Abstract
Obesity results from increased energy intake or defects in energy expenditure. Brown adipose tissue (BAT) is specialized for energy expenditure, a process called adaptive thermogenesis. Peroxisome proliferator–activated receptor γ coactivator 1α (PGC1α) controls BAT-mediated thermogenesis by regulating the expression of Ucp1. Inhibitor of differentiation 1 (Id1) is a helix-loop-helix transcription factor that plays an important role in cell proliferation and differentiation. We demonstrate a novel function of Id1 in BAT thermogenesis and programming of beige adipocytes in white adipose tissue (WAT). We found that adipose tissue–specific overexpression of Id1 causes age-associated and high-fat diet–induced obesity in mice. Id1 suppresses BAT thermogenesis by binding to and suppressing PGC1α transcriptional activity. In WAT, Id1 is mainly localized in the stromal vascular fraction, where the adipose progenitor/precursors reside. Lack of Id1 increases beige gene and Ucp1 expression in the WAT in response to cold exposure. Furthermore, brown-like differentiation is increased in Id1-deficient mouse embryonic fibroblasts. At the molecular level, Id1 directly interacts with and suppresses Ebf2 transcriptional activity, leading to reduced expression of Prdm16, which determines beige/brown adipocyte cell fate. Overall, the study highlights the existence of novel regulatory mechanisms between Id1/PGC1α and Id1/Ebf2 in controlling brown fat metabolism, which has significant implications in the treatment of obesity and its associated diseases, such as diabetes.
Introduction
Obesity is a significant risk factor for a vast number of diseases, such as cardiovascular disease, type 2 diabetes, hypertension, fatty liver disease, and numerous cancers (1,2). Obesity results from increased energy intake or defects in energy expenditure. The excessive energy is stored as triglycerides in the white adipose tissue (WAT). A second type of adipose tissue, known as brown adipose tissue (BAT), has been well known for a long time in rodents and newborn humans (3). BAT is specialized for energy expenditure, a process called adaptive thermogenesis, which is a physiological mechanism during which energy is dissipated to generate heat in response to cold temperatures and possibly diet (3,4). The very densely packed mitochondria in BAT produce heat through a unique protein called uncoupling protein-1 (UCP1). UCP1 uncouples mitochondrial oxidative phosphorylation from ATP production and dissipates chemical energy as heat, thereby strongly increasing energy expenditure (5). Peroxisome proliferator–activated receptor γ (PPARγ) coactivator 1α (PGC1α) promotes BAT-mediated thermogenesis by directly regulating the expression of Ucp1. Because of their critical role in thermogenesis, PGC1α and UCP1 expression and activity are tightly controlled by several other factors that either positively or negatively regulate PGC1α and UCP1. Some of the important factors that positively regulate PGC1α or UCP1 are FOXC2, SRC1, CREB, IRF4, SIRT3, and p38 MAPK, whereas RIP140, LXRα, Cidea, pRB, SRC2, Twist-1, and TRPV4 negatively regulate PGC1α and/or UCP1 (4,6–9).
In addition to WAT and BAT, a third type of adipocytes arises in the body in response to certain physiological stimuli. For example, in response to cold and β-androgen receptor or PPARγ agonists, pools of UCP1-expressing brown-like adipocytes are detected in mouse WAT (10,11). These adipocytes are called beige or brite cells. Beige and brown adipocytes share a number of thermogenic genes, such as Pgc1a, Ucp1, Cidea, and Prdm16; however, beige cells also express several unique genes such as CD40, CD137, Tmem26, and Tbx1 that apparently reflect their distinct developmental origin (12). Lineage-tracing studies revealed that the beige adipocytes are derived from adipose progenitor/precursor cells (13). In these cells, early B-cell factor-2 (Ebf2) and PPARγ cooperatively induce the expression of Prdm16, which determines the beige adipocyte cell fate (14–16). PRDM16 can bind to and promote the transcriptional activity of numerous factors, such as PPARα, PPARγ, and PGC1α, thus functioning as a critical activator of brown/beige adipocyte cell fate (15,17). Induced expression of Prdm16 or Ebf2 is sufficient to convert white adipose precursors into brown-like UCP1-expressing cells in vitro (14,17). Similarly, forced expression of Ebf2 or Prdm16 stimulates beige cell formation in subcutaneous WAT, and transgenic mice display increased energy expenditure and reduced weight gain in response to a high-fat diet (HFD) (18,19).
Inhibitor of differentiation 1 (Id1) is a helix-loop-helix transcription factor that lacks a DNA binding domain (DBD). Id1 directly binds to other transcription factors and suppresses their transcriptional activity, thereby playing an important role in a number of cellular processes, such as cell proliferation, differentiation, hematopoiesis, and cancer cell metabolic adaptation (20–22). However, the adipose tissue–specific function of Id1 and its relative contribution to body energy expenditure remains unclear. Moreover, although Id1 is expressed in both BAT and WAT, the specific involvement of Id1 in BAT-mediated thermogenesis or browning of WAT has not been fully established. To understand the adipose tissue–specific function of Id1, we have generated adipose tissue–specific Id1 transgenic mice (aP2-Id1Tg+). We discovered that adipose-specific overexpression of Id1 causes age- and HFD-associated obesity in male mice, whereas female mice are resistant to Id1-induced obesity. At the molecular level, Id1 directly binds to the central regulators of BAT thermogenesis and WAT browning, PGC1α, and Ebf2 (4,16,19) and suppresses their transcriptional activity. The results highlight a novel function of Id1 in BAT thermogenesis and WAT browning and, thus, in body energy homeostasis.
Research Design and Methods
Mice and Diet
The aP2-Id1Tg+ transgenic expression vector and mice were generated by using the services of Cyagen Biosciences (Santa Clara, CA). Of four transgenic lines generated, one line showed strong expression of Id1 in WAT and BAT and was used for this study. The following primers were used to distinguish the aP2-Id1Tg+ transgenic from control aP2-Id1Tg− mice: Tg F: ATCTTTAAAAGCGAGTTCCCT; Tg R: CTCCGACAGACCAAGTACCA; internal control F: ACTCCAAGGCCACTTATCACC; and internal control R: ATTGTTACCAACTGGGACGACA. Endogenous mouse β-actin was used as the internal control. C57BL/6J background aP2-Id1Tg+, aP2-Id1Tg−, Id1+/+, and Id1−/− mice were used for this study. Mice were housed in a barrier facility under standard conditions with a 12-h light-dark cycle. Mice were handled in compliance with National Institutes of Health guidelines for animal care and use. All animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee of Augusta University (Augusta, GA). Mice were fed a standard chow normal diet (ND) containing 6% crude fat (Harlan Teklad Rodent Diet 2918). For the HFD experiments, 1-month-old mice were switched from ND to HFD (60% kcal from fat, D12492; Research Diets, New Brunswick, NJ) and fed for 12 weeks. For cold exposure studies, mice were housed in standard cages without bedding, and the cages were placed in the cold room (4°C) for 4–12 h. Mice were then euthanized and tissues harvested.
Quantitative Real-Time PCR
Total RNA from cells and tissues was prepared by using TRIzol Reagent (15596-026; Life Technologies) according to the manufacturer’s instructions. Total RNA (2 μg) from each sample was reverse transcribed into cDNA by using a RevertAid Reverse Transcription Kit (K1691; Thermo Fisher Scientific) according to the manufacturer’s instructions. Quantitative PCR (qPCR) analysis was performed by using Power SYBR Green PCR Master Mix (4367659; Applied Biosystems) according to the manufacturer’s instructions in a 20-μL final reaction volume in 96-well plates (4346907; Applied Biosystems). The qPCR primers used are listed in Supplementary Table 1.
Statistical Analysis
The quantitative data for the experiments are presented as mean ± SD. Statistical analyses were performed by unpaired Student t test, and P < 0.05 was considered statistically significant. Oxymax metabolic data were analyzed by one-way repeated-measures ANOVA, and P < 0.01 was considered statistically significant. Detailed methods are provided in the Supplementary Data.
Results
Id1 Is Expressed in Adipose Tissues, and Its Expression Is Strongly Induced During Brown Adipocyte Differentiation
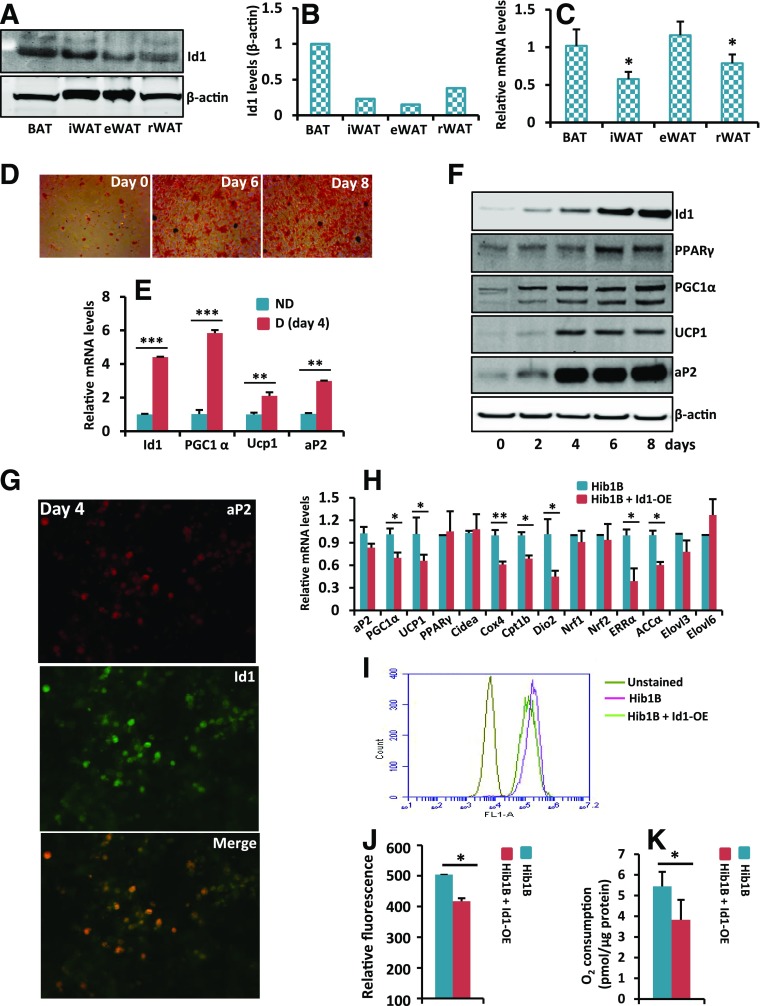
To investigate whether Id1 plays a role in adipose tissue metabolism, we analyzed the expression pattern of Id1 protein in mouse BAT, inguinal WAT (iWAT), epididymal WAT (eWAT), and retroperitoneal WAT (rWAT). Id1 was expressed in all the adipose tissues, and its expression was relatively stronger in BAT than in iWAT, eWAT, and rWAT (Fig. 1A and B). In addition, analysis of Id1 mRNA revealed a relatively higher Id1 mRNA in BAT and eWAT than in iWAT and rWAT (Fig. 1C). To investigate whether Id1 is required for brown adipocyte differentiation, we induced differentiation in the HIB1B brown preadipocyte cell line and detected a threefold induction of Id1 mRNA in day 4 differentiated cells compared with undifferentiated cells (Fig. 1D and E). The adipocyte differentiation marker aP2 and the brown adipocyte–specific thermogenic genes Pgc1α and Ucp1 also were induced in day 4 differentiated cells, as expected (Fig. 1E). Further analysis at the protein level revealed low levels of Id1 in undifferentiated HIB1B cells, and its expression was steadily elevated during the differentiation process (Fig. 1F). To further confirm that Id1 expression is induced in differentiated cells compared with nondifferentiated HIB1B cells, we performed Id1/aP2 costaining, which revealed increased Id1 staining in aP2-positive cells (Fig. 1G). These observations suggest that Id1 plays a role in brown adipocyte differentiation or in brown adipocyte–associated thermogenesis.
Figure 1.
Id1 expression is induced during brown adipocyte differentiation in HIB1B cells. A and B: Expression levels of Id1 and β-actin in various adipose tissues of 2-month-old C57BL/6J mice (A). After acquiring images, band intensities were measured and normalized to β-actin by the Li-Cor Odyssey system (B). C: Id1 mRNA transcript levels in various adipose tissues of 2-month-old C57BL/6J mice. Compared with BAT or eWAT, Id1 expression level is lower in iWAT and rWAT (n = 3). D: Oil Red O staining of nondifferentiated (day 0) and day 6 and day 8 differentiated HIB1B brown preadipocyte cells. E: Id1, aP2, PGC1α, and Ucp1 mRNA transcript levels in day 4 differentiated (D) HIB1B cells compared with nondifferentiated (ND) cells (n = 3). F: The expression patterns of Id1, PPARγ, PGC1α, UCP1, aP2, and β-actin during HIB1B brown adipocyte differentiation at the indicated time points. G: Immunofluorescence costaining of aP2 and Id1 on day 4 differentiated HIB1B cells showing strong expression of Id1 in aP2-positive cells. Original magnification ×20. H: Relative mRNA transcript levels of indicated genes in control and Id1-OE nondifferentiated HIB1B cells (n = 3). I: FACS analysis of unstained and nonyl acridine orange–stained control and Id1-OE nondifferentiated HIB1B cells. J: The relative fluorescence level (FL) of nonyl acridine orange in control and Id1-OE cells (n = 3). K: VO2 rate of control and Id1-OE nondifferentiated HIB1B cells (n = 4). Data are mean ± SD. *P < 0.05; **P < 0.005; ***P < 0.0005.
Id1 Is Not Required for Brown Adipocyte Differentiation
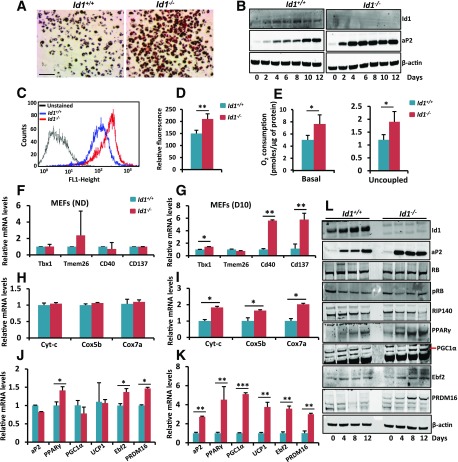
Strong induction of Id1 during brown adipocyte differentiation prompted us to ask whether Id1 promotes brown adipocyte differentiation. Id1 expression was strongly induced in differentiated HIB1B cells, whereas its expression was low in undifferentiated cells (Fig. 1F); therefore, we asked whether forced expression of Id1 in undifferentiated HIB1B cells accelerates differentiation. To this end, we generated an Id1-overexpressed (Id1-OE) HIB1B cell line (Supplementary Fig. 1A). Analysis of the expression levels of various mitochondrial and thermogenesis genes in nondifferentiated control and Id1-OE HIB1B cells revealed downregulation of Pgc1α, Ucp1, Cox4, Cpt1b, Dio2, and ERRα in Id1-OE compared with control HIB1B cells (Fig. 1H). In addition, mitochondrial content was reduced in Id1-OE cells compared with control cells (Fig. 1I and J). Consequently, Id1-OE cells displayed reduced respiration (VO2) compared with control nondifferentiated HIB1B cells (Fig. 1K). These results indicate that Id1 suppresses mitochondrial and thermogenesis genes and, thus, that Id1 could play a role in brown adipocyte differentiation. However, induction of brown adipogenesis in the control and Id1-OE HIB1B cells showed no significant difference as revealed by Oil Red O staining and the expression levels of the adipocyte differentiation marker aP2 and brown adipocyte markers PGC1α and Ucp1 in fully differentiated adipocytes (Supplementary Fig. 1B and C). To investigate whether loss of Id1 has any effect on brown adipocyte differentiation, we knocked down Id1 in HIB1B cells by short hairpin RNA (Supplementary Fig. 1D) and induced brown adipocyte differentiation. We did not observe significant differences in the differentiation between control and Id1 short hairpin RNA HIB1B cells as revealed by Oil Red O staining and the expression levels of aP2, PGC1α, and Ucp1 in fully differentiated adipocytes (Supplementary Fig. 1E and F). Together, these results suggest that Id1 is not required for brown adipocyte differentiation and that Id1 might have a different function in brown adipocytes.
Reduced Energy Expenditure and Increased Adiposity in aP2-Id1Tg+ Mice
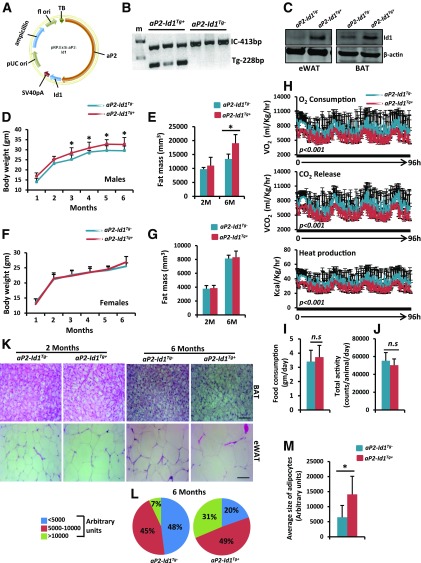
Previously, aP2 promoter–driven transgenic mice have been extensively used to study BAT thermogenesis and WAT browning (15,19,23). Therefore, to investigate the specific function of Id1 in BAT-mediated thermogenesis and adipose tissue metabolism in a more physiologically relevant in vivo system, we generated aP2-Id1Tg+ transgenic mice where Id1 is specifically overexpressed in adipose tissues (Fig. 2A–C). We found that Id1 expression is three- to fourfold higher in the adipose tissues of aP2-Id1Tg+ compared with aP2-Id1Tg− control mice (Fig. 2C). To investigate whether adipose-specific overexpression of Id1 causes metabolic alterations that lead to changes in body adiposity, we measured body weight and fat mass in aP2-Id1Tg+ and control mice fed an ND. The male aP2-Id1Tg+ mice steadily gained more body weight, and the body weight and fat mass were significantly increased in 6-month-old adult aP2-Id1Tg+ males compared with aP2-Id1Tg− males (Fig. 2D and E), whereas lean mass, bone mass, and interscapular BAT weight were unchanged between the two genotypes (Supplementary Fig. 2A–E). In contrast, females did not show differences in body weight and fat mass between the two genotypes in 2-month-old (young) and 6-month-old (adult) mice (Fig. 2F and G). These observations suggest that females are resistant to Id1-induced weight gain, whereas aP2-Id1Tg+ male mice accumulate more fat mass over time. To investigate whether increased fat mass and weight gain in aP2-Id1Tg+ mice are due to changes in metabolic activity, we measured various metabolic parameters in 2- and 6-month-old mice by indirect open-circuit calorimeter (Oxymax/Comprehensive Lab Animal Monitoring System [CLAMS]), and detected reduced VO2, VCO2, and heat production (thermogenesis) in aP2-Id1Tg+ compared with aP2-Id1Tg− mice (Fig. 2H and Supplementary Fig. 3A), indicating that body energy expenditure is reduced in aP2-Id1Tg+ male mice. Physical activity and food consumption were unchanged between the two genotypes (Fig. 2I and J and Supplementary Fig. 3B and C), further confirming that reduced energy expenditure is mainly responsible for increased body weight and fat mass gain in aP2-Id1Tg+ male mice. Consequently, we observed increased lipid accumulation in the BAT and white adipocyte hypertrophy in 6-month-old aP2-Id1Tg+ compared with aP2-Id1Tg− mice (Fig. 2K–M). These observations suggest that adipose tissue–specific overexpression of Id1 suppresses body energy expenditure and promotes weight gain in male mice.
Figure 2.
Adipose tissue–specific overexpression of Id1 reduces energy expenditure. A: The aP2-Id1 expression vector used to generate adipose tissue–specific aP2-Id1Tg+ transgenic mice. f1 ori, f1 single-strand RNA origin; pUC ori, pUC origin; TB, transcription blocker. B: Gel photograph showing the PCR bands of internal control (IC) (β-actin) and Id1 transgene. C: Expression levels of Id1 protein and β-actin in the eWAT and BAT of aP2-Id1Tg− control and aP2-Id1Tg+ mice. D: Body weight of male aP2-Id1Tg− and aP2-Id1Tg+ mice (n = 8–10) at the indicated time points. E: Fat mass density in 2- and 6-month-old aP2-Id1Tg− and aP2-Id1Tg+ male mice measured by multislice computed tomography scanner (n = 5–6). F: Body weight of female aP2-Id1Tg− and aP2-Id1Tg+ mice (n = 8–10) at the indicated time points. G: Fat mass density in 2- and 6-month-old aP2-Id1Tg− and aP2-Id1Tg+ female mice measured by multislice computed tomography scanner (n = 5). Data are mean ± SD. H–J: VO2 rate, VCO2, and heat production (H); food consumption (I); and physical activity (J) in aP2-Id1Tg− and aP2-Id1Tg+ male mice measured for 4 days with Oxymax/CLAMS (n = 5–6). K: Representative hematoxylin-eosin–stained BAT and eWAT sections. Scale bars = 10 μm. L and M: Adipocyte size distribution (L) and the average size of adipocytes (M) in the eWAT of 6-month-old aP2-Id1Tg− and aP2-Id1Tg+ male mice. Data are mean ± SD. *P < 0.05. bp, base pair; M, month; n.s, not significant.
aP2-Id1Tg+ Male Mice Are Prone to HFD-Induced Obesity
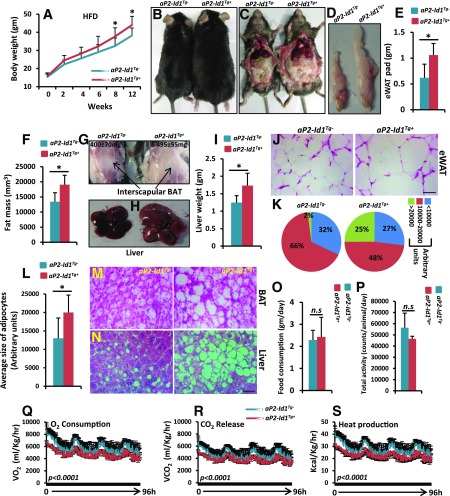
On an ND (6% kcal from fat), aP2-Id1Tg+ male mice gained more body weight over time and displayed reduced energy expenditure (Fig. 2D, E, and H). Therefore, we asked whether aP2-Id1Tg+ mice are prone to HFD-induced obesity. In response to 12 weeks of HFD (60% kcal from fat), aP2-Id1Tg+ mice gained more body weight than aP2-Id1Tg− mice (Fig. 3A and B). The aP2-Id1Tg+ mice showed an increased amount of visceral fat (Fig. 3C), larger eWAT pads (Fig. 3D and E), increased accumulation of body fat (Fig. 3F), and higher blood glucose, insulin, and leptin levels (Supplementary Table 2) than aP2-Id1Tg− mice. Consequently, aP2-Id1Tg+ male mice displayed increased accumulation of lipids in and around the interscapular BAT (Fig. 3G) and increased liver size and weight compared with aP2-Id1Tg− mice (Fig. 3H and I). Histological analysis of WAT from aP2-Id1Tg+ mice showed white adipocyte hypertrophy (Fig. 3J–L) and increased lipid accumulation in the BAT and liver compared with aP2-Id1Tg− mice (Fig. 3M and N). To investigate whether increased weight gain in aP2-Id1Tg+ mice in response to HFD is due to changes in metabolic activity, we measured various metabolic parameters. Food consumption and physical activity were unchanged between the two genotypes (Fig. 3O and P), but reduced VO2, VCO2, and thermogenesis in aP2-Id1Tg+ compared with aP2-Id1Tg− mice were detected (Fig. 3Q–S). Moreover, thermogenic and mitochondrial gene expression was reduced in the BAT of HFD-fed aP2-Id1Tg+ compared with aP2-Id1Tg− mice (Supplementary Fig. 3D and E). In contrast to males, aP2-Id1Tg+ females did not gain significant body weight compared with aP2-Id1Tg− controls in response to HFD (data not shown). Together, these results indicate that aP2-Id1Tg+ male mice are prone to HFD-induced obesity as a result of reduced energy expenditure.
Figure 3.
aP2-Id1Tg+ mice are prone to HFD-induced obesity. A: Body weights of HFD-fed male aP2-Id1Tg− and aP2-Id1Tg+ mice (n = 9) at the indicated time points. B–D: Representative photographs of aP2-Id1Tg− and aP2-Id1Tg+ male mice (B), visceral fat (C), and eWAT pads (D) after 12 weeks of HFD. E: Average weight of eWAT pads in aP2-Id1Tg− and aP2-Id1Tg+ mice (n = 6). F: Fat mass density in aP2-Id1Tg− and aP2-Id1Tg+ male mice after 12 weeks of HFD measured by multislice computed tomography scanner (n = 6). G and H: Representative photographs of interscapular BAT (G) and liver (H) of aP2-Id1Tg− and aP2-Id1Tg+ male mice. BAT weight is significantly higher in HFD-fed aP2-Id1Tg+ mice than in aP2-Id1Tg− mice as a result of higher lipid accumulation (n = 6). I: Average liver weight in aP2-Id1Tg− and aP2-Id1Tg+ mice after 12 weeks of HFD (n = 6). J–L: Representative hematoxylin-eosin–stained eWAT sections (J), adipocyte size distribution (K), and the average size of adipocytes (L) in the eWAT of HFD-fed aP2-Id1Tg− and aP2-Id1Tg+ male mice (n = 6). M and N: Representative hematoxylin-eosin–stained BAT and liver sections of HFD-fed aP2-Id1Tg− and aP2-Id1Tg+ male mice. O–S: Food consumption (O), physical activity (P), VO2 rate (Q), VCO2 (R), and heat production (S) in HFD-fed (12 weeks) aP2-Id1Tg− and aP2-Id1Tg+ male mice measured for 4 days by using Oxymax/CLAMS (n = 5). Data are mean ± SD. Scale bars = 10 μm. *P < 0.05. n.s, not significant.
Id1 Suppresses BAT Thermogenesis by Inhibiting PGC1α Transcriptional Activity
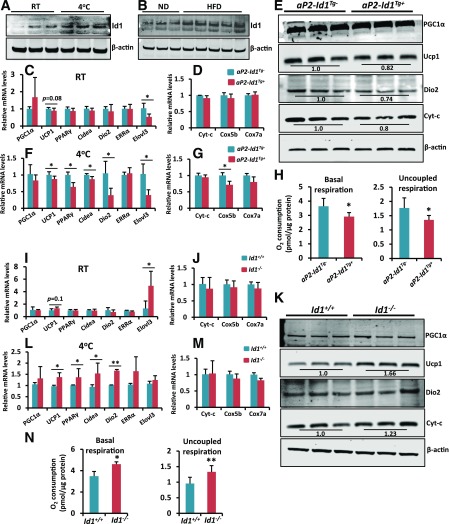
To investigate how Id1 is expressed in vivo in response to thermogenic stimuli such as cold exposure or HFD, we exposed wild-type mice to 4°C for 4 h, which led to a strong induction of Id1 in BAT (Fig. 4A). Similarly, 1 week of HFD feeding led to strong induction of Id1 in the BAT of wild-type mice (Fig. 4B), indicating that Id1 is involved in BAT thermogenesis. Therefore, we asked whether reduced energy expenditure in aP2-Id1Tg+ mice is due to downregulation of thermogenesis genes in BAT. At room temperature (RT) (23°C), we detected some reduction in the expression of Ucp1 in the BAT of aP2-Id1Tg+ mice, but it did not reach statistical significance. Elovl3, on the other hand, was significantly downregulated compared with aP2-Id1Tg− BAT (Fig. 4C and D). However, further expression analysis at the protein level revealed reduced levels of thermogenic proteins Ucp1, Dio2, and Cyt-c in aP2-Id1Tg+ compared with aP2-Id1Tg− BAT (Fig. 4E). Moreover, when mice were exposed to 4°C for 4 h, induction of thermogenic and mitochondrial genes was reduced in aP2-Id1Tg+ compared with aP2-Id1Tg− BAT (Fig. 4F and G). Consistent with reduced expression of thermogenic proteins, interscapular BAT harvested from aP2-Id1Tg+ mice displayed reduced basal and uncoupled respiration compared with aP2-Id1Tg− BAT (Fig. 4H). Because increased expression of Id1 suppressed the expression of thermogenic genes and reduced VO2, we asked whether loss of Id1 results in increased expression of thermogenic genes. To this end, we used Id1+/+ and Id1−/− mice and detected a slight upregulation of Ucp1 and a strong induction of Elovl3 at RT in Id1−/− compared with Id1+/+ BAT (Fig. 4I and J). However, at the protein level, Ucp1 and Cyt-c were increased in Id1−/− compared with Id1+/+ BAT (Fig. 4K). In addition, exposure of mice to 4°C for 4 h led to a significant upregulation of thermogenic genes, such as Ucp1, PPARγ, Cidea, and Dio2 in Id1−/− compared with Id1+/+ BAT (Fig. 4L and M). Moreover, interscapular BAT isolated from Id1−/− mice displayed increased basal and uncoupled respiration compared with Id1+/+ BAT (Fig. 4N). Together, these results indicate that Id1 suppresses thermogenic gene expression and lack of Id1 leads to increased expression of thermogenic genes.
Figure 4.
Thermogenic protein levels and VO2 rate are reduced in the BAT of aP2-Id1Tg+ mice. A: Expression levels of Id1 and β-actin in the BAT of 2-month-old wild-type mice at RT and after exposing mice to 4°C for 4 h. B: Expression levels of Id1 and β-actin in the BAT of 2-month-old ND- or HFD-fed (1 week) wild-type mice. C and D: Relative mRNA transcript levels of thermogenic and mitochondrial genes in the BAT of aP2-Id1Tg− and aP2-Id1Tg+ male mice at RT. E: Expression levels of indicated proteins in the BAT of 2-month-old aP2-Id1Tg− and aP2-Id1Tg+ male mice at RT. F and G: Relative mRNA transcript levels of thermogenic and mitochondrial genes in the BAT of aP2-Id1Tg− and aP2-Id1Tg+ male mice after exposure to 4°C for 4 h. H: Basal respiration and uncoupled respiration (after blocking ATP synthase with oligomycin) were determined in the BAT explants of aP2-Id1Tg− and aP2-Id1Tg+ male mice with an Oxygraph Plus System (n = 4). I and J: Relative mRNA transcript levels of thermogenic and mitochondrial genes in the BAT of Id1+/+ and Id1−/− male mice at RT. K: Expression levels of indicated proteins in the BAT of 2-month-old Id1+/+ and Id1−/− mice at RT. After acquiring images, band intensities were measured and normalized to β-actin by the Li-Cor Odyssey system. L and M: Relative mRNA transcript levels of thermogenic and mitochondrial genes in the BAT of Id1+/+ and Id1−/− male mice after exposure to 4°C for 4 h (n = 6–8). N: Basal respiration and uncoupled respiration were determined in the BAT explants of Id1+/+ and Id1−/− mice (n = 4). Data are mean ± SD. *P < 0.05; **P < 0.005.
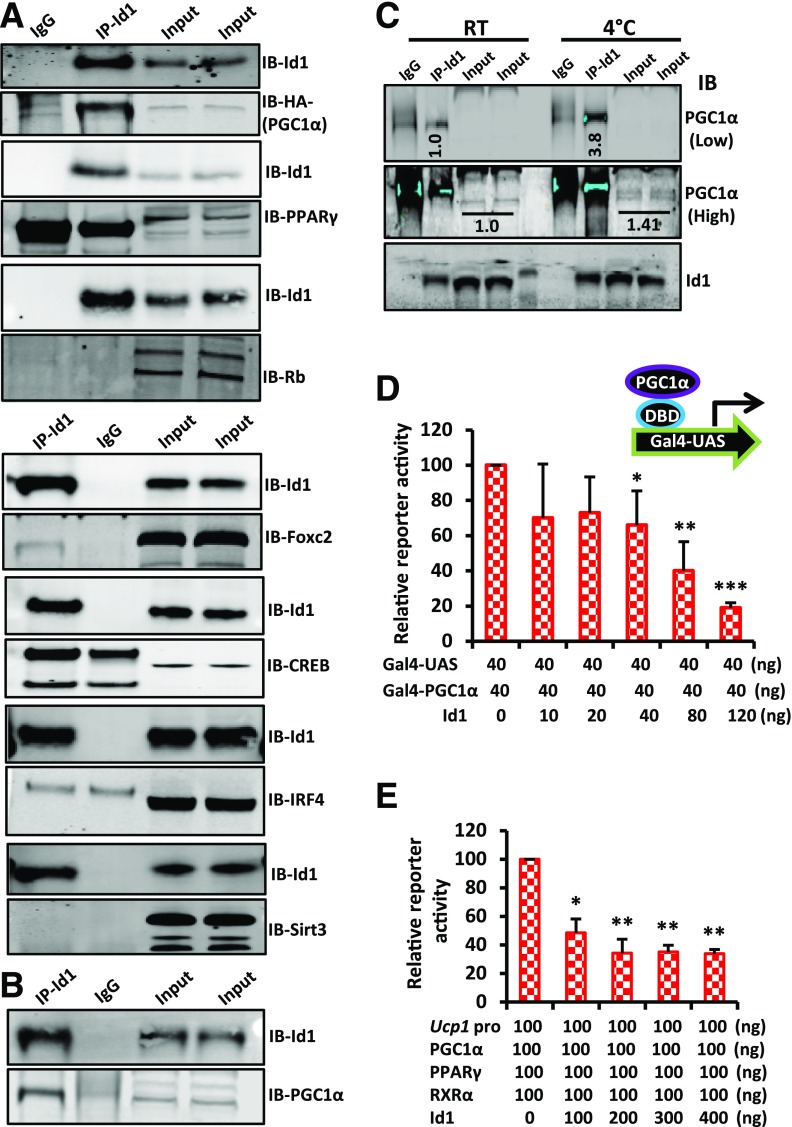
A number of factors activate or inhibit thermogenesis by either positively or negatively regulating the PGC1α/UCP1 thermogenesis pathway (4,6–8,24). Because Id1 lacks a DBD and regulates other transcription factors by direct interaction (20,25), we reasoned that Id1 directly binds to a transcriptional activator of thermogenesis and suppresses its transcriptional activity. Alternatively, Id1 directly binds to a negative regulator of thermogenesis and promotes its inhibitory action, leading to reduced energy expenditure. To investigate these possibilities, we coexpressed Id1 and some of the known positive and negative regulators of thermogenesis in HEK293 cells and performed a series of coimmunoprecipitations (co-IPs). These analyses revealed a direct interaction between Id1 and PGC1α (Fig. 5A), whereas Id1 did not interact with the other factors tested. Further analysis revealed a direct interaction between Id1 and PGC1α in day 6 differentiated HIB1B cells, indicating that Id1 indeed interacts with PGC1α endogenously (Fig. 5B). In response to cold exposure, Id1 expression was induced (Fig. 4A), and thermogenic gene expression was suppressed in aP2-Id1Tg+ and increased in Id1−/− mouse BAT compared with controls (Fig. 4F and L), Therefore, we asked whether a thermogenic stimulus promotes Id1 interaction with PGC1α to prevent excessive thermogenesis. We exposed wild-type mice to 4°C for 4 h and detected an increased interaction between Id1 and PGC1α in the BAT of mice exposed to 4°C compared with mice at RT (Fig. 5C). To determine whether the Id1/PGC1α interaction affects PGC1α transcriptional activity, we performed a luciferase reporter assay in Cos7 cells by using a luciferase reporter driven by the Gal-upstream activation sequence (UAS) and full-length PGC1α fused with the DBD of yeast Gal4 as previously described (26). We found that the activity of Gal4-PGC1α was strongly inhibited when Id1 was coexpressed (Fig. 5D). Moreover, coexpression of the Ucp1 promoter–driven luciferase reporter vector along with PPARγ/RXRα/PGC1α or PPARγ/RXRα/PGC1α/Id1 expression vectors revealed significant suppression of Ucp1 promoter–driven luciferase activity when Id1 was coexpressed (Fig. 5E), suggesting that binding of Id1 indeed suppresses PGC1α transcriptional activity. Together, these results suggest that Id1 suppresses BAT thermogenesis by binding to and inhibiting the transcriptional activity of PGC1α.
Figure 5.
Id1 suppresses PGC1α transcriptional activity. A: Co-IP followed by Western blot showing a direct interaction between Id1 and PGC1α (HA tag), whereas Id1 did not directly interact with other proteins tested. Input: 2% of IP reaction. B: Co-IP followed by Western blot showing a direct interaction between endogenous Id1 and PGC1α in day 6 differentiated HIB1B cells. Input: 5% of IP reaction. C: Co-IP followed by Western blot showing a direct interaction between endogenous Id1 and PGC1α in the BAT of mice at RT and after exposure to 4°C for 4 h. Input: 5% of IP reaction. The higher-exposure image also was included to show PGC1α input bands not visible under lower exposure. Band intensities were measured and normalized to PGC1α input bands by the Li-Cor Odyssey system. D: Transcriptional activity of PGC1α was assayed by cotransfecting Cos-7 cells with luciferase reporter driven by Gal-UAS and Gal4-PGC1α plasmid (full-length PGC1α fused with the DBD of yeast Gal4) with various concentrations of the Id1 expression vector (n = 3). E: Transcriptional activity of PGC1α was assayed by cotransfecting Cos-7 cells with luciferase reporter driven by Ucp1 promoter, PPARγ, RXRα (PPARγ binding partner), and various concentrations of Id1 plasmid (n = 2). Data are mean ± SD. *P < 0.05; **P < 0.005; ***P < 0.0005. IB, immunoblot.
Id1 Deficiency Results in Increased Beige/Thermogenic Gene Expression in iWAT
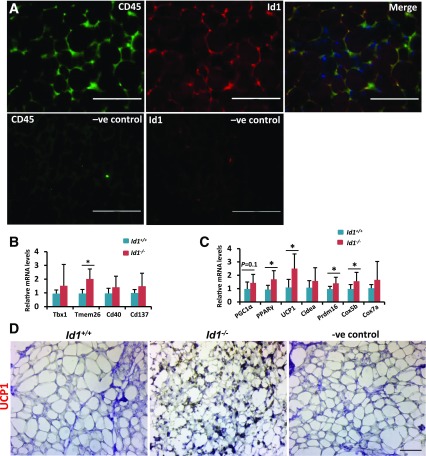
In addition to BAT, Id1 was expressed in WAT (Fig. 1A). WAT consists of adipocytes and the stromal vascular fraction (SVF) where the preadipocytes/progenitors reside (27). To investigate whether Id1 is expressed in the white adipocytes or in the SVF, we performed Id1 and CD45 (a general marker of SVF [28]) costaining on iWAT and detected a strong colocalization of Id1 and CD45 (Fig. 6A), indicating that Id1 is mainly localized in the SVF of WAT and thus possibly involved in adipose progenitor cell differentiation. To investigate whether Id1 enhances or inhibits cold-induced brown-like (beige) differentiation of white preadipocytes in iWAT, we exposed Id1+/+ and Id1−/− mice to 4°C for 12 h and analyzed the expression levels of the beige markers Tbx1, Tmem26, CD40, and CD137 (12) and thermogenic genes in iWAT. We detected significant upregulation of Tmem26, PPARγ, Ucp1, and Prdm16 in the iWAT of Id1−/− compared with Id1+/+ mice (Fig. 6B and C). Moreover, UCP1 staining on the iWAT of Id1+/+ and Id1−/− mice after 12 h of cold exposure revealed strong UCP1 staining on Id1−/− iWAT and is not detectable on Id1+/+ iWAT (Fig. 6D). These observations suggest that loss of Id1 enhances browning of iWAT.
Figure 6.
Increased beige/thermogenic gene expression in the iWAT Id1−/− mice. A: Immunofluorescence staining showing the localization pattern of CD45 and Id1 in the iWAT of 2-month-old wild-type mice. iWAT sections incubated only with fluorescent secondary antibodies served as negative (–ve) controls. Scale bars = 100 μm. B and C: Relative mRNA transcript levels of beige and thermogenic genes in the iWAT of Id1+/+ and Id1−/− male mice after exposure to 4°C for 12 h (n = 7). D: Representative immunohistochemistry staining showing the expression pattern of UCP1 in the iWAT of Id1+/+ and Id1−/− mice after exposure to 4°C for 12 h (n = 5; 3 of 5 Id1−/− mice showed strong UCP1 staining in the iWAT; no staining was detected in the Id1+/+ mice). Scale bar = 10 μm. iWAT sections incubated only with horseradish peroxidase–conjugated secondary antibody served as the –ve control. Data are mean ± SD. *P < 0.05.
Differentiation of Mouse Embryonic Fibroblasts Into Brown-Like Cells Is Increased in the Absence of Id1
To further investigate whether loss of Id1 increases differentiation of white preadipocytes into brown-like cells, we harvested the SVF from Id1+/+ and Id1−/− WAT and attempted to culture white adipose progenitors as previously described (16,29). However, Id1−/− progenitor cells failed to grow and showed a senescence-like phenotype (data not shown), which is consistent with previous observations that Id1 promotes cell proliferation and lack of Id1 results in premature senescence in certain cell types (30). Therefore, we were unable to perform in vitro differentiation experiments in Id1−/− progenitor cells. To overcome this obstacle, we used an alternative in vitro system: early passage (P2) mouse embryonic fibroblasts (MEFs). MEFs are unprogrammed cells, share several characteristics with mesenchymal stem cells, and can differentiate into various mesenchymal lineages (31). MEFs can be induced to differentiate into brown-like cells that express mitochondrial and thermogenesis genes (24,32). To test whether Id1 is involved in the differentiation of MEFs into brown-like cells, we treated Id1+/+ and Id1−/− E13.5 MEFs with brown adipocyte differentiation media that consisted of rosiglitazone as previously described (24). We detected increased adipocyte differentiation in Id1−/− compared with Id1+/+ cultures as revealed by enhanced expression of the adipocyte differentiation marker aP2 and Oil Red O staining (Fig. 7A and B). Expression of Id1 is induced during differentiation of MEFs into brown adipocytes in Id1+/+ cultures and is completely absent in Id1−/− cells as expected (Fig. 7A). Consistent with increased brown adipogenesis, differentiated Id1−/− cells displayed increased mitochondrial content and increased cellular respiration compared with Id1+/+ cells (Fig. 7C–E). At the molecular level, Id1−/− cells displayed increased expression of beige and mitochondrial genes in day 10 differentiated cells, whereas their expression was similar in undifferentiated MEFs (Fig. 7F–I). We also detected increased expression of thermogenic genes, such as Pparγ, Pgc1α, Ucp1, Ebf2, and Prdm16, in differentiated Id1−/− cells compared with Id1+/+ cells (Fig. 7J and K). Of note, the expression of Ebf2 and Prdm16, the major determinants of brown adipocyte identity, were increased even in the nondifferentiated Id1−/− MEFs (Fig. 7J). Further analysis at the protein level revealed elevated levels of aP2, PPARγ, PGC1α, Ebf2, and PRDM16 in Id1−/−-differentiating cells compared with Id1+/+ cells (Fig. 7L). We also analyzed RB and RIP140, which are known to suppress WAT browning (33). Although RB and pRB levels were unchanged, we found a detectable reduction in RIP140 levels in Id1−/− cells (Fig. 7L). These results suggest that lack of Id1 promotes differentiation of MEFs into brown-like cells because of induced expression of PPARγ, Ebf2, PRDM16, and PGC1α.
Figure 7.
Increased beige/thermogenic gene expression in Id1−/− MEFs. A: Representative Oil Red O staining of day 10 differentiated Id1+/+ and Id1−/− MEFs. Scale bar = 20 μm. B: Expression patterns of Id1, aP2, and β-actin in differentiating MEFs at the indicated time points after inducing brown adipogenesis. C and D: FACS analysis of unstained and nonyl acridine orange–stained day 10 differentiated Id1+/+ and Id1−/− MEFs (C) and the quantification of fluorescence level (FL) (D) (n = 4). E: Basal and uncoupled respiration (after blocking ATP synthase with oligomycin) rate of day 10 differentiated Id1+/+ and Id1−/− MEFs (n = 3). F–K: Relative mRNA transcript levels of beige (F and G), mitochondrial (H and I), and thermogenic (J and K) genes in nondifferentiated (ND) (F, H, and J) and day 10 differentiated (D10) (G, I, and K) Id1+/+ and Id1−/− MEFs (n = 3). L: Expression levels of indicated proteins in Id1+/+ and Id1−/− differentiating MEFs at the indicated time points after inducing brown adipogenesis. Data are mean ± SD. *P < 0.05; **P < 0.005; ***P < 0.0005.
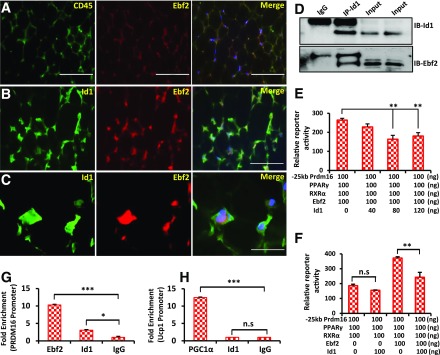
Id1 Directly Binds to and Suppresses Ebf2 Transcriptional Activity
Previously, it was demonstrated that by recruiting PPARγ to the Prdm16 promoter, Ebf2 induces the expression of Prdm16, which determines brown adipocyte cell fate (14–16,34). Ebf2 is a helix-loop-helix transcription factor, and Id proteins are known to interact with helix-loop-helix proteins with very high affinity (20,25), indicating the possibility of a direct interaction between Id1 and Ebf2. Therefore, we asked whether Id1 downregulates Prdm16 expression by directly interacting with and suppressing Ebf2 transcriptional activity. To this end, we first asked whether Id1 and Ebf2 colocalize in WAT. We stained for Id1 and Ebf2 on iWAT and detected colocalization of Id1 and Ebf2 (Fig. 8A–C), indicating the possibility of a direct interaction between Id1 and Ebf2. Next, we coexpressed Id1 and Ebf2 in HEK293 cells and performed co-IP, which revealed a direct interaction between Id1 and Ebf2 (Fig. 8D). To determine whether Id1 binding to Ebf2 affects the transcriptional activity of Ebf2, we coexpressed a Prdm16 promoter–driven luciferase reporter vector along with PPARγ/RXRα/Ebf2 or PPARγ/RXRα/Ebf2/Id1 expression vectors. We detected a significant suppression of Prdm16 promoter–driven luciferase activity when Id1 is coexpressed with PPARγ/RXRα/Ebf2 (Fig. 8E), whereas in the absence of Ebf2, Id1 did not suppress Prdm16 promoter–driven luciferase activity (Fig. 8F), suggesting that Id1 downregulates Prdm16 expression by binding to and suppressing the transcriptional activity of Ebf2. Because Id1 does not have a DBD and directly binds to Ebf2 and PGC1α, we wondered whether Id1 colocalizes with Ebf2 and PGC1α on their regulatory regions, such as the Prdm16 and Ucp1 promoters. Chromatin IP-qPCR assays in day 4 differentiated HIB1B cells revealed that Id1 is localized to the Prdm16 promoter region but not to the Ucp1 promoter region (Fig. 8G and H). These observations indicate that Id1 can associate with Ebf2 at the Ebf2 regulatory regions, whereas Id1/PGC1α interaction precludes PGC1α from associating with its regulatory regions. As a result, Id1 is not associated with the Ucp1 promoter.
Figure 8.
Id1 suppresses Ebf2 transcriptional activity. A–C: Immunofluorescence staining showing the localization patterns of CD45/Ebf2 and Id1/Ebf2 in the iWAT of 2-month-old wild-type mice. Scale bars = 100 μm (A and B) and 20 μm (C). D: Co-IP followed by Western blot showing a direct interaction between Id1 and Ebf2. Input: 2% of IP reaction. E: Relative reporter activity of Prdm16 promoter–driven luciferase activity in the presence of PPARγ, RXRα (PPARγ binding partner), Ebf2, and different concentrations of Id1 plasmid (n = 2). F: Relative reporter activity of Prdm16 promoter–driven luciferase activity in the presence of PPARγ and RXRα and in the presence and absence of Ebf2 and Id1 (n = 2). Id1 suppressed Prdm16 promoter–driven luciferase activity only when Ebf2 was present. G and H: Chromatin IP-qPCR analysis of Ebf2 and Id1 binding to the Prdm16 promoter (G) and PGC1α and Id1 binding to the Ucp1 promoter (H) in day 4 differentiated HIB1B cells after normalizing to 18S DNA binding (n = 2). Data are mean ± SD. *P < 0.05; **P < 0.005; ***P < 0.0005. IB, immunoblot; kb, kilobase; n.s, not significant.
Discussion
Id1 has been known to play critical roles in cell proliferation, cellular differentiation, and tumorigenesis (35,36). Previously, we and others demonstrated that Id1 whole-body knockout mice (Id1−/−) are lean and protected from HFD-induced insulin resistance and hepatosteatosis (37,38). However, the adipose tissue–specific function of Id1 and its relative contribution to body energy expenditure remains unclear because it is a whole-body Id1-deficient mouse. In the current work, we discovered a novel function of Id1 in BAT-mediated thermogenesis and its potential involvement in the programming of preadipocytes into brown-like adipocytes. During HIB1B brown adipocyte differentiation, the Id1 expression pattern closely mimicked the expression pattern of PPARγ, PGC1α, and UCP1. However, unlike PPARγ, which is absolutely required for both white and brown adipocyte differentiation (39), Id1 is dispensable for brown adipocyte differentiation. Previously, it was demonstrated that Id1 is not required for white adipocyte differentiation (37), indicating that Id1 is dispensable for both white and brown adipocyte differentiation. The aP2-Id1Tg+ mice displayed reduced energy expenditure, indicating that Id1 suppresses BAT thermogenesis. In aP2-Id1Tg BAT, thermogenic gene expression is mildly reduced compared with controls at RT, but over time, this can have a significant effect on body weight because it slowly but steadily reduces body energy expenditure. For this possible reason, aP2-Id1Tg mice gained weight slowly during aging. Id1 lacks a DBD; therefore, the only way Id1 could inhibit thermogenesis is by directly binding to and suppressing the transcriptional activity of a factor that promotes thermogenesis. Our attempt to identify a possible interaction between Id1 and some of the known activators/suppressors of thermogenesis unexpectedly revealed a direct interaction between Id1 and PGC1α with a concomitant suppression of PGC1α transcriptional activity. If Id1 suppresses PGC1α transcriptional activity, then why is the PGC1α downstream target UCP1 only slightly reduced when Id1 levels are strongly increased during HIB1B differentiation? A possible explanation could be that PPARγ, PGC1α, and UCP1 are highly expressed in fully differentiated HIB1B cells and in the BAT where the majority of the cells are mature, fully differentiated brown adipocytes. The transcription factors Twist-1 and Cidea, which inhibit PGC1α and UCP1, also are highly expressed in BAT (26,40). Therefore, both the activators and the suppressors of thermogenesis appear to coexist in the BAT and in fully differentiated brown adipocytes. When thermogenesis is strongly activated, it triggers a negative feedback loop that further induces the expression of the thermogenic suppressors such as Id1, which in turn prevents excessive thermogenesis by inhibiting PGC1α and/or UCP1. Consistent with this explanation, we found that cold exposure enhanced the expression of Id1, which in turn resulted in increased interaction between Id1 and PGC1α. Conversely, cold exposure results in increased expression of UCP1 in the BAT of Id1−/− mice (37).
In the adipose progenitor cells, Ebf2 and PPARγ cooperatively induce the expression of Prdm16, which controls beige adipocyte determination. We found that both Id1 and Ebf2 are mainly localized in the SVF of iWAT, where the adipose progenitors reside. Furthermore, Id1 directly interacted with Ebf2 and suppressed its transcriptional activity, leading to impaired expression of Prdm16. Consequently, in Id1−/− iWAT, the expression of beige and thermogenic genes, including Prdm16, was increased in response to prolonged cold exposure. In addition, the differentiation of MEFs into brown-like cells is significantly increased in the absence of Id1, which contrasts with our observation that Id1 is dispensable for HIB1B brown adipocyte differentiation. A possible reason for the knockdown or overexpression of Id1 not affecting differentiation of HIB1B cells could be that HIB1B cells comprise a preprogrammed brown adipocyte cell line, and upon brown adipogenic stimulation, they differentiate into brown adipocytes. Therefore, HIB1B cell differentiation may no longer be influenced by the factors involved in the earlier cell fate determination. Id1 appears to participate in the earlier programming of preadipocytes into beige adipocytes by regulating Ebf2 transcriptional activity. Ebf2 determines not only beige but also brown adipocyte cell fate, and loss of Ebf2 results in a significant reduction of BAT and an almost total loss of brown fat–specific gene expression in Ebf2−/− embryos (16). If Id1 functions as an upstream inhibitor of Ebf2, then why does elevated expression of Id1 (aP2-Id1Tg+) or loss of Id1 (Id1−/−) have no detectable effect on BAT development? A possible explanation is that in aP2-Id1Tg+ mice, Id1 expression is directed by the aP2 promoter/enhancer, which may not be active in the adipose progenitors during embryonic development. Therefore, Id1 may not have been induced during brown adipocyte cell fate determination in aP2-Id1Tg+ mice and therefore did not interfere with Ebf2/Prdm16-mediated brown adipocyte programming and BAT development. In Id1−/− mice, Ebf2 is free from Id1-mediated suppression and induces Prdm16 expression, leading to normal BAT development. In contrast to brown adipocytes, beige adipocytes are generated in response to a substantial physiological stimulus, such as prolonged cold exposure or β3-androgen receptor-adrenergic agonists (12,41,42). Of note, Id1 expression is also strongly induced in response to cold exposure. Once induced, Id1 functions as an upstream transcriptional suppressor of Ebf2, and in the absence of Id1, Ebf2-mediated browning of iWAT is enhanced. Together, the current results suggest that Id1 regulates two pathways—PGC1α/Ucp1 thermogenic and Ebf2/Prdm16 adipose progenitor cell programming—by inhibiting PGC1α in the BAT and Ebf2 in the progenitor cells. Nevertheless, we cannot exclude the possibility that Id1 also interacts with PGC1α in the progenitor cells. However, Id1/PGC1α interaction may not prevent the progenitors from differentiating into brown-like cells because neither PGC1α nor Id1 is required for brown adipocyte differentiation.
In contrast to males, female aP2-Id1Tg+ mice are protected from age- and HFD-associated obesity. Sex-dependent differences in energy intake, storage, and expenditure are well known, and these physiological mechanisms are under the control of sex hormones (43). For example, ovarian estrogens control energy homeostasis (44), and estradiol activates thermogenesis in BAT through the sympathetic nervous system (45). In the current study, Id1-mediated suppression of energy expenditure caused obesity in males but not in females, suggesting that female sex hormones can counteract the effects of some of the negative regulators of energy expenditure. Therefore, future studies should focus on exploring which of these factors are under the control of female sex hormones. In the current study, we used aP2-Id1Tg mice, and other studies demonstrated that aP2 promoter–driven expression is not highly specific to adipose tissues and can have certain low, nonspecific expression in other tissues, such as the lung, heart, muscle, liver, and testes (46–48). In the current aP2-Id1Tg mouse model, we did not detect any difference in lean mass and bone density between control and aP2-Id1Tg+ mice. Moreover, female aP2-Id1Tg+ mice did not show detectable phenotypes in adipose or other tissues, suggesting that aP2 promoter–driven Id1 expression did not cause significant side effects.
In conclusion, this study demonstrates that by suppressing PGC1α-mediated BAT thermogenesis and Ebf2-mediated beige adipocyte programming, Id1 promotes energy storage and obesity, which is a significant risk factor for insulin resistance and diabetes. Therefore, Id1 could potentially function as a molecular target to reverse obesity. Id1−/− mice are viable and fertile and live normally, suggesting that most cell types do not require Id1 for normal functioning. Therefore, targeting Id1 in vivo could be a relatively safe strategy to increase energy expenditure, reduce adiposity, and treat obesity and its associated diseases, such as diabetes.
Supplementary Material
Article Information
Acknowledgments. The authors thank Dr. Jonathan Keller (Mouse Cancer Genetics Program, National Cancer Institute, Frederick, MD) for providing Id1−/− mice, Dr. Bruce Spiegelman (Department of Cell Biology, Harvard Medical School) for providing Gal4-PGC1α vectors and HIB1B cells, Dr. Patrick Seale (Department of Cell and Developmental Biology, Perelman School of Medicine, University of Pennsylvania) for providing Prdm16 luciferase reporter vectors, and Dr. Mark Christian (Division of Metabolic and Vascular Health, University of Warwick) for providing Ucp1 luciferase vector. The authors also thank Dr. Ali Arbab, Roxan Ara, and Chris Middleton (Tumor Angiogenesis Section, Georgia Cancer Center, Augusta University) for technical help with computed tomography scans; Dr. Jenfeng Pang (Molecular Oncology Program, Georgia Cancer Center, Augusta University) for help with Oxymax readings; and Dr. Rhea-Beth Markowitz (Georgia Cancer Center, Augusta University) for reviewing and editing the manuscript.
Funding. This research was supported by the National Cancer Institute (grant K22-CA-168828 to A.S.) and the National Institute of Diabetes and Digestive and Kidney Diseases (grant DP2-DK-105565 to A.S.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. M.P., B.K.S., S.E., J.C., and S.K. performed experiments and collected and analyzed data. J.Y. maintained and provided animals for experiments. A.S. supervised the study. M.P. and A.S. analyzed and interpreted the data and wrote the manuscript. M.P. and A.S. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-1079/-/DC1.
References
- 1.Vucenik I, Stains JP. Obesity and cancer risk: evidence, mechanisms, and recommendations. Ann N Y Acad Sci 2012;1271:37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poirier P, Eckel RH. Obesity and cardiovascular disease. Curr Atheroscler Rep 2002;4:448–453 [DOI] [PubMed] [Google Scholar]
- 3.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev 2004;84:277–359 [DOI] [PubMed] [Google Scholar]
- 4.Seale P, Kajimura S, Spiegelman BM. Transcriptional control of brown adipocyte development and physiological function—of mice and men. Genes Dev 2009;23:788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak LP, Anunciado-Koza R. UCP1: its involvement and utility in obesity. Int J Obes 2008;32(Suppl. 7):S32–S38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest 2006;116:615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong X, Banks A, Liu T, et al. . IRF4 is a key thermogenic transcriptional partner of PGC-1α. Cell 2014;158:69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Picard F, Géhin M, Annicotte J, et al. . SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell 2002;111:931–941 [DOI] [PubMed] [Google Scholar]
- 9.Sharma BK, Patil M, Satyanarayana A. Negative regulators of brown adipose tissue (BAT)-mediated thermogenesis. J Cell Physiol 2014;229:1901–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vitali A, Murano I, Zingaretti MC, Frontini A, Ricquier D, Cinti S. The adipose organ of obesity-prone C57BL/6J mice is composed of mixed white and brown adipocytes. J Lipid Res 2012;53:619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbatelli G, Murano I, Madsen L, et al. . The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am J Physiol Endocrinol Metab 2010;298:E1244–E1253 [DOI] [PubMed] [Google Scholar]
- 12.Wu J, Boström P, Sparks LM, et al. . Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012;150:366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med 2013;19:1338–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seale P, Bjork B, Yang W, et al. . PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008;454:961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seale P, Kajimura S, Yang W, et al. . Transcriptional control of brown fat determination by PRDM16. Cell Metab 2007;6:38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajakumari S, Wu J, Ishibashi J, et al. . EBF2 determines and maintains brown adipocyte identity. Cell Metab 2013;17:562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kajimura S, Seale P, Kubota K, et al. . Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature 2009;460:1154–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seale P, Conroe HM, Estall J, et al. . Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest 2011;121:96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stine RR, Shapira SN, Lim HW, et al. . EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Mol Metab 2015;5:57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci 2000;113:3897–3905 [DOI] [PubMed] [Google Scholar]
- 21.Perry SS, Zhao Y, Nie L, Cochrane SW, Huang Z, Sun XH. Id1, but not Id3, directs long-term repopulating hematopoietic stem-cell maintenance. Blood 2007;110:2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma BK, Kolhe R, Black SM, Keller JR, Mivechi NF, Satyanarayana A. Inhibitor of differentiation 1 transcription factor promotes metabolic reprogramming in hepatocellular carcinoma cells. FASEB J 2016;30:262–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vernochet C, Mourier A, Bezy O, et al. . Adipose-specific deletion of TFAM increases mitochondrial oxidation and protects mice against obesity and insulin resistance. Cell Metab 2012;16:765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen JB, Jørgensen C, Petersen RK, et al. . Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc Natl Acad Sci U S A 2004;101:4112–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Toole PJ, Inoue T, Emerson L, et al. . Id proteins negatively regulate basic helix-loop-helix transcription factor function by disrupting subnuclear compartmentalization. J Biol Chem 2003;278:45770–45776 [DOI] [PubMed] [Google Scholar]
- 26.Pan D, Fujimoto M, Lopes A, Wang YX. Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell 2009;137:73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell 2008;135:240–249 [DOI] [PubMed] [Google Scholar]
- 28.Bourin P, Bunnell BA, Casteilla L, et al. . Stromal cells from the adipose tissue-derived stromal vascular fraction and culture expanded adipose tissue-derived stromal/stem cells: a joint statement of the International Federation for Adipose Therapeutics and Science (IFATS) and the International Society for Cellular Therapy (ISCT). Cytotherapy 2013;15:641–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aune UL, Ruiz L, Kajimura S. Isolation and differentiation of stromal vascular cells to beige/brite cells. J Vis Exp 2013. (73):e50191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alani RM, Young AZ, Shifflett CB. Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proc Natl Acad Sci U S A 2001;98:7812–7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yusuf B, Gopurappilly R, Dadheech N, Gupta S, Bhonde R, Pal R. Embryonic fibroblasts represent a connecting link between mesenchymal and embryonic stem cells. Dev Growth Differ 2013;55:330–340 [DOI] [PubMed] [Google Scholar]
- 32.Sellayah D, Bharaj P, Sikder D. Orexin is required for brown adipose tissue development, differentiation, and function. Cell Metab 2011;14:478–490 [DOI] [PubMed] [Google Scholar]
- 33.Scimè A, Grenier G, Huh MS, et al. . Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metab 2005;2:283–295 [DOI] [PubMed] [Google Scholar]
- 34.Timmons JA, Wennmalm K, Larsson O, et al. . Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A 2007;104:4401–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zebedee Z, Hara E. Id proteins in cell cycle control and cellular senescence. Oncogene 2001;20:8317–8325 [DOI] [PubMed] [Google Scholar]
- 36.Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell 2003;3:525–530 [DOI] [PubMed] [Google Scholar]
- 37.Satyanarayana A, Klarmann KD, Gavrilova O, Keller JR. Ablation of the transcriptional regulator Id1 enhances energy expenditure, increases insulin sensitivity, and protects against age and diet induced insulin resistance, and hepatosteatosis. FASEB J 2012;26:309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y, Ling F, Griffin TM, et al. . Up-regulation of the sirtuin 1 (Sirt1) and peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) genes in white adipose tissue of Id1 protein-deficient mice: implications in the protection against diet and age-induced glucose intolerance. J Biol Chem 2014;289:29112–29122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev 2000;14:1293–1307 [PubMed] [Google Scholar]
- 40.Zhou Z, Yon Toh S, Chen Z, et al. . Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat Genet 2003;35:49–56 [DOI] [PubMed] [Google Scholar]
- 41.Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARγ agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab 2012;15:395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med 2013;19:1252–1263 [DOI] [PubMed] [Google Scholar]
- 43.Shi H, Seeley RJ, Clegg DJ. Sexual differences in the control of energy homeostasis. Front Neuroendocrinol 2009;30:396–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mauvais-Jarvis F, Clegg DJ, Hevener AL. The role of estrogens in control of energy balance and glucose homeostasis. Endocr Rev 2013;34:309–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martínez de Morentin PB, González-García I, Martins L, et al. . Estradiol regulates brown adipose tissue thermogenesis via hypothalamic AMPK. Cell Metab 2014;20:41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeffery E, Berry R, Church CD, et al. . Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte 2014;3:206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mullican SE, Tomaru T, Gaddis CA, Peed LC, Sundaram A, Lazar MA. A novel adipose-specific gene deletion model demonstrates potential pitfalls of existing methods. Mol Endocrinol 2013;27:127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee KY, Russell SJ, Ussar S, et al. . Lessons on conditional gene targeting in mouse adipose tissue. Diabetes 2013;62:864–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.