 mM EDTA, and 0.3% Tween 20) at 0°C for 15 min and washed with the same buffer. Whole-cell lysates of 1.5 × 106 cells were incubated with 10-μl aliquots of ProTα-Sepharose beads in 200 μl of buffer A at 0°C overnight, the beads were washed with the same buffer, and the bound proteins were eluted with 1% SDS in buffer A at 37°C for 10 min. Eluates were subjected to SDS-8% polyacrylamide gel electrophoresis (PAGE) and analyzed by Western blotting with anti-Keap1(1-139) and anti-Keap1(308-624) polyclonal antibodies.
Reporter gene assays.
Luciferase and β-Gal were used as reporters. Cotransfection with β-Gal-expressing plasmid pcDNA4/HisMax/lacZ (Invitrogen) was employed to normalize for transfection efficiencies. Cells were lysed, 36 h after transfection, in reporter lysis buffer (Promega), and luciferase and β-Gal assays were performed with the Luciferase Assay System (Promega) according to the protocol supplied. All experiments were performed in triplicate.
RNA isolation and Northern blotting.
Total cellular RNA was isolated from 5 × 105 transfected HeLa cells with the RNAqueous total RNA isolation kit (Ambion) according to the protocol supplied by the manufacturer. RNA samples, in 20-μg aliquots, were electrophoresed on 1.5% agarose-2.2% formaldehyde gels, transferred onto Hybond N+ membranes (Amersham Pharmacia Biotech), and hybridized sequentially with [32P]DNA probes obtained by random-priming labeling of the indicated cDNAs. Quantification was performed with ImageQuant software after exposure of the membranes to a PhosphorImager (Molecular Dynamics).
ProTα overproduction and small interfering RNA (siRNA) knockdown in human cells.
ProTα lacking artificially added sequences was overproduced in HeLa cells transfected with pcDNA4-enh-wt ProTα and pcDNA4-enh-MProTα(44,50) as described previously (13). Cells were treated with 100 μM DEM or with diluent (dimethyl sulfoxide) only for 22 h prior to harvesting. For ProTα mRNA interference experiments, HeLa cells were grown on 35-mm-diameter dishes to 80% confluence and transfected twice, at a 2-day interval, with 6 μg of pSuper or pSuper-RNAi. Total RNA was isolated from cells 48 h after the last transfection step and subjected to Northern blot analysis. Aliquots of the same cells were used for Western blot analysis with the indicated antibodies and for partial purification of ProTα followed by 7 M urea-8% PAGE (13).
RESULTS
Yeast two-hybrid screen for ProTα-interacting proteins.
A search for human proteins interacting with ProTα was performed with the yeast two-hybrid system. cDNA libraries from HB and HBM cloned downstream from the Gal4 activation domain (AD)-coding sequence served as prey, whereas full-length human ProTα fused to LexA served as bait. The bait construct alone failed to activate transcription of the lacZ, HIS3, and URA3 reporter genes when introduced into S. cerevisiae L40ccU cells despite the synthesis of easily detectable amounts of the LexA-ProTα protein (data not shown). Yeast L40ccU cells cotransformed with the bait and the library prey constructs were selected for the gained ability to grow in the absence of uracil and histidine and were further screened with the β-galactosidase filter assay. Of 107 transformants tested, three positive clones from the HB library (with identical inserts) and one positive clone from the HBM library (with a larger insert) were identified. The library plasmids rescued from these clones encoded putative ProTα interactors, as they were able, upon the retransformation into the yeast strain, to activate expression of the reporter genes in the presence of the bait construct encoding LexA-ProTα but not in the absence of the bait or in the presence of unrelated proteins fused to LexA. Sequencing of the inserts from the rescued library plasmids (one plasmid for each library) revealed that they encode portions of the same protein, known as Keap1 (KIAA0132), a 624-amino-acid protein inhibitor of the transcription factor Nrf2. One plasmid (pACT2-HB) contained the Keap1 ORF starting from residue 308, and another (pACT2-HBM) contained the Keap1 ORF starting from residue 50. We then restored the complete ORF of human Keap1 and demonstrated that, in the yeast two-hybrid assays, full-length Keap1 binds ProTα efficiently (data not shown).
Delineating a Keap1-binding region in ProTα.
To learn which region in ProTα is responsible for binding to Keap1, a set of ProTα deletion mutants was constructed (Fig. 1A). The ability of these mutants to bind full-length Keap1 was assessed with the yeast two-hybrid system. A short central region of ProTα (residues 32 to 52) turned out to be both necessary and sufficient to provide the interaction (Fig. 1A). Furthermore, testing the ProTα E(44)G,E(50)G double mutant [MProTα(44,50)] from our collection (34) in the yeast two-hybrid assay demonstrated the loss of interaction of this ProTα mutant with Keap1 (Fig. 1A). The effect was not due to instability of either the ProTα or Keap1 derivatives in yeast cells (Fig. 1B and C) and provides a further argument for high specificity of the ProTα-Keap1 binding.
FIG. 1.
Keap1 is a ProTα interactor: evidence from yeast two-hybrid analysis. (A) Identification of a Keap1-binding region (black) in ProTα. + and −, relative β-Gal levels above 300 and below 10 U, respectively. wt, wild type. (B and C) Lack of detectable interaction between the MProTα(44,50) mutant and Keap1 is not due to instability of either protein. Shown is Western blot analysis of lysates of yeast cells producing the following: lanes 1, LexA-ProTα plus AD-Keap1; lanes 2, LexA-MProTα(44,50) plus AD-Keap1; lanes 3, LexA plus AD-Keap1; lane 4, LexA-MProTα(44,50) plus AD. The primary antibodies used were anti-LexA (B) and anti-Keap1 (C). (D) An intact Kelch (diglycine, GG) repeat domain of Keap1 is required for ProTα binding. Individual repeats are numbered from 1 to 6. BTB/POZ, BTB/POZ domain of Keap1.
Domain in Keap1 involved in binding to ProTα.
We then identified a region in Keap1 that is involved in the interaction with ProTα. A set of Keap1 deletion mutants was constructed (Fig. 1D) and assayed for ProTα binding by using the yeast two-hybrid system. The carboxyl-terminal half of Keap1 turned out to be the shortest Keap1 fragment capable of ProTα binding (Fig. 1D). This area of Keap1 comprises six Kelch repeats which together form a tertiary structure known as the β-propeller (1). Deletion of even one Kelch repeat in Keap1 resulted in the elimination of ProTα binding (Fig. 1D), suggesting that the intact β-propeller structure in Keap1 is essential for ProTα recognition.
Evidence for the ProTα-Keap1 interaction in human cell lysates.
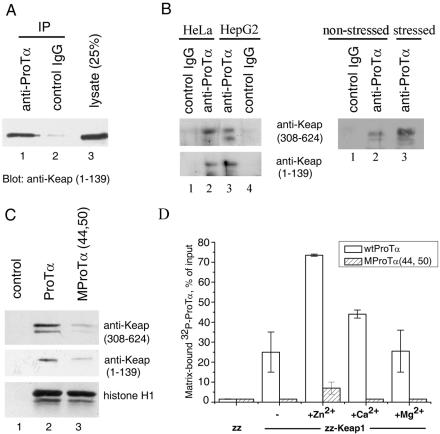
To learn whether Keap1 could bind to ProTα in human cells, a whole-cell lysate of HeLa cells ectopically expressing full-length Keap1 was treated with anti-ProTα 2F11 monoclonal antibody or with control mouse IgG, and the antigen-antibody complex was collected with protein A-Sepharose and analyzed by Western blotting with anti-Keap1 polyclonal antibodies. Figure 2A demonstrates that Keap1 coprecipitated with endogenous ProTα when anti-ProTα antibody was used (lane 1), whereas control mouse IgGs were inactive (lane 2). The possibility that anti-ProTα antibody could recognize Keap1 directly was excluded by Western blot analysis of Keap1 with anti-ProTα 2F11 monoclonal antibody (data not shown). Coimmunoprecipitation of the ectopically produced Keap1 with ProTα was then confirmed with lysates of HepG2 cells (data not shown).
FIG. 2.
Keap1 interacts with ProTα in human cell lysates (A, B, and C) and in vitro (D). (A) Coimmunoprecipitation of the ectopically produced Keap1 with ProTα from a whole-cell lysate of HeLa cells. Precipitation was performed with anti-ProTα 2F11 monoclonal antibody (lane 1) or with control mouse IgG (lane 2). Lane 3, whole-cell lysate. Detection of Keap1 was performed by Western blot analysis with anti-Keap1 polyclonal antibodies. (B) Endogenous Keap1 associates with ProTα. Left panel: Coprecipitation of endogenous Keap1 with ProTα from whole-cell lysates of HeLa (lanes 1 and 2) and HepG2 (lanes 3 and 4) cells. Precipitation and detection were performed as described for (A). Right panel: Coprecipitation of endogenous Keap1 from lysates of HeLa cells either untreated (lanes 1 and 2) or treated with 100 μM DEM for 4 h (lane 3). Note that in panels A and B the membranes were exposed to X-ray film for 30 s and 30 min, respectively. (C) Binding of endogenous Keap1 to Sepharose-immobilized ProTα is impaired by the (44,50) mutation in ProTα. Lane 1, resin without ProTα; lane 2, wild-type ProTα; lane 3, MProTα(44,50) mutant. The amount of Keap1 from HeLa whole-cell lysates bound to each resin was evaluated as described for panel A with two different anti-Keap1 antibodies. Bottom row: histone H1 binding to the same affinity resins, verifying equal amounts of immobilized ProTα. (D) Interaction of the purified recombinant ProTα and Keap1 in vitro. IgG-Sepharose-immobilized zz-Keap1 (150 ng) or zz alone (30 ng) was incubated with 32P-radiolabeled wild-type (wt) ProTα or the MProTα(44,50) mutant, either in the absence or in the presence of the indicated metal cations (100 μM). After incubation, the unbound ProTα was removed by washing the resin, and the amount of 32P-ProTα bound to each affinity resin was determined by measuring the radioactivity. Error bars indicate standard deviations.
Likewise, endogenous Keap1 could be coimmunoprecipitated with endogenous ProTα from whole-cell lysates of HeLa and HepG2 cells, as determined by Western blot analysis with two different anti-Keap1 polyclonal antibodies (Fig. 2B, left panel). Furthermore, even larger amounts of ProTα-associated Keap1 were detectable in HeLa cells treated with DEM, an oxidative stress inducer, relative to the nonstressed sample (Fig. 2B, right panel). Of note, Keap1 coimmunoprecipitated from the DEM-treated cell sample displayed a slightly lower electrophoretic mobility and appeared to be more heterogenous (Fig. 2B, right panel, lane 3 versus lane 2), probably due to posttranslational modification of the protein.
Therefore, the ProTα-Keap1 interaction may occur in the homologous cell system, under both normal and oxidative stress conditions.
To further substantiate this conclusion and to check for the specificity of the interaction, recombinant human ProTα and its MProTα(44,50) mutant were immobilized on BrCN-activated Sepharose and used for isolation of endogenous Keap1 from HeLa and HepG2 cell lysates. The amount of Keap1 bound to each resin was evaluated by Western blotting with two different types of anti-Keap1 antibodies. Binding of Keap1 to wild-type ProTα was observed (Fig. 2C), whereas only trace amounts of Keap1 could bind to the MProTα(44,50) mutant, thus confirming our data on the specificity of the ProTα-Keap1 interaction obtained with the use of the yeast two-hybrid system.
Binding of Keap1 to ProTα is direct.
To test whether the interaction between ProTα and Keap1 is direct or is mediated by some “bridging” component of cell lysates, a pull-down assay with purified recombinant human ProTα and Keap1 proteins was employed. Human Keap1 was overproduced as a fusion with the zz domain (z is an IgG-binding domain of staphylococcal protein A) and immobilized on IgG-Sepharose. For accurate and quantitative determination of ProTα binding to this affinity matrix, human wild-type ProTα and MProTα(44,50) were fused at their N termini to a short peptide comprising the protein kinase A recognition sequence and 32P radiolabeled. Binding of the labeled wild-type ProTα to the immobilized zz-Keap1 was observed in this in vitro system (Fig. 2D), whereas binding of ProTα to the zz domain alone was negligible. Recombinant unlabeled ProTα lacking artificially added sequences could compete with the binding of the radiolabeled probe (data not shown), indicating that binding to Keap1 is specific for the ProTα sequence. Furthermore, the MProTα(44,50) mutant failed to bind Keap1 (Fig. 2D).
Previously, we have demonstrated that ProTα binds Zn2+ and Ca2+ (but not Mg2+) (5). Therefore, we tested whether these divalent cations could affect ProTα biding to Keap1. In the presence of 100 μM Zn2+ or Ca2+, this binding was markedly enhanced (Fig. 2D). Substitution of Mg2+ for these cations did not result in ProTα binding over the level seen in the absence of divalent cations (Fig. 2D).
Thus, ProTα interacts with Keap1 directly and specifically, and the divalent cations that bind to ProTα significantly enhance the ProTα-Keap1 interaction.
How can ProTα and Keap1 meet each other?
The in vivo and in vitro data presented above seem to strongly suggest that Keap1 might be a genuine partner of ProTα. One major objection to this idea is that ProTα and Keap1 have been thought to be localized to different subcellular compartments. While ProTα is known to be an exclusively nuclear protein, Keap1 was reported to be cytoplasmic and excluded from the nucleus. Moreover, such a localization is consistent with the postulated role of Keap1 as a cytoplasmic anchor for the Nrf2 transcription factor.
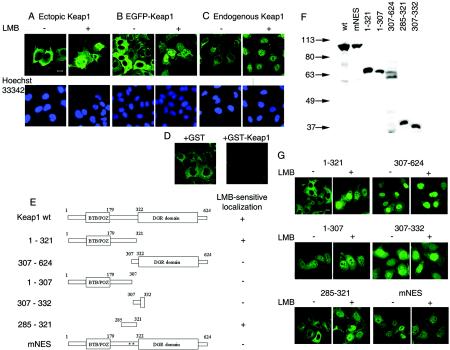
To test an idea that Keap1 might be a shuttling protein, we localized Keap1 in HeLa cells before and after treatment with LMB, a known and specific inhibitor of nuclear export of proteins possessing leucine-rich NESs. HeLa cells ectopically expressing Keap1 or EGFP-Keap1 fusion protein or containing endogenous Keap1 only were examined by fluorescence microscopy with anti-Keap1 monoclonal antibodies or the intrinsic fluorescence of EGFP. In the absence of LMB, both the endogenous and ectopically produced Keap1 were predominantly cytoplasmic (Fig. 3A to C). Upon treatment with LMB, a dramatic relocation of all three proteins from the cytoplasm to the nucleus occurred (Fig. 3A to C). The specificity of detection of endogenous Keap1 with the anti-Keap1 2H5 monoclonal antibody was confirmed by using an exogenously added GST-Keap1 competition assay (Fig. 3D) and a different anti-Keap1 monoclonal antibody, 2E4 (data not shown). A very similar nuclear translocation of Keap1 in response to LMB treatment was observed in another human cell line, HepG2 (data not shown). Our data thus imply that Keap1 is indeed a shuttling protein likely equipped with a leucine-rich NES. Importantly, an ability of Keap1 to enter the nucleus lends further support to our conclusion that Keap1 may be a true ProTα interactor.
FIG. 3.
Keap1 is a NES-containing nuclear-cytoplasmic shuttling protein. (A to C) Treatment of HeLa cells with LMB induces nuclear translocation of the ectopically produced full-length Keap1 (A), of the EGFP-Keap1 fusion protein (B), and of endogenous Keap1 (C). Confocal images were obtained by using anti-Keap1 2H5 monoclonal antibody (A and C) or intrinsic fluorescence of EGFP (B). (D) Recognition of endogenous Keap1 in HeLa cells is prevented by preincubation of the anti-Keap1 2H5 monoclonal antibody with recombinant GST-Keap1(308-624) (+GST-Keap1 panel) but not with GST alone (+GST panel). Images were acquired with identical confocal microscope settings. (E) Schematic representation of the Keap1 mutants used for the identification of the NES of human Keap1. DGR, diglycine repeat; wt, wild type. The indicated proteins were fused to the C terminus of EGFP. (F) Western blot analysis of the production of the EGFP-fused Keap1 mutants shown in panel E in transfected HeLa cells. Anti-EGFP antibody was used for detection of the respective proteins. (G) Confocal images showing intracellular localization of the Keap1 mutants fused to EGFP in HeLa cells either untreated or treated with LMB for 4 h. The results presented are also summarized in panel E, right column. Bars, 10 μm.
Identification of a nuclear export signal in Keap1.
To identify a putative NES of Keap1, a set of deletion mutants of Keap1 fused to the C terminus of EGFP reporter protein was constructed (Fig. 3E). HeLa cells transfected with the respective plasmids and producing EGFP-Keap1 derivatives of the expected size (Fig. 3F) were inspected for intracellular localization of the fusion proteins by fluorescence microscopy both before and after LMB treatment (Fig. 3G). Only the Keap1 derivatives encompassing the central “intervening” region of the protein displayed LMB-sensitive subcellular localization, being excluded from the nucleus in the absence of LMB and relocating into the nucleus after LMB treatment [Keap1(1-321) and Keap1(285-321)] (Fig. 3G).
Within the Keap1(285-321) region, there is the amino acid sequence L301VKIFEELTL310, which conforms to the consensus of the leucine-rich NES, H-2/3X-H-2X-H-X-H (where H stands for hydrophobic [frequently Leu] residues). Within this putative Keap1 NES, we replaced two leucine residues (L308 and L310) that are known to be the most essential for NES function with alanines to produce an EGFP-linked full-length Keap1 mutant (Keap1mNES). In contrast to the wild-type Keap1 (Fig. 3B), the mutant protein was no longer excluded from the nuclei of HeLa (Fig. 3G, mNES) and HepG2 (data not shown) cells in the absence of the LMB treatment. Since our results indicate that the identified sequence is both necessary and sufficient for the LMB-sensitive nuclear exclusion of Keap1, we conclude that it represents a genuine NES in Keap1 and that Keap1 is equipped with a single NES.
Coordinated nuclear-cytoplasmic shuttling of Nrf2 with Keap1.
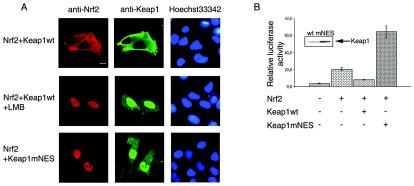
To learn whether shuttling of Keap1 between the nucleus and cytoplasm may be of functional significance, an effect of dysfunction of the NES of Keap1 on the subcellular localization of Nrf2 was studied by confocal laser scanning microscopy. In HeLa cells cotransfected with the Nrf2- and the wild-type Keap1-encoding plasmids, both proteins were essentially cytoplasmic (Fig. 4A, top row). LMB treatment of these cells resulted in the expected nuclear translocation of Keap1, similar to that observed without Nrf2 overproduction, which was mimicked by nuclear accumulation of Nrf2 (Fig. 4A, middle row). Alternatively, in cells cotransfected with the Nrf2- and Keap1mNES-encoding plasmids efficient nuclear accumulation of both proteins was observed even in the absence of LMB (Fig. 4A, bottom row). Thus, nuclear translocation of Nrf2 triggered by the dysfunction of the Keap1 NES is indicative of the nuclear-cytoplasmic shuttling of the Nrf2-Keap1 complex.
FIG. 4.
Impairment of the Keap1 NES function results in coordinated nuclear translocation of Keap1 and Nrf2 (A) and in transcriptional activation (B). (A) HeLa cells were cotransfected with the Nrf2- and either with the wild-type (wt) Keap1- or Keap1mNES-encoding plasmids. In the former case, transfected cells were either treated with 2 nM LMB for 4 h or left untreated. For simultaneous detection of Keap1 and Nrf2, fixed cells were incubated with mouse anti-Keap1 and rabbit anti-Nrf2 antibodies, followed by treatment with fluorescein isothiocyanate-labeled anti-mouse and Texas Red-labeled anti-rabbit secondary antibodies. The microscope settings used excluded cross-fluorescence between the red and green channels. Right column, nuclei stained with Hoechst 33342. Bar, 10 μm. (B) The pARE/luc reporter plasmid was transfected into HepG2 cells together with expression plasmids encoding Nrf2, wt Keap1, or Keap1mNES, as indicated. A β-Gal-expressing plasmid was used as a transfection control. The total amount of DNA used for transfection was normalized with an empty vector. Cells were harvested at 48 h after transfection, and the luciferase activity of each sample was measured and normalized for β-Gal activity. The intactness of wt Keap1 and Keap1mNES was verified by Western blotting analysis of the lysates with anti-Keap1 antibody and is shown in the inset. Error bars indicate standard deviations.
NES-less Keap1 is an activator, rather than an inhibitor, of Nrf2-mediated transcription.
Alteration of the subcellular localization of Nrf2 and Keap1 due to destruction of Keap1's NES might be expected to influence Nrf2-mediated transcriptional activation. To monitor Nrf2-dependent transcription, the reporter plasmid pARE/luc was constructed by inserting a synthetic Nrf2-responsive ARE of the human NQO1 gene into the luciferase-expressing pGL3-Promoter plasmid. Expression of this luciferase reporter construct, upon transfection into HepG2 cells, was confirmed to be up-regulated by ectopic production of Nrf2 and down-regulated by the coexpression of wild-type Keap1 together with Nrf2, as expected (Fig. 4B). However, coexpression of the Keap1mNES mutant together with Nrf2 not only failed to suppress luciferase gene expression but resulted in a very pronounced enhancement of Nrf2-driven transcription (Fig. 4B). This activating effect was reproduced in HeLa cells (not shown). Thus, destruction of the NES by point mutations converted Keap1 from an inhibitor to an activator of Nrf2-dependent gene expression.
ProTα competes with Nrf2 for binding to Keap1.
As demonstrated above, ProTα binds to the C-terminal half of Keap1 comprising six Kelch (diglycine) repeats. Nrf2 was shown previously to interact with the C-terminal half of Keap1 as well (19). To test whether ProTα and Nrf2 could compete with each other for Keap1 binding, an in vitro system utilizing purified recombinant proteins was employed. A Keap1-ProTα complex was preformed by binding 32P-ProTα to zz-Keap1 immobilized on IgG-Sepharose. This complex was challenged with recombinant human Nrf2. A parallel sample was treated with buffer lacking Nrf2 to serve as a control. The amounts of 32P-ProTα shifted from the matrix-bound complex to the supernatant were determined at several time points. As shown in Fig. 5A, addition of Nrf2 resulted in the efficient dissociation of ProTα from Keap1. In the absence of Nrf2, dissociation of the Keap1-ProTα complex was much slower (Fig. 5A), indicating that it was Nrf2 that forced ProTα displacement from Keap1.
FIG. 5.
ProTα competes with Nrf2 for binding to Keap1 in vitro. (A) Recombinant zz-Keap1 (50 ng) was immobilized on IgG-Sepharose and charged with 32P-ProTα. Unbound ProTα was removed, and the immobilized Keap1-ProTα complex was incubated with buffer alone (solid line) or with the same buffer containing recombinant Nrf2 (250 ng) (dotted line). At the indicated time intervals, the amount of 32P-ProTα in the pulled-down fractions was determined. The amount of matrix-bound ProTα at zero time was taken as 100%. (B) Recombinant zz-Keap1 (50 ng) immobilized on IgG-Sepharose was charged with recombinant Nrf2 (250 ng). Unbound Nrf2 was removed, and the immobilized Keap1-Nrf2 complex was incubated in parallel with recombinant wild-type ProTα (lanes 2 and 5), with the MProTα(44,50) mutant (lanes 3 and 6) (50 μg each), or with buffer alone (lanes 1 and 4) for 1 h. The amount of Nrf2 in the pulled-down (Bound to Keap1, lanes 1 to 3) and supernatant (Displaced, lanes 4 to 6) fractions was determined by Western blotting analysis of each fraction with anti-Nrf2 antibody. Lane 7, Nrf2 used as a marker. (C) Displacement of Nrf2 from the Nrf2-Keap1 complex by ProTα is dose dependent. zz-Keap1-Nrf2 complex was formed as in panel B and challenged with increasing amounts of wild-type ProTα for 1 h. Amounts of displaced Nrf2 were determined by immunoblotting as in panel B. Lanes 1 to 4, 0.05, 0.5, 5.0, and 50 μg of ProTα, respectively. Lane 5, Nrf2 used as a marker.
To verify the competition, a reciprocal experiment was performed. IgG-Sepharose-immobilized zz-Keap1 was charged with Nrf2 and challenged, in three experiments performed in parallel, with either wild-type ProTα, the MProTα(44,50) mutant that fails to bind Keap1, or buffer alone. After 1 h of incubation, the amounts of Nrf2, both displaced from the complex with immobilized Keap1 and remaining bound after this treatment, were monitored by immunoblotting with anti-Nrf2 antibodies. Displacement of Nrf2 from its complex with Keap1 occurred upon challenge with wild-type ProTα, with a concomitant decrease in the amount of Nrf2 remaining bound to Keap1 (Fig. 5B, lanes 2 and 5). Only trace amounts of displaced Nrf2 were observed when the Keap1-Nrf2 complex was challenged with the MProTα(44,50) mutant or with buffer alone (Fig. 5B, lanes 4 and 6 versus lanes 1 and 3). Displacement of Nrf2 by wild-type ProTα was dose dependent (Fig. 5C). These findings indicate that ProTα, when competent for Keap1 binding, is able to specifically liberate Nrf2 from its complex with Keap1. It should be noted, however, that despite the use of a high molar excess of the competitor, ProTα was able to displace Nrf2 only partially (Fig. 5B, lane 5 versus lane 2).
ProTα contributes to Nrf2-dependent gene expression.
We then assessed the physiological relevance of the ProTα-Keap1 interaction. Binding of ProTα to Keap1 may indicate involvement of ProTα in regulation of the Nrf2-mediated transcription of the stress-protective genes. Because ProTα was able to liberate Nrf2 from its complex with Keap1, the latter being an inhibitor of the transcription factor, we proposed a role for ProTα that consists of dissociation of the Nrf2-Keap1 complex and thus of up-regulation of the expression of the Nrf2-dependent genes. If this was true, then alteration of the intracellular ProTα level should influence the Nrf2 transcriptional activity.
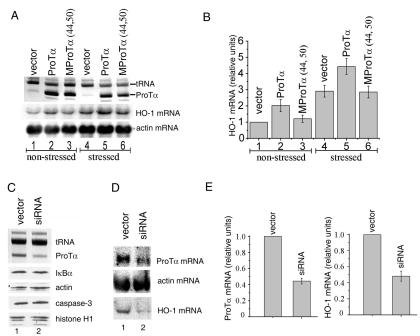
To test this idea, the effect of ProTα overproduction on Nrf2-driven transcription was assessed first. RNA samples from HeLa cells transfected with either the wild-type ProTα- or the MProTα(44,50)-encoding plasmid or with an empty vector were analyzed by Northern blot hybridization with an HO-1 probe. The HO-1 gene contains an ARE, and its expression is known to be regulated in an Nrf2-dependent fashion (2, 17). A β-actin probe served as a control. Overproduction of the wild-type ProTα (Fig. 6A, top panel) resulted in a concomitant increase in the HO-1 mRNA level, relative to the empty vector control (Fig. 6A, middle panel), both in nonstressed cells (lanes 1 to 3) and in cells treated with DEM to induce oxidative stress (lanes 4 to 6). The β-actin mRNA level remained unaffected (Fig. 6A, bottom panel). Importantly, overproduction of the MProTα(44,50) mutant, which has an impaired ability to bind Keap1 in vivo and in vitro and to liberate Nrf2 from the Nrf2-Keap1 complex (Fig. 5B), failed to up-regulate HO-1 gene expression (Fig. 6A). Thus, in agreement with our model, an increase in the ProTα level could stimulate transcription of an Nrf2-dependent gene, and this effect appeared to be specifically mediated by the ProTα binding to Keap1 (see Fig. 6B for quantification).
FIG. 6.
HO-1 gene expression in HeLa cells is directly proportional to the ProTα level. (A) ProTα overproduction enhances HO-1 transcription. Lysates of HeLa cells transfected with the wild-type ProTα- or with the MProTα(44,50)-encoding plasmid or with vector alone were analyzed 48 h after transfection for the ProTα content (top panel) and for relative levels of expression of HO-1 mRNA (Nrf2 dependent, middle panel), and β-actin mRNA (Nrf2 independent, bottom panel). Lanes 1 to 3, nonstressed cells; lanes 4 to 6, cells treated with 100 μM DEM for 22 h prior to harvesting. Top panel: Partially purified ProTα was fractionated in a 7 M urea-8% polyacrylamide gel and visualized by methylene blue staining. tRNA present in the partially purified ProTα samples verifies equal loading. Middle and bottom panels: Northern blot analysis of lysates with HO-1 and β-actin probes, respectively. The same membrane was used with both probes. (B) Quantitative analysis of the data presented in panel A. In panels B and E, all transfection experiments were performed a minimum of four times before calculating means and standard deviations. The amount of the corresponding mRNA from vector-transfected cells was taken as 1.0. panel C Down-regulation of ProTα level through ProTα mRNA interference. HeLa cells were transfected with either ProTα siRNA-expressing plasmids or with empty vector, as described in Materials and Methods. Two days after the last transfection step, cell lysates were analyzed for the ProTα content (top panel) as described for panel A. In parallel, the lysates were analyzed for the levels of several unrelated proteins by Western blotting with antibodies to actin, histone H1, IκBα, and procaspase-3 (bottom panels). (D) ProTα mRNA interference down-regulates HO-1 gene expression. Lysates of HeLa cells transfected as described for panel C were analyzed by Northern blotting with ProTα, HO-1, and β-actin probes. (E) Quantitative analysis of the data presented in panel D.
To further verify our model, a reciprocal, ProTα mRNA interference approach was taken. Plasmid-driven production of a hairpin siRNA targeted to the ProTα mRNA region encoding amino acid residues 30 to 36 of the protein (ProTα siRNA) led to a significant reduction in the intracellular ProTα level relative to the vector-transfected sample, as revealed by PAGE upon partial purification of the protein (Fig. 6C, top panel) and confirmed by sandwich ELISA with crude cell lysates and a pair of anti-ProTα monoclonal antibodies (data not shown). No difference from the control sample was observed when the lysates were probed for actin, histone H1, IκBα, and procaspase-3 contents by Western blotting (Fig. 6C, bottom panels), indicating that the effect of RNA interference was ProTα specific.
At the RNA level, expression of the ProTα siRNA resulted in a marked decrease in the amount of ProTα mRNA detected, relative to the empty vector control (Fig. 6D, top panel), while it had no effect on the level of β-actin mRNA (Fig. 6D, middle panel). To determine whether the reduction in the ProTα level could influence Nrf2-dependent gene expression, the RNA blot depicted in Fig. 6D was rehybridized with the HO-1 probe. Down-regulation of the expression of HO-1 mRNA was observed in cells expressing ProTα siRNA (Fig. 6D, bottom panel; see also Fig. 6E for quantification). Thus, the level of Nrf2-dependent gene expression was correlated with the intracellular level of ProTα by using both up- and down-regulation of ProTα synthesis.
DISCUSSION
Interaction of the Nrf2 transcription factor with its inhibitor Keap1 is central in regulation of expression of the genes defending cells against oxidative stress and electrophilic attack. According to current thinking, in the absence of stress, actin-bound Keap1 serves as a cytoplasmic anchor for Nrf2, preventing nuclear uptake of the transcription factor and activation of expression of Nrf2-dependent genes and targeting Nrf2 for proteosome-mediated degradation. Upon induction of stress, the Nrf2-Keap1 interaction is destabilized, most probably due to posttranslational modification of Nrf2, Keap1, or perhaps both constituents, thus liberating Nrf2 from sequestration and permitting its nuclear translocation to induce expression of stress-preventing genes.
In this study, we searched for protein partners of the small nuclear protein ProTα. ProTα is an essential protein involved in the proliferation of mammalian cells and in their protection against apoptosis. We provided in vivo and in vitro evidence that Keap1 is a genuine partner of ProTα and that ProTα contributes to Nrf2-dependent gene expression. The notion that ProTα might be involved in up-regulation of expression of genes protecting cells from oxidative stress was not anticipated earlier and illuminates a novel function of this small but evidently multifunctional protein. However, our results appear to provide clues to the functioning of the Nrf2-Keap1 system as well.
First, we demonstrated that Keap1 is a shuttling protein that migrates dynamically between the nucleus and the cytoplasm. Nuclear export of Keap1 is Crm1 (exportin-1) dependent and is mediated by a functional leucine-rich NES positioned in the central intervening region of Keap1 (residues 301 to 310). Besides, because Keap1, even when overproduced, is rapidly translocated to the nucleus upon LMB- and point mutation-mediated NES dysfunction, Keap1 is expected to possess a nuclear localization signal as well. Mutating the NES in Keap1 results in a profound nuclear accumulation of both Keap1 and Nrf2, implying that, besides shuttling by itself, Keap1 confers nuclear-cytoplasmic shuttling on the Nrf2-Keap1 complex. The shuttling model proposed here may have several advantages over the accepted cytoplasmic anchoring model, as follows. (i) The shuttling Nrf2-Keap1 complex could gain an ability to sense stress conditions in both compartments, nuclear and cytoplasmic. (ii) The appearance of Nrf2, albeit in complex with Keap1, in the nuclei of nonstressed cells is in line with the occurrence of the basal ARE-mediated transcription, which is not readily explicable in terms of the cytoplasmic anchoring model. (iii) Shuttling of Keap1 could provide a mechanism for termination of the induced expression of the stress-protective genes by withdrawing Nrf2 from the nucleus when the insult is surmounted.
Second, our in vivo and in vitro data indicate that interaction of ProTα with Keap1 is highly specific (e.g., suppressed by point mutations in ProTα) and functionally relevant [Nrf2-dependent transcription was found to be directly proportional to the intracellular level of wild-type ProTα but not to that of MProTα(44,50)]. Clues to the mechanism of ProTα involvement in Nrf2-Keap1 functioning came from the identification of the ProTα-binding domain in Keap1. We showed that ProTα binds to the C-terminal half of Keap1 comprising six Kelch repeats, as Nrf2 does. Consistent with this finding, ProTα competed with Nrf2 for binding to Keap1. Furthermore, challenging the Nrf2-Keap1 complex with ProTα leads to displacement of Nrf2. Significantly, the ProTα mutant with an impaired ability to bind Keap1 failed to liberate Nrf2. A high molar excess of ProTα required for partial displacement of Nrf2 may, of course, be considered to represent a limitation of the in vitro assay. However, we think that this situation is close to the natural one, as ProTα is a very abundant nuclear protein. Limited release of Nrf2 triggered by ProTα is likely to provide basal ARE-mediated transcription. Upon stress induction, destabilization of the Nrf2-Keap1 complex should facilitate its dissociation by ProTα and result in a far larger amount of liberated Nrf2, leading to enhanced expression of the ARE-containing genes.
Our results attribute a role to ProTα as the intranuclear dissociator of the Nrf2-Keap1 complex. According to our model, the Nrf2-Keap1 complex shuttles constantly between the cytoplasm and the nucleus. Once in the nucleus, the complex is attacked by ProTα present in large excess, resulting in ProTα binding to Keap1 and concomitant displacement of Nrf2, with the latter being directed to the target genes to activate their expression. This liberation of Nrf2 is partial in nonstressed cells, providing a basal level of transcription, and is far more pronounced in stress-induced cells due to predestabilization of the Nrf2-Keap1 complex. After completion of stress, an excess of free Nrf2 is withdrawn from the nucleus by binding to the shuttling Keap1 because dissociation of the stabilized (nonmodified) Nrf2-Keap1 complex by ProTα again becomes inefficient.
In conclusion, our data have revealed an unexpected function of ProTα and contribute to understanding of molecular mechanisms of expression of oxidative stress-protecting genes.
Acknowledgments
We thank R. Agami for providing pSuper and Y. Lazebnik for providing anti-caspase-3 antibodies. We are grateful to Tatyana Pestova and Christopher Hellen for their constant support.
This work was supported in part by the Ludwig Institute for Cancer Research (to A.B.V.) and by grants from the Russian Foundation for Basic Research (to N.V.C., A.G.E., and A.B.V.), the U.S. Civilian Research and Development Foundation for the Independent States of the Former Soviet Union (CRDF) (to A.B.V.), and the Russian Frontiers in Genetics Program (to A.B.V.).
REFERENCES
- 1.Adams, J., R. Kelso, and L. Cooley. 2000. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 10:17-24. [DOI] [PubMed] [Google Scholar]
- 2.Alam, J., D. Stewart, C. Touchard, S. Boinapally, A. M. Choi, and J. L. Cook. 1999. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 274:26071-26078. [DOI] [PubMed] [Google Scholar]
- 3.Bloom, D. A., and A. K. Jaiswal. 2003. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J. Biol. Chem. 278:44675-44682. [DOI] [PubMed] [Google Scholar]
- 4.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 5.Chichkova, N. V., A. G. Evstafieva, I. G. Lyakhov, A. S. Tsvetkov, T. A. Smirnova, R. N. Karapetian, E. M. Karger, and A. B. Vartapetian. 2000. Divalent metal cation binding properties of human prothymosin alpha. Eur. J. Biochem. 267:4745-4752. [DOI] [PubMed] [Google Scholar]
- 6.Clinton, M., L. Graeve, H. el-Dorry, E. Rodriguez-Boulan, and B. L. Horecker. 1991. Evidence for nuclear targeting of prothymosin and parathymosin synthesized in situ. Proc. Natl. Acad. Sci. USA 88:6608-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullinan, S. B., D. Zhang, M. Hannink, E. Arvisais, R. J. Kaufman, and J. A. Diehl. 2003. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 23:7198-7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhakshinamoorthy, S., and A. K. Jaiswal. 2001. Functional characterization and role of INrf2 in antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene 20:3906-3917. [DOI] [PubMed] [Google Scholar]
- 9.Dinkova-Kostova, A. T., W. D. Holtzclaw, R. N. Cole, K. Itoh, N. Wakabayashi, Y. Katoh, M. Yamamoto, and P. Talalay. 2002. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 99:11908-11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominguez, F., C. Magdalena, E. Cancio, E. Roson, J. Paredes, L. Loidi, J. Zalvide, M. Fraga, J. Forteza, B. J. Regueiro, and J. L. Puente. 1993. Tissue concentrations of prothymosin alpha: a novel proliferation index of primary breast cancer. Eur. J. Cancer 29A:893-897. [DOI] [PubMed] [Google Scholar]
- 11.Eschenfeldt, W. H., and S. L. Berger. 1986. The human prothymosin alpha gene is polymorphic and induced upon growth stimulation: evidence using a cloned cDNA. Proc. Natl. Acad. Sci. USA 83:9403-9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evstafieva, A. G., G. A. Belov, M. Kalkum, N. V. Chichkova, A. A. Bogdanov, V. I. Agol, and A. B. Vartapetian. 2000. Prothymosin alpha fragmentation in apoptosis. FEBS Lett. 467:150-154. [DOI] [PubMed] [Google Scholar]
- 13.Evstafieva, A. G., G. A. Belov, Y. P. Rubtsov, M. Kalkum, B. Joseph, N. V. Chichkova, E. A. Sukhacheva, A. A. Bogdanov, R. F. Pettersson, V. I. Agol, and A. B. Vartapetian. 2003. Apoptosis-related fragmentation, translocation, and properties of human prothymosin alpha. Exp. Cell Res. 284:211-223. [DOI] [PubMed] [Google Scholar]
- 14.Gomez-Marquez, J., F. Segade, M. Dosil, J. G. Pichel, X. R. Bustelo, and M. Freire. 1989. The expression of prothymosin alpha gene in T lymphocytes and leukemic lymphoid cells is tied to lymphocyte proliferation. J. Biol. Chem. 264:8451-8454. [PubMed] [Google Scholar]
- 15.Hayes, J. D., and M. McMahon. 2001. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer Lett. 174:103-113. [DOI] [PubMed] [Google Scholar]
- 16.Huang, H. C., T. Nguyen, and C. B. Pickett. 2002. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 277:42769-42774. [DOI] [PubMed] [Google Scholar]
- 17.Ishii, T., K. Itoh, S. Takahashi, H. Sato, T. Yanagawa, Y. Katoh, S. Bannai, and M. Yamamoto. 2000. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 275:16023-16029. [DOI] [PubMed] [Google Scholar]
- 18.Itoh, K., T. Chiba, S. Takahashi, T. Ishii, K. Igarashi, Y. Katoh, T. Oyake, N. Hayashi, K. Satoh, I. Hatayama, M. Yamamoto, and Y. Nabeshima. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236:313-322. [DOI] [PubMed] [Google Scholar]
- 19.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, T. O'Connor, and M. Yamamoto. 2003. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8:379-391. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, X., H. E. Kim, H. Shu, Y. Zhao, H. Zhang, J. Kofron, J. Donnelly, D. Burns, S. C. Ng, S. Rosenberg, and X. Wang. 2003. Distinctive roles of PHAP proteins and prothymosin-alpha in a death regulatory pathway. Science 299:223-226. [DOI] [PubMed] [Google Scholar]
- 22.Kang, M. I., A. Kobayashi, N. Wakabayashi, S. G. Kim, and M. Yamamoto. 2004. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Natl. Acad. Sci. USA 101:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karetsou, Z., A. Kretsovali, C. Murphy, O. Tsolas, and T. Papamarcaki. 2002. Prothymosin alpha interacts with the CREB-binding protein and potentiates transcription. EMBO Rep. 3:361-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levonen, A. L., A. Landar, A. Ramachandran, E. K. Ceaser, D. A. Dickinson, G. Zanoni, J. D. Morrow, and V. M. Darley-Usmar. 2004. Cellular mechanisms of redox cell signaling: the role of cysteine modification in controlling antioxidant defenses in response to electrophilic lipid oxidation products. Biochem. J. 378:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manrow, R. E., A. R. Sburlati, J. A. Hanover, and S. L. Berger. 1991. Nuclear targeting of prothymosin alpha. J. Biol. Chem. 266:3916-3924. [PubMed] [Google Scholar]
- 26.Martini, P. G., R. Delage-Mourroux, D. M. Kraichely, and B. S. Katzenellenbogen. 2000. Prothymosin alpha selectively enhances estrogen receptor transcriptional activity by interacting with a repressor of estrogen receptor activity. Mol. Cell. Biol. 20:6224-6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMahon, M., K. Itoh, M. Yamamoto, S. A. Chanas, C. J. Henderson, L. I. McLellan, C. R. Wolf, C. Cavin, and J. D. Hayes. 2001. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 61:3299-3307. [PubMed] [Google Scholar]
- 28.McMahon, M., K. Itoh, M. Yamamoto, and J. D. Hayes. 2003. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 278:21592-21600. [DOI] [PubMed] [Google Scholar]
- 29.Motohashi, H., F. Katsuoka, J. D. Engel, and M. Yamamoto. 2004. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc. Natl. Acad. Sci. USA 101:6379-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orre, R. S., M. A. Cotter II, C. Subramanian, and E. S. Robertson. 2001. Prothymosin alpha functions as a cellular oncoprotein by inducing transformation of rodent fibroblasts in vitro. J. Biol. Chem. 276:1794-1799. [DOI] [PubMed] [Google Scholar]
- 31.Ramos-Gomez, M., M. K. Kwak, P. M. Dolan, K. Itoh, M. Yamamoto, P. Talalay, and T. W. Kensler. 2001. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. USA 98:3410-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez, P., J. E. Vinuela, L. Alvarez-Fernandez, M. Buceta, A. Vidal, F. Dominguez, and J. Gomez-Marquez. 1998. Overexpression of prothymosin alpha accelerates proliferation and retards differentiation in HL-60 cells. Biochem. J. 331:753-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez, P., J. E. Vinuela, L. Alvarez-Fernandez, and J. Gomez-Marquez. 1999. Prothymosin alpha antisense oligonucleotides induce apoptosis in HL-60 cells. Cell Death Differ. 6:3-5. [DOI] [PubMed] [Google Scholar]
- 34.Rubtsov, Y. P., A. S. Zolotukhin, I. A. Vorobjev, N. V. Chichkova, N. A. Pavlov, E. M. Karger, A. G. Evstafieva, B. K. Felber, and A. B. Vartapetian. 1997. Mutational analysis of human prothymosin alpha reveals a bipartite nuclear localization signal. FEBS Lett. 413:135-141. [DOI] [PubMed] [Google Scholar]
- 35.Rushmore, T. H., M. R. Morton, and C. B. Pickett. 1991. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 266:11632-11639. [PubMed] [Google Scholar]
- 36.Sburlati, A. R., R. E. Manrow, and S. L. Berger. 1991. Prothymosin alpha antisense oligomers inhibit myeloma cell division. Proc. Natl. Acad. Sci. USA 88:253-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sukhacheva, E. A., A. G. Evstafieva, T. V. Fateeva, V. R. Shakulov, N. A. Efimova, R. N. Karapetian, Y. P. Rubtsov, and A. B. Vartapetian. 2002. Sensing prothymosin alpha origin, mutations and conformation with monoclonal antibodies. J. Immunol. Methods 266:185-196. [DOI] [PubMed] [Google Scholar]
- 38.Tsitsiloni, O. E., J. Stiakakis, A. Koutselinis, J. Gogas, C. Markopoulos, P. Yialouris, S. Bekris, D. Panoussopoulos, V. Kiortsis, W. Voelter, and A. A. Haritos. 1993. Expression of alpha-thymosins in human tissues in normal and abnormal growth. Proc. Natl. Acad. Sci. USA 90:9504-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vareli, K., M. Frangou-Lazaridis, I. van der Kraan, O. Tsolas, and R. van Driel. 2000. Nuclear distribution of prothymosin alpha and parathymosin: evidence that prothymosin alpha is associated with RNA synthesis processing and parathymosin with early DNA replication. Exp. Cell Res. 257:152-161. [DOI] [PubMed] [Google Scholar]
- 40.Wakabayashi, N., K. Itoh, J. Wakabayashi, H. Motohashi, S. Noda, S. Takahashi, S. Imakado, T. Kotsuji, F. Otsuka, D. R. Roop, T. Harada, J. D. Engel, and M. Yamamoto. 2003. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 35:238-245. [DOI] [PubMed] [Google Scholar]
- 41.Wakabayashi, N., A. T. Dinkova-Kostova, W. D. Holtzclaw, M. I. Kang, A. Kobayashi, M. Yamamoto, T. W. Kensler, and P. Talalay. 2004. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. USA 101:2040-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wanker, E. E., C. Rovira, E. Scherzinger, R. Hasenbank, S. Walter, D. Tait, J. Colicelli, and H. Lehrach. 1997. HIP-I: a huntingtin interacting protein isolated by the yeast two-hybrid system. Hum. Mol. Genet. 6:487-495. [DOI] [PubMed] [Google Scholar]
- 43.Wu, C. L., A. L. Shiau, and C. S. Lin. 1997. Prothymosin alpha promotes cell proliferation in NIH3T3 cells. Life Sci. 61:2091-2101. [DOI] [PubMed] [Google Scholar]
- 44.Xue, F., and L. Cooley. 1993. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72:681-693. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, D. D., and M. Hannink. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 23:8137-8151. [DOI] [PMC free article] [PubMed] [Google Scholar]