Abstract
Activation of thermogenic beige adipocytes has recently emerged as a promising therapeutic target in obesity and diabetes. Relevant human models for beige adipocyte differentiation are essential to implement such therapeutic strategies. We report a straightforward and efficient protocol to generate functional human beige adipocytes from human induced pluripotent stem cells (hiPSCs). Without overexpression of exogenous adipogenic genes, our method recapitulates an adipogenic developmental pathway through successive mesodermal and adipogenic progenitor stages. hiPSC-derived adipocytes are insulin sensitive and display beige-specific markers and functional properties, including upregulation of thermogenic genes, increased mitochondrial content, and increased oxygen consumption upon activation with cAMP analogs. Engraftment of hiPSC-derived adipocytes in mice produces well-organized and vascularized adipose tissue, capable of β-adrenergic–responsive glucose uptake. Our model of human beige adipocyte development provides a new and scalable tool for disease modeling and therapeutic screening.
Introduction
The role of adipose tissue in the regulation of energy metabolism has been recently revisited with the discovery of brown and beige fat in human adults (1). Whereas white adipocytes store triglycerides, brown adipocytes carry out efficient thermogenesis through mitochondrial uncoupling. A third type of adipocytes, named “brite” or “beige,” was recently shown to also dissipate energy upon induction by thermogenic stimuli (2,3). Beige adipocytes display a distinct molecular signature and may constitute the majority of energy-dissipating cells in human adults. As thermogenic activity of adipose tissue inversely correlates with the risk of obesity and diabetes (4), therapies aiming to activate beige adipocytes may offer perspectives to face metabolic diseases.
As prerequisite, robust knowledge of developmental pathways leading to brown or beige adipocytes in humans is paramount (5). Human induced pluripotent stem cells (hiPSCs) could provide highly relevant models to achieve this goal; however, current protocols of hiPSC differentiation into adipocytes are relatively inefficient. These rely on derivation of mesenchymal stem cells (MSCs) or embryoid bodies before applying an adipogenic stimulus (6) or on overexpression of adipogenic genes (7). These protocols however bypass key adipogenic signaling pathways, hampering developmental and physiological studies.
We report an efficient and scalable protocol to differentiate hiPSCs into beige adipocytes involving successive mesodermal and adipogenic induction steps. This unlimited source of human adipocytes, able to produce well-organized fat after engraftment in vivo, represents a powerful tool to model adipose tissue pathophysiology and develop new therapeutic approaches.
Research Design and Methods
hiPSCs and Adipogenic Differentiation
We used hiPSC lines reprogrammed from fibroblasts of three individuals (Supplementary Table 1). Mesoderm differentiation was induced on day 0 (D0) in STEMPro34 with 2 mmol/L GlutaMAX (Life Technologies), 50 µg/mL ascorbic acid (Sigma-Aldrich), 10 ng/mL bone morphogenic protein-4 (BMP4), and 25 ng/mL activin A (R&D Systems). On D4, adipose differentiation was induced in DMEM/F12 (Life Technologies) with 10% FCS, 10 μg/mL insulin, 500 μmol/L isobutylmethylxanthine (IBMX), 1 μmol/L dexamethasone, and 50 μmol/L indomethacin (Sigma-Aldrich). Adipocytes were cultured in DMEM/F12 with 10% FCS and 1 μg/mL insulin (D10 to D20).
Transcriptome Analysis
RNA was isolated from hiPSC#1 on D0, D4, D8, D12, and D20 of differentiation and hybridized onto Affymetrix Human Gene 2.0 ST arrays.
Immunofluorescence microscopy, immunohistochemistry, RT-quantitative PCR (RT-qPCR), and Western blot were performed using standard procedures (Supplementary Tables 2 and 3). Protein and mRNA extracts from PAZ6 cells, before and/or after 20 days of differentiation, were obtained from Antonio Vidal-Puig (Institute of Metabolic Science, Cambridge, U.K.).
Oxygen Consumption Measurements
Cells were harvested on D20 and transferred to Oxoplate OP96C (PreSens). PO2 was measured using FlexStation3 (Molecular Devices) supplied with SoftMax ProMicroplate Data Acquisition and Analysis Software.
Adipocyte Transplantation
Ten million hiPSC#1 were harvested on D18 of differentiation (TrypLE Express; Life Technologies), resuspended in a DMEM/F12/Matrigel solution (1:1) containing 10 μg/mL insulin, 100 μmol/L IBMX, 1 μmol/L dexamethasone, and 50 μmol/L indomethacin, and subcutaneously injected in the back of 6-week-old FoxN1Nu athymic mice (Taconic Biosciences). As controls, 3 × 107 hiPSC-derived MSCs differentiated as previously described (8) and characterized by FACS (Supplementary Table 2), or Matrigel only, were injected subcutaneously in the sternum region of the same mice. Engrafted cells were excised after 30 days.
In Vivo Stimulation and 18Fluorodeoxyglucose Uptake Analysis of Neoformed Fat Pads
Mice were subcutaneously injected daily with 100 μL isoproterenol (5 μmol/L; Sigma-Aldrich) or PBS for 7 days, fasted for 6 h with free access to water, and then injected with 100 μL isoproterenol and 5 MBq 18fluorodeoxyglucose (18FDG) in the retro-orbital sinus. After 75 min, mice were euthanized and fat pads removed and weighed. Counts per minute of 18F were measured (gamma counter Wizard 1480; PerkinElmer). The percentage of injected 18FDG per gram of tissue was calculated after correction (18F decay).
Statistical Analyses
Mean ± SEM of at least three independent experiments are shown. Significance (P < 0.05) was tested with nonparametric unpaired Mann-Whitney U test unless indicated otherwise.
Results
Adipogenic Differentiation of hiPSCs via Small Molecule–Driven Mesodermal Induction
Adipogenesis and angiogenesis are tightly interdependent during embryonic development and adulthood (9), and human beige adipocyte progenitors proliferate in association with capillary networks (10). We therefore rationalized that inducing mesodermal differentiation of hiPSCs in a medium supporting hematopoietic differentiation would enhance subsequent white and/or beige adipose differentiation.
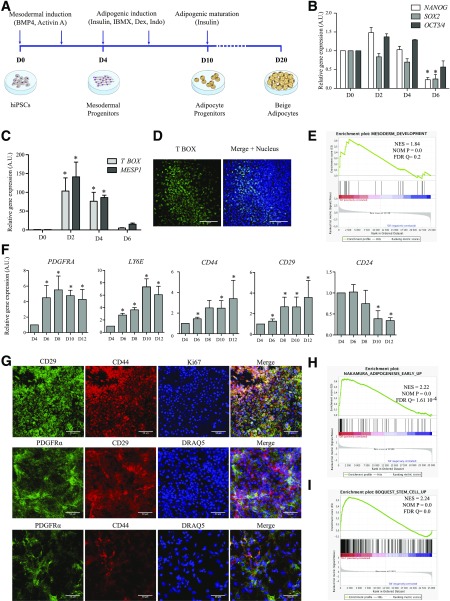
To test this hypothesis, we subjected three hiPSC lines (Supplementary Table 1 and Supplementary Fig. 1A–E) to a three-step adipogenic differentiation procedure recapitulating mesodermal induction, adipogenic stimulation, and adipogenic maturation (Fig. 1A). First, BMP4 and activin A, two mesodermal inducers, elicit differentiation of subconfluent plated hiPSCs. In addition to the decreased expression of pluripotency genes from D0 onwards (Fig. 1B and Supplementary Fig. 1F), mesodermal induction is shown by transient expression of transcripts of the mesodermal markers TBOX (T-brachyury homolog) and MESP1 (Fig. 1C and Supplementary Fig. 1G), expression of T BOX protein (Fig. 1D), and transcriptome analysis revealing significant correlations of genes directly implicated in mesoderm development on D4 (Fig. 1E and Supplementary Fig. 3C).
Figure 1.
hiPSC-derived mesodermal progenitors efficiently differentiate into adipose progenitors. A: Experimental scheme for the differentiation of hiPSCs into beige adipocytes. RT-qPCR analysis of expression of indicated pluripotency markers (B) and mesodermal transcription factors (C) in hiPSC#1 clone from D0 to D6 (mean ± SEM arbitrary units [A.U.] relative to D0; n ≥ 3 independent experiments; *P < 0.05 relative to D0, Mann-Whitney U test). D: T BOX immunostaining of mesodermal progenitors issued from hiPSC#1 (D4). Nuclei were stained with DRAQ5 (blue). Scale bars = 100 µm. E: GSEA of hiPSC-derived cells on D4 vs. D0 using the mesoderm development Gene Ontology gene set (GO:0007498); n ≥ 3 experiments. F: Time-course RT-qPCR analysis of expression of indicated genes after adipogenic induction of hiPSC#1 (D4 to D12; mean ± SEM A.U. relative to D4; n ≥ 3 experiments; *P < 0.05 relative to D4; Mann-Whitney U test). G: Immunodetection of markers of adipose progenitors and of cell proliferation (Ki67) on D10. Nuclei were stained with DRAQ5. Scale bars = 50 μm. H and I: GSEA of hiPSC-derived cells using published data sets, with early adipogenesis transcriptomic signatures on D8 vs. D4 (H) and human adult adipose stromal cell signature on D12 vs. D4 (I) (n = 3 independent experiments).
Second, D4 mesodermal precursors are subjected to an adipogenic cocktail without thiazolidinedione. From D4 to D12, mRNA expression of the adipose progenitor cell markers PDGFRA, LY6E (human homolog of murine Sca1), CD44, and CD29 increases, whereas CD24 expression decreases, in agreement with an adipogenic commitment (11) (Fig. 1F and Supplementary Fig. 2A). PDGFRα, CD44, and CD29 proteins are coexpressed, and cells are positive for the proliferation marker Ki67 (Fig. 1G). Transcriptome analysis reveals consecutive waves of transcriptional activation and repression consistent with recent transcriptome profiling by RNA sequencing in differentiating human primary adipose progenitors (Supplementary Fig. 3A and B) (12). Gene set enrichment analysis (GSEA) further indicates that on D8 relative to D4, genes upregulated in hiPSC-derived cells strongly correlate with gene sets upregulated during early adipogenic differentiation (13) (Fig. 1H and Supplementary Fig. 3D). On D12, the transcriptome of hiPSC-derived adipose progenitors is remarkably similar to that of adult human primary adipose stem cells (Fig. 1I and Supplementary Fig. 3E) (14). Thus, adipogenic induction of hiPSC-derived mesodermal precursors drives the emergence of proliferating adipose progenitors.
High-Yield Adipogenic Differentiation
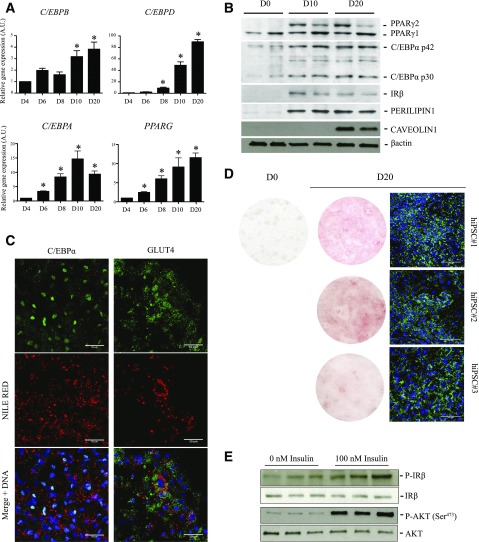
The third step of differentiation consists of culturing hiPSC-derived adipose progenitors (from D10 onwards) under conditions supporting adipogenic differentiation and maturation. We find that transcripts of CCAAT/enhancer-binding proteins CEBPA, CEBPB, and CEBPD and of the master adipogenic transcription factor PPARG are robustly induced from D10 to D20 (Fig. 2A and Supplementary Fig. 2B). In accordance, the adipose-specific protein isoforms PPARγ2 and C/EBPα p30/p42 and the mature adipose proteins insulin receptor (IR), perilipin 1, caveolin 1, and GLUT4 are induced (Fig. 2B and C). hiPSC-derived adipocytes store triglycerides, as shown by Nile Red, Oil Red O, and BODIPY staining, and respond to insulin through phosphorylation of the IR-β subunit and its downstream target AKT/PKB (Fig. 2C–E). Collectively, these results demonstrate that hiPSCs efficiently differentiate into functional, hormone-responsive adipocytes within 20 days.
Figure 2.
hiPSCs differentiate into adipocytes. A: Time-course mRNA expression of indicated adipocyte transcription factors during differentiation of hiPSC#1 (mean ± SEM arbitrary units [A.U.] relative to D4; n ≥ 3 experiments; *P < 0.05 relative to D4, Mann-Whitney U test). B: Detection of PPARγ1, PPARγ2, C/EBPα p30/p42, IR-β, perilipin 1, and caveolin 1 in hiPSC#1 on D0, D10, and D20. β-Actin was used as loading control. Duplicate lanes shown are from duplicate differentiation wells. C: Immunodetection of C/EBPα and GLUT4 in lipid-containing (Nile Red) adipocytes (hiPSC#1) on D20. Scale bars = 50 µm. D: Oil Red O (left panels) and BODIPY (right panel) staining of adipocyte-differentiated hiPSC#1, hiPSC#2, and hiPSC#3 on D0 and/or D20. Cells were fixed in 3.2% paraformaldehyde for 20 min and incubated for 1 h in Oil Red O (O0625; Sigma-Aldrich) diluted in isopropanol. Scale bars = 100 μm. E: Short-term insulin-mediated phosphorylation of IR-β and AKT/PKB evaluated using phosphospecific and pan antibodies in hiPSC#1 on D20. Differentiated adipocytes were deprived in insulin and serum for 12 h before 10 min incubation in 100 nmol/L insulin (Sigma-Aldrich), and then immediately lysed and processed for Western blotting. Triplicate lanes are from three independent stimulation experiments.
hiPSC-Derived Adipocytes Display a Beige Phenotype In Vitro
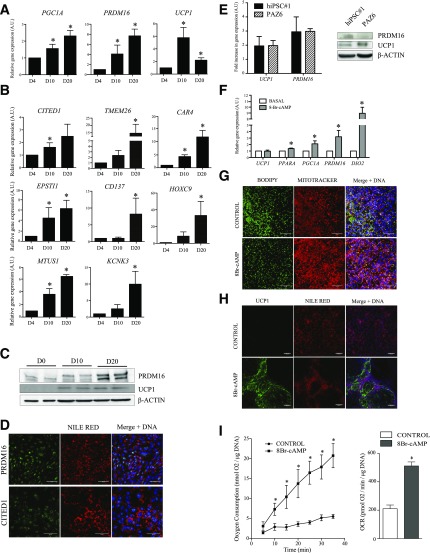
Lineage tracing studies identified PDGFRα+ cells as bipotential adipose progenitors able to differentiate toward white and beige adipocytes in vivo (15). Therefore, we examined beige/brown markers expressed from D4 to D20 in hiPSC-derived cells. Remarkably, we note the mRNA induction of uncoupling protein-1 (UCP1) and of the transcription factors PGC1A and PRDM16 that characterize beige and brown adipocytes (5) (Fig. 3A and Supplementary Fig. 2C). However, progenitor or mature brown adipocyte markers MYF5 or ZIC1 are not detected throughout differentiation (not shown), suggesting that hiPSC-derived cells are not of the brown adipocyte lineage. In addition, the beige adipocyte markers CITED1, TMEM26, CD137, HOXC9, EPSTI1, CAR4, MTUS1, and KCNK3 (16) are upregulated during differentiation (Fig. 3B and Supplementary Fig. 2D) and we confirm PRDM16 and UCP1 protein expression by Western blotting (Fig. 3C). Moreover, PRDM16 and CITED1 proteins are clearly detected by immunofluorescence in nuclei of Nile Red–positive cells (Fig. 3D). We also compared the expression of the thermogenic markers UCP1 and PRDM16 in hiPSC-derived adipocytes and in PAZ6 cells, an established model of human thermogenic adipocytes (17). Protein and mRNA expression of these markers were comparable in both cell models, studied at baseline and/or after 20 days of differentiation (Fig. 3E). These results confirm the thermogenic phenotype of hiPSC-derived adipocytes and collectively indicate that hiPSC-derived differentiated cells harbor a beige phenotype.
Figure 3.
hiPSC-derived adipocytes display beige properties. Time-course mRNA expression of beige/brown adipocyte markers (A) and beige adipocyte-specific genes (B) (mean ± SEM arbitrary units [A.U.] relative to D4; n ≥ 3 experiments; *P < 0.05 relative to D4, Mann-Whitney U test). C: PRDM16 and UCP1 protein expression on D0, D10, and D20. D: PRDM16 and CITED1 immunostaining on D20 in lipid-containing (Nile Red) adipocytes. Scale bars = 50 µm. E: Comparison of PRDM16 and UCP1 mRNA and protein expression in hiPSC-derived adipocytes and in PAZ6 cells, at baseline and/or after 20 days of differentiation. F: Relative levels of expression of indicated genes in hiPSC#1-derived adipocytes at D20 treated or not with 1 mmol/L 8Br-cAMP (Sigma-Aldrich) for 48 h (mean ± SEM A.U. relative to D4; n ≥ 3 experiments; *P < 0.05 relative to D4; Mann-Whitney U test). Mitochondrial content (MitoTracker) (G) and UCP1 immunostaining (H) in lipid-containing cells (BODIPY or Nile Red, as indicated) analyzed as in F. Cells were incubated with 1 μmol/L MitoTracker Red CMXRos (Life Technologies) for 45 min and fixed with 3.2% paraformaldehyde for 15 min. Scale bars = 50 µm. I: Time-course analysis of oxygen consumption (left) and oxygen consumption rates (OCR; right) in hiPSC#1-derived adipocytes at D20 stimulated or not with 8Br-cAMP (mean ± SEM; n ≥ 3; *P < 0.05 relative to control, Mann-Whitney U test). PO2 values were plotted over time to calculate OCR from the slope.
A key functional feature of beige adipocytes is their capacity to activate a thermogenic program under long-term β-adrenergic stimulation (3). Accordingly, treatment of D20 hiPSC-derived adipocytes with the cAMP analog 8Br-cAMP for 48 h significantly upregulates thermogenic genes, including PGC1A, PRDM16, PPARA, and DIO2 (Fig. 3F), increases intracellular mitochondrial content and UCP1 staining, and induces a shrinking of lipid droplets (Fig. 3G and H). We did not observe any increase in the mRNA expression of UCP1 after 8Br-cAMP stimulation in our model (Fig. 3F). However, it was previously shown that UCP1 mRNA levels do not consistently correlate with the thermogenic capacity of beige adipocytes (18). In addition, hiPSC-derived adipocytes display a 2.5-fold increase in oxygen consumption in response to acute 8Br-cAMP stimulation (Fig. 3I). Thus, hiPSC-derived adipocytes show typical beige functional properties.
hiPSC-Derived Adipocytes Give Rise to Adipose Tissue In Vivo
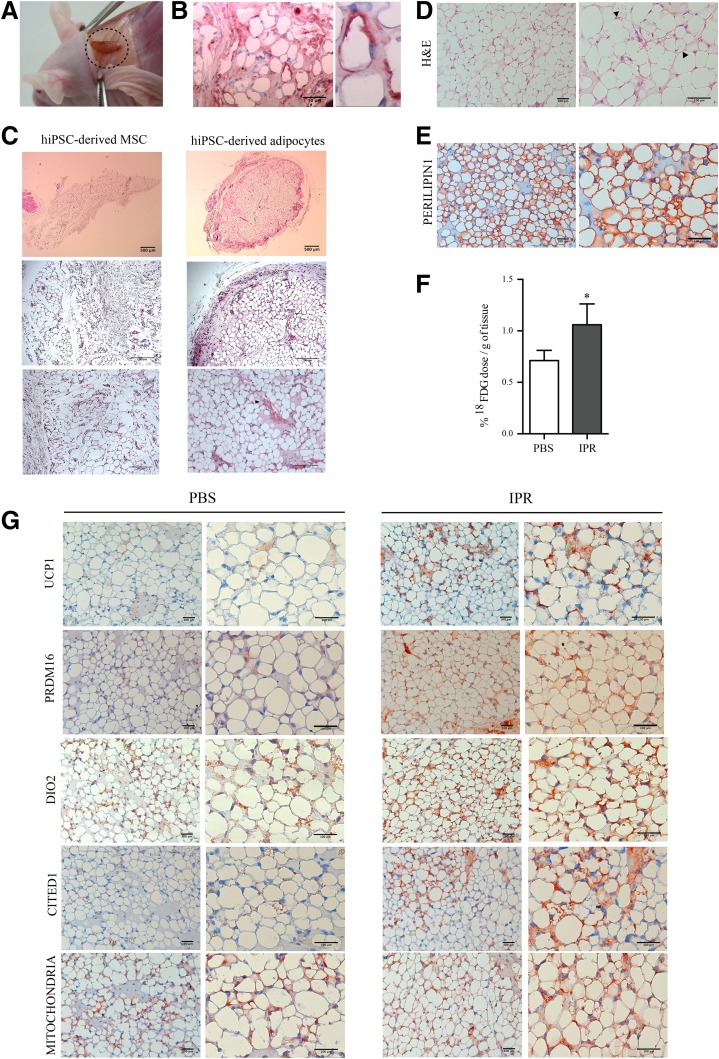
Subcutaneous injection of hiPSC-derived adipocytes specifically results in the neo-formation of fat pads positively stained for human-specific antibodies (Fig. 4A, B, and G and data not shown). hiPSC-derived adipocytes were organized in a fully differentiated and vascularized adipose tissue, in contrast to tissues formed from hiPSC-derived MSCs (Fig. 4C). Moreover, hiPSC-derived adipose tissue contains clusters of terminally differentiated beige multilocular adipocytes, as shown by perilipin 1, UCP1, PRDM16, DIO2, and CITED1 staining (Fig. 4D, E, and G).
Figure 4.
hiPSC-derived adipocytes generate adipose tissue in vivo. A: Macroscopic view of a neoformed fat pad developed from hiPSC-derived adipocytes. B: Human cytoplasmic staining of neoformed adipose tissue (n ≥ 3). C: Hematoxylin-eosin (H&E) staining of fat pads developed from hiPSC-derived MSCs or hiPSC-derived adipocytes. Arrowheads indicate blood vessels. Scale bars = 500 μm (top), 200 μm (middle), 100 μm (bottom) (n ≥ 3). Staining of a neoformed fat pad developed from hiPSC-derived adipocytes with H&E (D) or antibodies against perilipin 1 (E). Arrowheads indicate multilocular adipocytes. Scale bars = 100 μm (n ≥ 3 independent experiments). F: 18FDG uptake in neoformed fat pads after hiPSC-derived adipocyte engraftment in immunodeficient mice treated with vehicle (PBS) or isoproterenol (IPR; mean ± SEM; n ≥ 3 independent experiments; *P < 0.05 relative to PBS control, Wilcoxon signed rank test). G: Immunostaining of a neoformed fat pad developed from hiPSC-derived adipocytes for UCP1, PRDM16, DIO2, CITED1, and a human-specific mitochondrial marker (targeted by the MAB1273 antibody) in basal (PBS) and stimulated (IPR) conditions. Scale bars = 100 μm.
In vivo, beige and brown adipose tissues respond to β-adrenergic stimulation by increasing glucose uptake (1). Accordingly, after acute isoproterenol stimulation, 18FDG uptake is increased in neoformed pads pretreated with isoproterenol for 7 days (Fig. 4F). Importantly, in these long-term stimulated pads, UCP1, PRDM16, CITED1, and DIO2 levels are enhanced while the diameter of most adipocytes is reduced and their mitochondrial content is increased (Fig. 4G), consistent with a β-adrenergic–mediated increase in thermogenic activity.
We describe a straightforward and efficient protocol to derive from hiPSC adipocytes able to form bona fide human adipose tissue of dual white/beige phenotype. hiPSC-derived adipocytes are capable of responding to a β-adrenergic stimulation in vivo, indicative of their functional relevance for developmental and metabolic pathophysiological studies of adipose tissue.
Discussion
Metabolically active adipose tissues arise from mesoderm, and intrinsic properties of discrete body fat depots suggest distinct mesodermal origins (5). A key feature of the adipose differentiation protocol reported here is the derivation of human beige adipocytes after an initial mesodermal induction. Importantly, our protocol is distinct from existing pluripotent cell–based adipogenic protocols by its approach and end point: 1) it relies on a developmental mesodermal induction of hiPSCs rather than a differentiation through the previously described derivation of MSCs (7,19); 2) adipocytes are obtained without overexpression of adipogenic transcription factors, which have been used to force and accelerate adipocyte differentiation by others (7,20); 3) it requires a simple bidimensional cell culture, avoiding the initial derivation of embryoid bodies, which necessitates further selection and cellular dissociation, as performed classically (7,19,21–25); 4) it is fast, allowing differentiated adipocytes to be obtained within 20 days, whereas other protocols require at least 48 days (19–25); 5) it leads to functional adipocytes with a beige phenotype and respiratory function; and 6) hiPSC-derived adipocytes can form well-organized and vascularized adipose tissue in vivo, capable of β-adrenergic–responsive glucose uptake.
Adipogenic induction of mesodermal cells give rise to a progenitor population closely related to adult primary adipose stromal cells (14) and expressing markers such as PDGFRA, LY6E, CD29, and CD44 (26). Of relevance to the markers examined here, CD29+ human adipose progenitors were recently shown to display strong potential to differentiate toward thermogenic adipocytes in vitro (27). A Sca1(LY6E)+/PDGFRα+ population has also been shown to give rise to both UCP1+ and UCP1− cells in vivo in mice (28), and a PDGFRα+/CD44+ progenitor population can be differentiated into beige or white adipocytes in response to β3 agonists or to a high-fat diet (15). These studies suggest that white and beige adipocytes could derive from a PDGFRα+ common progenitor, although they may also potentially originate from bidirectional differentiation depending on the fat depot (29,30). Accordingly, in vitro and in vivo differentiation of hiPSC-derived adipose progenitors gives rise to adipocytes displaying beige markers and a functional beige adipocyte response. Mesodermal induction of hiPSCs in hematopoietic medium introduced in our protocol may favor a thermogenic adipose phenotype. In accordance, human beige adipocytes develop in association with capillary networks, and angiogenesis regulates beige adipocyte differentiation via a paracrine mechanism (10,31). In addition, BMP4, used for mesodermal induction in our protocol, may direct adipocytes toward a beige phenotype, as demonstrated for human adult preadipocytes (32). Beige adipocytes are likely present, albeit in small amounts, in various human adipose depots and may constitute an important subset of thermogenic cells in adults (2,3). The scalability of our protocol makes it a valuable tool to provide an unlimited source of cells to model beige adipose tissue development and physiology, and for metabolic investigations, including therapeutic screening.
Supplementary Material
Article Information
Acknowledgments. The authors thank C. Cowan (Harvard Department of Stem Cell and Regenerative Biology) for the hiPSC#1 cell line, A. Vidal-Puig and S. Carobbio (Institute of Metabolic Science, University of Cambridge) for the protein and mRNA extracts from PAZ6 cells, L. Douay and L. Kobari (Centre de Recherche Saint-Antoine, INSERM UMR_S938) for cDNA from hES cells, T. Ledent (Centre de Recherche Saint-Antoine, INSERM UMR_S938) for help with in vivo experiments, and S. Dumont and F. Merabtene (Unité Mixte de Service d'Imagerie et de Cytométrie [UMS30 LUMIC], INSERM/Université Pierre et Marie Curie) for help with immunohistochemistry.
Funding. This work was supported by the Institute of Cardiometabolism and Nutrition (ANR-10-IAHU-05), INSERM, Université Pierre et Marie Curie, Société Francophone du Diabète/Pierre Fabre Médicament, and SATT-Lutech. A.-C.G. was funded by the Institute of Cardiometabolism and Nutrition, and N.B. by DIM-Biotherapies/STEM Pole and Fondation de France. R.J.H. is supported by grants from the National Institutes of Health National Heart, Lung, and Blood Institute (NHLBI) (R01 HL117505, HL119046, and P50 HL112324) and an NHLBI Program of Excellence in Nanotechnology (PEN) Award (contract HHSN268201000045C). P.C. is supported by the Research Council of Norway and the University of Oslo.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. A.-C.G. designed the study, developed protocols, analyzed transcriptomic data, and wrote the manuscript. N.B. designed the study, developed protocols, analyzed data, and wrote the manuscript. E.C. maintained and characterized hiPSCs and performed differentiation experiments and histology. F.D. analyzed transcriptomic data. R.M. performed image acquisitions. C.P. performed 18FDG experiments. F.S. and D.J. provided hiPSC#2 and hiPSC#3, respectively, and helped with characterization. J.-P.S. performed karyotyping. R.J.H., B.F., J.-S.H., and P.C. contributed to the manuscript. J.C. contributed to study design and to the manuscript. C.V. contributed to study design and wrote the manuscript. A.-C.G. and N.B. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-1107/-/DC1.
References
- 1.Cypess AM, Lehman S, Williams G, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med 2009;360:1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lidell ME, Betz MJ, Dahlqvist Leinhard O, et al. Evidence for two types of brown adipose tissue in humans. Nat Med 2013;19:631–634 [DOI] [PubMed] [Google Scholar]
- 3.Cereijo R, Giralt M, Villarroya F. Thermogenic brown and beige/brite adipogenesis in humans. Ann Med 2015;47:169–177 [DOI] [PubMed] [Google Scholar]
- 4.Chondronikola M, Volpi E, Børsheim E, et al. Brown adipose tissue improves whole-body glucose homeostasis and insulin sensitivity in humans. Diabetes 2014;63:4089–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kajimura S, Spiegelman BM, Seale P. Brown and beige fat: physiological roles beyond heat generation. Cell Metab 2015;22:546–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hafner AL, Dani C. Human induced pluripotent stem cells: a new source for brown and white adipocytes. World J Stem Cells 2014;6:467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahfeldt T, Schinzel RT, Lee YK, et al. Programming human pluripotent stem cells into white and brown adipocytes. Nat Cell Biol 2012;14:209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denis JA, Rochon-Beaucourt C, Champon B, Pietu G. Global transcriptional profiling of neural and mesenchymal progenitors derived from human embryonic stem cells reveals alternative developmental signaling pathways. Stem Cells Dev 2011;20:1395–1409 [DOI] [PubMed] [Google Scholar]
- 9.Tang W, Zeve D, Suh JM, et al. White fat progenitor cells reside in the adipose vasculature. Science 2008;322:583–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Min SY, Kady J, Nam M, et al. Human ‘brite/beige’ adipocytes develop from capillary networks, and their implantation improves metabolic homeostasis in mice. Nat Med 2016;22:312–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nat Cell Biol 2013;15:302–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah A, Oldenburg A, Collas P. A hyper-dynamic nature of bivalent promoter states underlies coordinated developmental gene expression modules. BMC Genomics 2014;15:1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura T, Shiojima S, Hirai Y, et al. Temporal gene expression changes during adipogenesis in human mesenchymal stem cells. Biochem Biophys Res Commun 2003;303:306–312 [DOI] [PubMed] [Google Scholar]
- 14.Boquest AC, Shahdadfar A, Frønsdal K, et al. Isolation and transcription profiling of purified uncultured human stromal stem cells: alteration of gene expression after in vitro cell culture. Mol Biol Cell 2005;16:1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab 2012;15:480–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Jong JM, Larsson O, Cannon B, Nedergaard J. A stringent validation of mouse adipose tissue identity markers. Am J Physiol Endocrinol Metab 2015;308:E1085–E1105 [DOI] [PubMed] [Google Scholar]
- 17.Kazantzis M, Takahashi V, Hinkle J, et al. PAZ6 cells constitute a representative model for human brown pre-adipocytes. Front Endocrinol (Lausanne) 2012;3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nedergaard J, Cannon B. UCP1 mRNA does not produce heat. Biochim Biophys Acta 2013;183:943–949 [DOI] [PubMed]
- 19.Xiong ZM, LaDana C, Wu D, Cao K. An inhibitory role of progerin in the gene induction network of adipocyte differentiation from iPS cells. Aging (Albany NY) 2013;5:288–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moisan A, Lee YK, Zhang JD, et al. White-to-brown metabolic conversion of human adipocytes by JAK inhibition. Nat Cell Biol 2015;17:57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hafner AL, Contet J, Ravaud C, et al. Brown-like adipose progenitors derived from human induced pluripotent stem cells: identification of critical pathways governing their adipogenic capacity. Sci Rep 2016;6:32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishio M, Yoneshiro T, Nakahara M, et al. Production of functional classical brown adipocytes from human pluripotent stem cells using specific hemopoietin cocktail without gene transfer. Cell Metab 2012;16:394–406 [DOI] [PubMed] [Google Scholar]
- 23.Taura D, Noguchi M, Sone M, et al. Adipogenic differentiation of human induced pluripotent stem cells: comparison with that of human embryonic stem cells. FEBS Lett 2009;583:1029–1033 [DOI] [PubMed] [Google Scholar]
- 24.Noguchi M, Hosoda K, Nakane M, et al. In vitro characterization and engraftment of adipocytes derived from human induced pluripotent stem cells and embryonic stem cells. Stem Cells Dev 2013;22:2895–2905 [DOI] [PubMed] [Google Scholar]
- 25.Mohsen-Kanson T, Hafner AL, Wdziekonski B, et al. Differentiation of human induced pluripotent stem cells into brown and white adipocytes: role of Pax3. Stem Cells 2014;32:1459–1467 [DOI] [PubMed] [Google Scholar]
- 26.Berry DC, Jiang Y, Graff JM. Emerging roles of adipose progenitor cells in tissue development, homeostasis, expansion and thermogenesis. Trends Endocrinol Metab 2016;27:574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue R, Lynes MD, Dreyfuss JM, et al. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nat Med 2015;21:760–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee YH, Granneman JG. Seeking the source of adipocytes in adult white adipose tissues. Adipocyte 2012;1:230–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyvönen MT, Spalding KL. Maintenance of white adipose tissue in man. Int J Biochem Cell Biol 2014;56:123–132 [DOI] [PubMed] [Google Scholar]
- 30.Rosenwald M, Wolfrum C. The origin and definition of brite versus white and classical brown adipocytes. Adipocyte 2014;3:4–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seki T, Hosaka K, Lim S, et al. Endothelial PDGF-CC regulates angiogenesis-dependent thermogenesis in beige fat. Nat Commun 2016;7:12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gustafson B, Hammarstedt A, Hedjazifar S, et al. BMP4 and BMP antagonists regulate human white and beige adipogenesis. Diabetes 2015;64:1670–1681 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.