Abstract
While collisions between replication and transcription in bacteria are deemed inevitable, the fine details of the interplay between the two machineries are poorly understood. In this study, we evaluate the effects of transcription on the replication fork progression in vivo, by using electrophoresis analysis of replication intermediates. Studying Escherichia coli plasmids, which carry constitutive or inducible promoters in different orientations relative to the replication origin, we show that the mutual orientation of the two processes determines their mode of interaction. Replication elongation appears not to be affected by transcription proceeding in the codirectional orientation. Head-on transcription, by contrast, leads to severe inhibition of the replication fork progression. Furthermore, we evaluate the mechanism of this inhibition by limiting the area of direct contact between the two machineries. We observe that replication pausing zones coincide exactly with transcribed DNA segments. We conclude, therefore, that the replication fork is most likely attenuated upon direct physical interaction with the head-on transcription machinery.
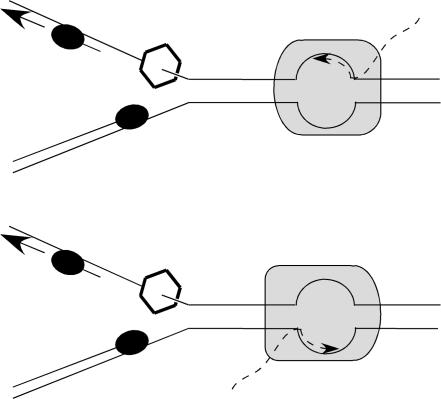
In bacteria, DNA replication and transcription continue throughout the life cycle. The speed of the replication fork progression in Escherichia coli is ∼1,000 bp per s (16), while the elongation rate for RNA polymerase is just 50 nucleotides (nt) per s (15); i.e., replication is approximately 20-fold faster than transcription. Since the two processes proceed simultaneously, frequent collisions between replication and transcription seem unavoidable (3, 38). Given that both processes are polar, the collisions can occur either head-on or codirectionally (Fig. 1). In the case of head-on collisions, the front edge of RNA polymerase meets the hexameric DNA helicase DnaB that moves along the lagging strand template. In the codirectional case, by contrast, the front edge of the leading strand DNA polymerase collides with the rear edge of RNA polymerase.
FIG. 1.
Schematic representation of the head-on and codirectional collisions between replication and transcription. The replication fork (left) and RNA polymerase (right) are shown with leading- and lagging-strand DNA polymerases as ovals and DNA helicase DnaB as a hexagon. Solid lines, DNA strands; broken lines, RNA strands.
While it is unclear a priori which type of collision is more damaging for DNA metabolism, the data on the organization of bacterial genomes point to selection against head-on collisions. Sequencing of the E. coli genome revealed that there is a bias towards codirectional alignment of transcription units with replication (2). Most strikingly, all seven ribosomal operons face the direction of their replichores. For other genes, however, this bias is much less pronounced: ∼62% of tRNA genes and ∼55% of protein-coding genes are aligned codirectionally with replication. Similar principles of gene arrangement were observed for other bacteria, such as Bacillus subtilis, Borrelia burgdorferi, Treponema pallidum, Haemophilus influenzae, Helicobacter pylori, Mycoplasma genitalium, and Mycoplasma pneumoniae (36), as well as for bacteriophages T7 and lambda (3).
Experimental data on transcription-replication collision are relatively scarce. This problem was addressed in vitro by studying interactions between bacterial RNA polymerases and phage replisomes. Phage T4 replication fork appeared to pause more strongly when encountering the stalled E. coli RNA polymerase head-on rather than codirectionally (25, 26). When RNA polymerase was allowed to move slowly, replication fork pausing became less pronounced. Phage φ29 replisome, in contrast, halted upon encountering stalled B. subtilis RNA polymerase in both orientations (12, 13). Once RNA polymerase movement was resumed, DNA synthesis continued with its normal speed in the head-on case but was much slower for the codirectional alignment.
The first evidence for transcription-replication collision in vivo came from studies of an inducible replication origin placed on either side of the rrnB ribosomal operon into the E. coli chromosome (14). Replication codirectional with the ribosomal operon proceeded with its typical high speed but was significantly slower when it faced transcription head-on. The mechanisms responsible for the slowing of replication in the head-on scenario remained unclear. Besides the obvious physical collision (Fig. 1), the cause of the slowing could be topological constraints. Both elongating RNA polymerase (27, 47) and the replication fork (40) generate positive supercoils in the downstream DNA. Consequently, frontal movement of the two machineries could generate highly positively supercoiled DNA domain, restraining both processes prior to the direct encounter (3, 11, 14). The fraction of DNA knots appeared to be greater in those plasmids where replication collided with transcription head-on, rather than codirectionally, supporting the formation of such positively supercoiled domains (39). This finding was explained by the migration of positive supercoils, which accumulated between the replisome and RNA polymerase, to the newly synthesized DNA behind the fork.
Whether replication and transcription actually collide in eukaryotic genomes is even less clear. The original support for collisions came from the discovery of the so-called replication fork barrier (RFB) at the 3′ end of the Saccharomyces cerevisiae rRNA genes (6, 24). It turned out, however, that RFB function did not depend on transcription per se (7), but was caused by a polar contrahelicase activity of the DNA-binding protein Fob1 (18). Only when the FOB1 gene was deleted and the number of rRNA gene repeats was reduced was transcription-replication collision at the ribosomal locus actually detected (46). Similarly, in mammals, RFB activity at the 3′ ends of ribosomal genes is caused by the transcription termination factor TTF-1 that happens to be a polar contrahelicase, as well (41). The mere existence of such contrahelicases strongly suggests a requirement to protect transcription of important genes from DNA replication. Replication fork barriers were also observed in such diverse eukaryotes as the pea (28), the mouse (29), Xenopus laevis (33), Tetrahymena thermophila (49), and fission yeast (44).
Another indication for transcription-replication collisions came from the detection of polar replication fork pause sites at tRNA genes in S. cerevisiae (11). The replication fork stalls when it encounters tRNA genes transcribed head-on, but not when it encounters those transcribed codirectionally. Since replication fork pause site activity was dependent on the functionality of both the gene promoter and RNA polymerase III, the direct involvement of transcription was plausible. Quite recently, however, it was argued that the presence of the transcription initiation complex, rather than transcription elongation, could be responsible for replication slowing at these genes (17).
Given the limited amount of data on transcription-replication collisions in vivo and the lack of understanding of their mechanisms, we decided to revisit this matter by assaying E. coli plasmid replication via electrophoresis analysis of replication intermediates. We show that codirectional transcription has no effect on the replication fork progression, suggesting that the replication fork efficiently bypasses RNA polymerase in vivo. Head-on transcription, in contrast, severely impedes replication fork progression. In the case of head-on transcription, the replication fork is slowed as the result of direct physical interaction with the transcription machinery rather than by propagation of superhelical stress.
MATERIALS AND METHODS
Oligonucleotides.
The P7 promoter was cloned with the following oligonucleotides: 5′-AGTCACTTTCGAGCAATTTTCCTTGAAAAAGAGGTTGACGCTGCAAGGCTCTATACGCATAATGCGCCCCGCAAC-3′ and 5′-GTTGCGGGGCGCATTATGCGTATAGAGCCTTGCAGCGTCAACCTCTTTTTCAAGGAAAATTGCTCGAAAGTGACT-3′. They were flanked either by the HindIII and EcoRI cohesive ends for cloning in the codirectional orientation or by the EcoRV and PstI ends for cloning in the head-on orientation.
The trc promoter was cloned with the following oligonucleotides: 5′-AGCTTCTGCAGAAATGAGCTGTTGACAATTAATCATCCGGCTCGTATAATGTGTGGAATTGTGAGCGGATAACAATTTCACACAGGAAACTGCA-3′ and 5′-GTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACATTATACGAGCCGGATGATTAATTGTCAACAGCTCATTTCTGCAGA-3′.
Plasmids.
Plasmids pP7-CD, pP7-HO, and pTrc-CD were constructed by cloning annealed primers containing P7, P7, and trc promoters, respectively, into the polylinker of the pTrcΔ vector, which was constructed by us earlier (43). Plasmid pTrc-HO was made by cloning the trc promoter, obtained by PCR from the pTrcCat/Pst plasmid (20), between the EcoRI and PstI sites of the pTrcΔ polylinker.
Plasmids pP7-CDΔPlac, pP7-HOΔPlac, pTrc-CDΔPlac, and pTrc-HOΔPlac were constructed in two steps. First, the PstI-PvuI fragment, containing the promoterless part of the bla gene, in pP7-CD, pP7-HO, pTrc-CD, and pTrc-HO, respectively, was replaced with the same part of the bla gene but with the promoter. The latter fragment was obtained by PCR by using pTrcCat/Pst plasmid as a template. Second, the MluI-BfrBI fragment, containing the lacIq promoter, was deleted from the resultant plasmids.
Plasmid pTrc-HO/LacIq was constructed by cloning the missing 3′ part of the lacIq gene into the Bsp120I and HindIII sites of the pTrc-HO plasmid.
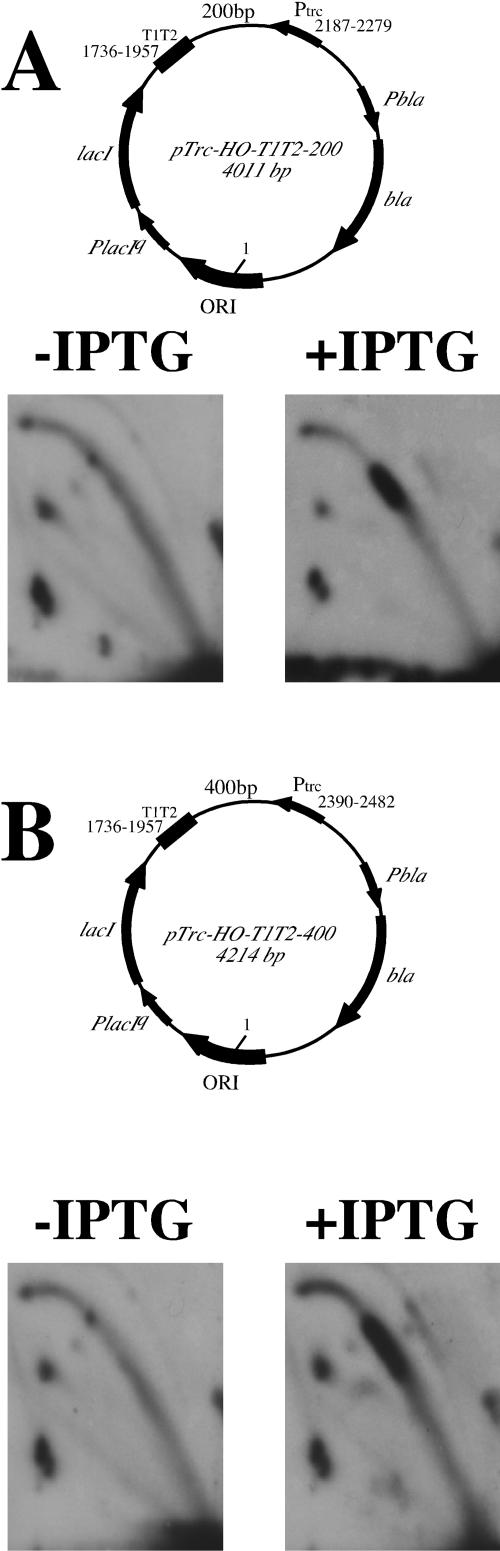
Plasmids pTrc-HO/T1T2-200 and pTrc-HO/T1T2-400 were constructed in two steps. First, the bla gene promoter was inserted into the pTrc-HO/LacIq plasmid. Second, T1T2 transcription terminators, along with either 200- or 400-bp fragments of the cat gene, were cloned between the EcoRI and HindIII sites of the resultant plasmid. Terminators containing fragments were obtained by PCR of the pTrcCat/Pst plasmid.
Electrophoresis analysis of replication intermediates.
Isolation of replication intermediates from bacteria, their separation by neutral/neutral two-dimensional gel electrophoresis, and mapping of the replication stop zones were performed as previously described (21). All replication intermediates were digested with AlwNI prior to loading onto the gel.
RESULTS
Promoter positioned head-on to replication in the E. coli chromosome impedes the replication fork progression in its natural orientation.
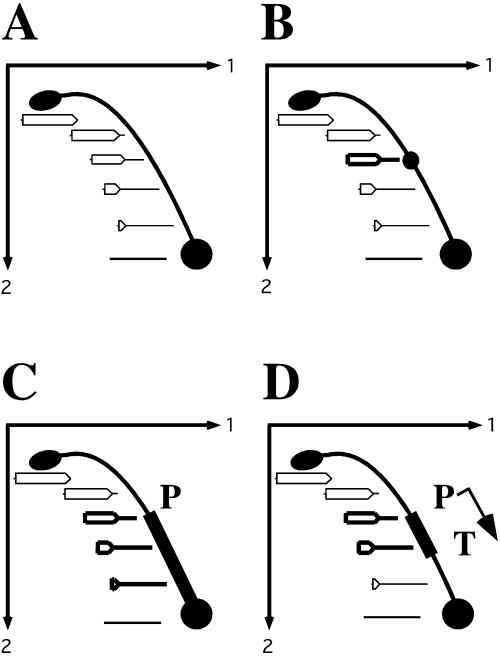
To study the effects of transcription on the replication fork progression, we chose electrophoresis analysis of replication intermediates (5), which is schematically illustrated for our E. coli plasmid system in Fig. 2. ColE1-derived plasmids replicate unidirectionally. Thus, replication intermediates, isolated from cells and cleaved immediately upstream of the origin, are bubble-shaped. The sizes of bubble intermediates increase twofold in the course of replication. These differences in sizes and shapes allow one to separate replication intermediates in two dimensions in the agarose gel, where the progression of the replication fork is manifested by the so-called bubble arc (34). If the replication fork progresses regularly throughout the whole plasmid, the bubble arc is fairly smooth (Fig. 2A). Replication stalling at a plasmid site results in the preferential accumulation of an intermediate of defined size and shape, i.e., the appearance of a distinct bulge on the otherwise smooth arc (Fig. 2B). We have previously applied this approach for detecting replication pauses caused by various DNA repeats (21, 22, 43) and have found it to be extremely reliable and sensitive.
FIG. 2.
Schematic representation of the two-dimensional electrophoresis analysis of replication intermediates. (A) Each dot on the bubble arc represents a replication intermediate, starting from the small bubble at the origin of replication (bottom) and going upward to the biggest, fully replicated bubble (top). The smooth bubble arc reflects the uniform speed of replication throughout the plasmid. (B) If replication progression is slowed down at a particular spot in the plasmid (bold replication intermediate), a distinct bulge appears on the arc. (C) Transcription from the head-on-oriented promoter (P) can inhibit replication progression in the whole DNA segment, separating the promoter and the origin. (D) Insertion of the transcription terminator (T) limits the area of replication inhibition.
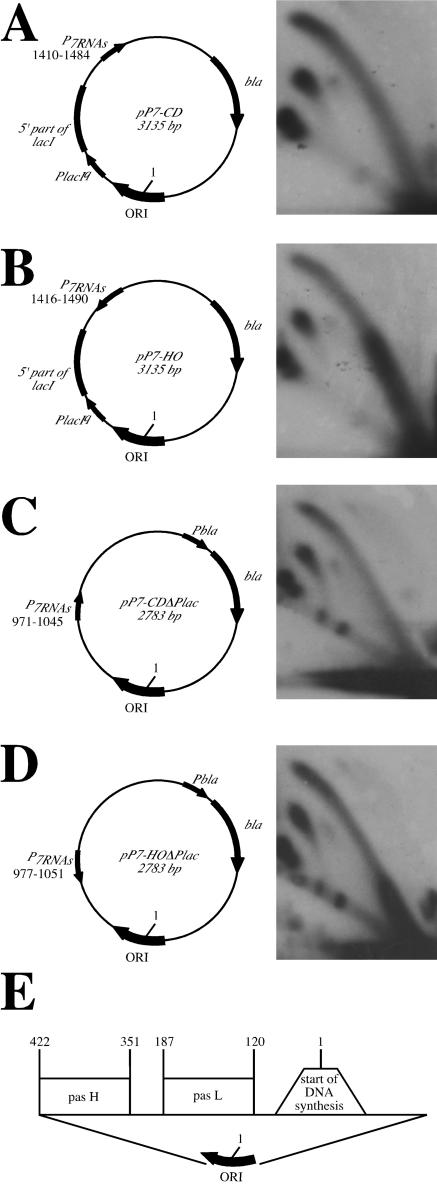
We first chose a constitutive E. coli promoter that drives the expression of the cluster of seven tRNA genes, positioned on minute 15 of the E. coli chromosome (37). This promoter, which we designated P7, is oriented head-on to the direction of replication in its normal chromosomal position (2). This promoter was recloned in two orientations into the pTrcΔ plasmid (43). The latter plasmid, derived from the pTrc99A vector (1), contains the entire replication origin from the pBR322 plasmid, including the pasL and pasH sites (Fig. 3E) that are required for switching from DNA polymerase I- to DNA polymerase III-mediated replication (32). As one can see from Fig. 3A and B, the P7 promoter is positioned approximately 1,000 bp downstream from the pasH site in those plasmids, i.e., in the area replicated by the DNA polymerase III holoenzyme. The effects of this promoter on DNA replication were studied by two-dimensional electrophoresis of replication intermediates (Fig. 3). One can clearly see that when transcription from the P7 promoter is codirectional with replication, replication was not affected (Fig. 3A). Head-on transcription, in contrast, imposed severe constraints on the replication fork progression (Fig. 3B). Note that replication was slowed down in a very broad zone, basically corresponding to the whole DNA interval between the P7 promoter and the replication origin. Schematically, this situation is presented in Fig. 2C. It is quite different from the repeat-caused replication stalling described by us previously (21, 22, 43), where distinct stalling sites were observed.
FIG. 3.
Replication inhibition by the head-on-oriented promoter for the seven tRNAs. In plasmids pP7-CD (A) and pP7-CDΔPlac (C), the P7 promoter faces the direction of replication. In the plasmids pP7-HO (B) and pP7-HOΔPlac (D), the P7 promoter faces replication head-on. The structure of the plasmid replication origin is shown (E). In all of the plasmids, nucleotide position 1 corresponds to the replication start site. Positions of the P7 promoter relative to the replication start site in each plasmid are indicated. Replication is inhibited when transcription faces replication head-on.
Bacterial plasmids, including ours, contain more than one promoter. In plasmids shown in Fig. 3A and B, there is a remnant of the lacIq gene, with its strong constitutive promoter, positioned between the origin and P7 promoter head-on to the p7 promoter. It was, thus, conceivable that replication slowing in this case was, in fact, a consequence of the transcription-transcription collision from the head-on P7 and lacIq promoters rather than replication-transcription collision per se. To rule out this possibility, we deleted the lacIq promoter from both P7-containing plasmids and looked at the replication fork progression in the resultant constructs (Fig. 3C and D). The P7 promoter is now located 550 bp downstream from the pasH site, yet again in the area replicated by the DNA polymerase III holoenzyme. One can still see a profound slowing of replication when the P7 promoter faced replication head-on. The area of replication inhibition was smaller than in the previous case, as expected, since the lacIq deletion moved the P7 promoter closer to the origin. We conclude, therefore, that replication is slowed as a result of head-on collision with transcription originated at the P7 promoter.
Mechanisms of transcription-replication collisions.
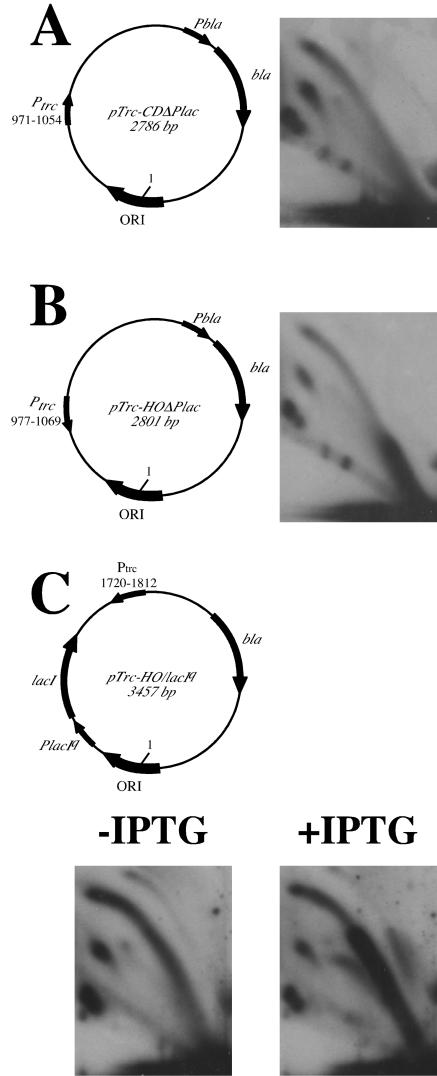
To confirm that transcription is indeed required for the replication fork stalling, we studied the effect of an inducible promoter, trc, on the fork progression. This promoter is an artificial hybrid consisting of the 5′ portion of the trp promoter and the 3′ portion of the lacUV5 promoter. Thus, it is regulated by the lactose repressor.
When cloned into a multicopy plasmid without the repressor gene, this promoter led to replication blockage in the head-on orientation, but not the codirectional orientation, even in the absence of IPTG (isopropyl-β-d-thiogalactopyranoside) (Fig. 4A and B). This result is due to the fact that the amount of the repressor expressed from a single chromosomal lacI gene in the DH5α strain is not sufficient to repress copious plasmid-encoded trc promoters. Note that our data for the trc promoter are virtually identical to those for the P7 promoter (compare Fig. 4B to Fig. 3D). Since very different promoters cause similar inhibitory effects on replication, it is likely that the act of transcription, rather than the nature of a promoter, is responsible for this effect. When the trc promoter was cloned into a plasmid carrying the lacIq gene (Fig. 4C), it was completely repressed in the absence of IPTG, while being strongly activated in its presence (20). Figure 4C shows that replication stalling in this case was evident only when transcription from the head-on-oriented trc promoter was active. We have already ruled out transcription-transcription collision for a similar plasmid (see Fig. 3C and D). Thus, it is active transcription from the head-on promoter that impedes the replication fork progression in vivo.
FIG. 4.
Active transcription is required for the replication inhibition. In the plasmid pTrc-CDΔPlac, the trc promoter faces the direction of replication (A). In the plasmid pTrc-HOΔPlac, the trc promoter faces replication head-on (B). Replication of the pTrc-HO/LacIq plasmid in the absence or presence of IPTG when the promoter is off or on, respectively, is shown in panel C. In all plasmids, nucleotide position 1 corresponds to the replication start site. Positions of the trc promoter relative to the replication start site in each plasmid are indicated. Replication inhibition is evident when transcription is on and faces replication head-on.
Two possible mechanisms are commonly discussed with regard to transcription-replication collisions (3, 11, 14). First, physical interaction between the two complexes could be at fault. Different outcomes for the head-on and codirectional collisions in this scenario could be due to the asymmetry of both complexes (Fig. 1). Second, topological constraints generated by replication and transcription complexes could be involved. In this case, the head-on collision is much more detrimental, since it generates high positive superhelical tension between the replication and transcription complexes moving towards each other.
To distinguish between these opportunities, we limited the area of direct contact between transcription and replication by inserting strong transcription terminators at various distances from the trc promoter. If a physical collision with transcription causes replication slowing, one would expect discrete stops on replication arcs, which are proportional in length to transcribed regions. This result is not expected if replication slowing was caused by topological problems, since superhelical stress rapidly propagates beyond the immediately transcribed areas (20). Our replication data for such constructs are presented in Fig. 5. Limiting transcription from the trc promoter to either 200- (Fig. 5A) or 400-bp (Fig. 5B) segments leads to the appearance of discrete protuberances on replication arcs, contrary to the extensive stalling zones observed without transcription terminators (Fig. 4C). This situation is shown schematically in Fig. 2D. It is also evident that the lengths of the replication stalling zones are proportional to the lengths of the transcribed DNA segments.
FIG. 5.
Insertion of transcription termination signals limits the area of replication inhibition. Replication of the plasmids, where transcription from the head-on trc promoter is limited to either 200- (A) or 400-bp (B) DNA segments, in the absence or presence of IPTG, is shown. In these plasmids, nucleotide position 1 corresponds to the replication start site. Positions of the trc promoter and transcription terminators relative to the replication start site are indicated.
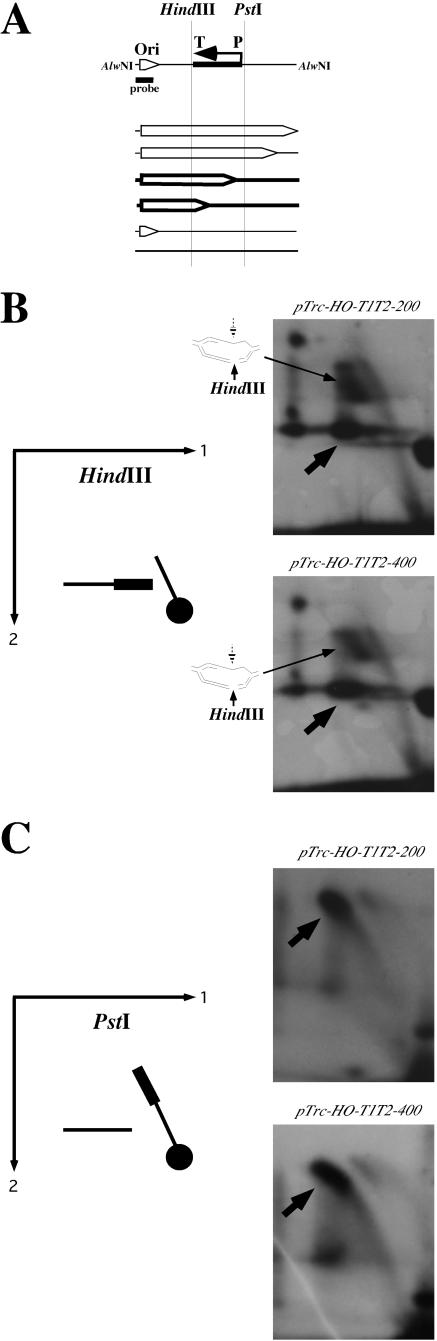
To provide further evidence that the replication stall zones coincide with the transcribed segments in our plasmids, we used a modification of the two-dimensional gel electrophoresis of replication intermediates, where an additional in-gel restriction digest was performed upon the first dimension of the gel electrophoresis. This approach, which is presented schematically in Fig. 6A, was used previously by us for mapping replication stall sites (21, 43). Upon in-gel digest, a fraction of replication intermediates was converted into identical Y-shaped molecules that migrate as a horizontal line in the second dimension.
FIG. 6.
Mapping of the replication stop zones. (A) Schematic representation of the in-gel digest of the replication intermediates after the first dimension of the two-dimensional gel electrophoresis. The vertical lines show the positions of the restriction sites immediately upstream (PstI) and downstream (HindIII) of the transcribed area, situated between the trc promoter (P) and transcription terminator (T). Specifically, the HindIII site is located in nucleotide position 1708 relative to the replication start site in both plasmids, while the PstI site is located in position 2280 in the pTrc-HO/T1T2-200 plasmid and in the position 2483 in the pTrc-HO/T1T2-400 plasmid. Replication intermediates are shown as bubbles, and those drawn with thick lines reflect replication stalling. Upon HindIII digestion, stalled intermediates become Y shaped, while after PstI digestion they remain bubble shaped. Replication intermediates of plasmids pTrc-HO/T1T2-200 and pTrc-HO/T1T2-400, isolated upon growth in the presence of IPTG, were digested with either HindIII (B) or PstI (C) after the first dimension of the electrophoresis. Note the arc-to-line transition of stalled replication intermediates, shown by thick arrows, in panel B but not in panel C. The partially digested underreplicated intermediates, migrating between the arc and the line in panel B, are shown by thin arrows (see text for details).
When digestion occurred immediately downstream of the transcribed segment, a bulk of stalled replication intermediates shifted from the bubble arc to the horizontal line, which means that replication stall zone does not spread beyond the transcription terminator (Fig. 6B). A fraction of stalled intermediates migrated between the bubble arc and the line. The shape of these intermediates is, therefore, less compact than the Y shape but more compact than the bubble. This migration pattern could be explained by assuming that the lagging strand at the HindIII site was underreplicated, resulting in incomplete digestion (21, 43).
In contrast, when digestion was made immediately upstream of the transcribed segment, the stalled replication intermediates remained on the bubble arc (Fig. 6C). Altogether, these data definitively show that replication fork stalls within the transcribed DNA segment, not outside of it. We conclude, therefore, that it is the physical collision with the head-on transcription machinery that causes replication fork to stall.
DISCUSSION
In E. coli, DNA replication proceeds throughout the whole life cycle. Furthermore, the speed of the replication fork movement is an order of magnitude faster than that of RNA polymerase. The combination of these two factors makes collisions between replication and transcription inevitable (3). Depending on a gene's orientation within its replichore, the replication fork would collide with RNA polymerase codirectionally or head-on. Since the E. coli chromosome seems to prefer the codirectional arrangement (2), it was proposed that head-on collisions are more detrimental for the chromosomal replication than codirectional ones (3). While some in vitro and in vivo experiments supported this idea (14, 25, 26, 39), other studies observed replication stalling for both types of collisions (12, 13) or neither of them (34). Here, we revisited this matter by using electrophoresis analysis of replication intermediates for ColE1-type plasmids in vivo. Our data explicitly demonstrate that progression of the replication fork is profoundly slowed when it collides with transcription head-on, but not when it collides codirectionally.
How relevant are the plasmid data to the replication of the bacterial chromosome? During replication of ColE1-type plasmids, the first 400 nt of the leading strand are synthesized by the DNA polymerase I, followed by switching to the θ-type replicative synthesis of both DNA strands by the DNA polymerase III holoenzyme (reviewed in reference 31). This switch depends on two cis-elements, pasL and pasH (32). pasL, which is situated on the lagging strand template 150 bp downstream from ori, is exposed as a consequence of the DNA polymerase I progression to serve as an assembly site for the lagging strand DNA synthesis machinery. pasH, which is positioned on the leading strand template another 250 bp downstream, mediates the PolI-PolIII switch during the leading strand synthesis. While the existence of the PolI-PolIII switching during the leading strand synthesis was established long ago (45) and supported by the numerous data (reviewed in reference 19), few studies question its value. For example, pUC vectors and their derivatives lacking pasH sites are highly replication proficient (48). Replication of the ColE1 plasmid might resist, at least to some extent, the inactivation of DNA polymerase III in a polCts mutant (10). Recently, an error-prone version of DNA polymerase I was shown to elevate the mutation rate as far as several kbp away from the origin in a plasmid lacking both pas sites (9). Note that none of these data are, in fact, definitive. In the absence of the original pasH site in a plasmid, polymerase switching in the leading strand might still occur at a fortuitous DNA sequence(s). Sustained replication of the ColE1 plasmid in the polCts mutant could be due to incomplete inactivation of the DNA polymerase III (10). Finally, in plasmids lacking both pas sites, the extended replication by the DNA polymerase I (8) could obviously be due to the inefficient polymerase switching. Yet these data raise a question on the efficiency of DNA polymerase switching for ColE1-type plasmids. Based on the mutational analysis in a strain with the error-prone DNA polymerase I, polymerase switching in plasmids lacking pas sites was estimated to occur with 70 to 95% efficiency (8). One would expect this percentage to be much higher in pas-containing plasmids.
Still, the mere notion that polymerase switching may not be 100% efficient for ColE1-type plasmids should be taken into account for the interpretation of our results. Do we detect head-on collisions between the RNA polymerase and the genuine replication fork, containing DNA polymerase III? Alternatively, could these collisions happen between transcription and DNA polymerase I, which extended its synthesis beyond the pas sites? We believe that our data make the latter a slim possibility. First, all our plasmids contain both pas sites; thus, polymerase switching should be highly efficient. Second, replication stalling takes place in the broad area between the replication origin and a promoter in plasmids shown in Fig. 3B and 4C. This area consists of a 400-bp-long DNA segment, always replicated by DNA polymerase I, and either 1,000- (Fig. 3B) or 1,300-bp-long (Fig. 4C) DNA segments, normally replicated by the DNA polymerase III. If DNA polymerase I alone were responsible for head-on collisions with transcription, one would expect a much stronger stalling within the first 400 bp from the ori than in the rest of the transcribed area. Evidently, this is not the case. Third, in plasmids, shown in Fig. 5, collisions between replication and transcription start at 1,736 bp downstream from the ori, i.e., in the area replicated by the DNA polymerase III in most instances. If these collisions were due to the occasional escape of Pol I-mediated DNA synthesis beyond pas sites, they should be much weaker than those observed (Fig. 3 and 4). Again, this is clearly not the case. We believe, therefore, that we observe collisions between the genuine replication fork and head-on transcription in vivo. Thus, our conclusions could be expanded to the chromosomal replication in E. coli.
For codirectional collision, it has been suggested that the replication fork simply follows the elongating RNA polymerase until transcription is complete (3). If this were true, the normal speed of the replication fork progression of ∼1,000 bp per s (16) would have been reduced to the speed of transcription elongation, i.e., 50 nt per s (15). A significant reduction in the replication rate due to codirectional collision with RNA polymerase was indeed observed in one study (12), but not in others (14, 26). In the present study, we failed to detect any reduction in the replication fork rate upon codirectional collisions with transcription. As discussed above, our plasmids are largely replicated by the DNA polymerase III holoenzyme. Consequently, the speed of the replication elongation in our case is more than sufficient to catch up with RNA polymerases moving codirectionally. One can argue that collisions between codirectionally moving replisomes and RNA polymerases occur in a small fraction of plasmid molecules that are undetectable by our analysis. We do not think that this possibility is very likely. We would expect the fraction of plasmids with replication-transcription collisions to be roughly the same in both codirectional and head-on situations. Based on our results, head-on collisions are easily detected. We believe, therefore, that the replication fork manages to bypass the codirectionally moving RNA polymerase or displace it from the DNA template. Direct data on the fate of RNA polymerase upon collision with the replication machinery are needed to support this hypothesis.
While it is generally believed that replication is inhibited upon head-on collision with transcription in vivo (11, 14, 46), such inhibition was not immediately evident in all of the studies (4, 34). Furthermore, the nature of transcription-caused replication inhibition was also under debate. Two possible scenarios have been proposed: the physical interaction with the transcription machinery or excessive positive superhelicity generated by the two head-on processes. We found that replication pausing zones are, in fact, strictly limited to transcribed DNA areas. This finding strongly indicates that the physical interaction with transcription is responsible for the replication inhibition. Note that our data do not argue with the accumulation of positive supercoils upon the head-on transcription-replication collisions (39), but they exclude these topological constrains as the cause for replication inhibition.
Interestingly, even in the case of transcriptional repression, one can still see a weak but defined replication stall site situated right at the promoter area (Fig. 5). This finding may suggest that the front edge of RNA polymerase may serve as a contrahelicase. Alternatively, some sort of abortive transcription at the repressed promoter could be responsible for this replication blockage. The existence of such abortive transcription at the repressed lac promoter was first suggested in reference 23.
It would clearly be of great interest to see what happens with transcription when it collides with the replication fork. In the previous in vivo study on this matter, RNA polymerase was dislodged from DNA when it encountered the replication fork in both orientations (14). Studies in vitro, by contrast, demonstrate that RNA polymerase resumes transcription after the replication fork has passed by in either orientation (12, 13, 25, 26). In light of our data, we would like to speculate that RNA polymerase is probably displaced upon both codirectional and head-on collisions with replication in vivo, though it takes much longer in the head-on event. Further studies are needed to test this hypothesis.
In a recent study of gene distribution in bacteria, it was found that it is likely the essentiality rather than the expressiveness of a gene that determines its orientation relative to the replication (42). Ninety percent of essential genes in B. subtilis and 70% of essential genes in E. coli are transcribed codirectionally with replication. The authors believe that the deleterious consequences of the head-on transcription-replication collision could be due to the formation of truncated transcripts and, consequently, truncated proteins, serving as dominant-negative forms of essential proteins. Note, however, that the amount of such truncated peptides should be negligible, given the short time that the replication fork spends at a given gene. Second, our data are indicative of RNA polymerase displacement during codirectional collisions, which should lead to the formation of truncated peptides as well. Therefore, we think that a different explanation is possible. Blockage of DNA replication by various barriers, including but not limited to DNA damage, could lead to the fork stalling that might require subsequent restart. Although the PriA- or RecG-mediated fork restarts in bacteria should be error-proof (30, 35), in the case of head-on transcription-replication collision there is a fair chance that the restarted fork would encounter another elongating RNA polymerase. We believe, therefore, that these multiple replication restarts could lead to mutations and thus should be avoided within essential genes.
Acknowledgments
We thank Vadim Nikiforov for encouragement; Maria Krasilnikova for help with experiments; Nick Cozzarelli, Mike O'Donnell, Hiroshi Hiasa, Patrick Higgins, Joan and Ron Conaway, and Michael Leffak for their critical comments; and Randal Cox for editorial help.
This work was supported by NIH grant GM60987 to S.M.M.
REFERENCES
- 1.Aman, E., B. Ochs, and K.-J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Brewer, B. J. 1988. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 53:679-686. [DOI] [PubMed] [Google Scholar]
- 4.Brewer, B. J., and W. L. Fangman. 1994. Initiation preference at a yeast origin of replication. Proc. Natl. Acad. Sci. USA 91:3418-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. [DOI] [PubMed] [Google Scholar]
- 6.Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55:637-643. [DOI] [PubMed] [Google Scholar]
- 7.Brewer, B. J., D. Lockshon, and W. L. Fangman. 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71:267-276. [DOI] [PubMed] [Google Scholar]
- 8.Camps, M., and L. A. Loeb. 2004. When Pol I goes into high gear: processive DNA synthesis by Pol I in the cell. Cell Cycle 3:116-118. [PubMed] [Google Scholar]
- 9.Camps, M., J. Naukkarinen, B. P. Johnson, and L. A. Loeb. 2003. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc. Natl. Acad. Sci. USA 100:9727-9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, J., P. Williams, and D. R. Helinski. 1975. Plasmid ColE1 DNA replication in Escherichia coli strains temperature-sensitive for DNA replication. Mol. Gen. Genet. 136:273-289. [DOI] [PubMed] [Google Scholar]
- 11.Deshpande, A. M., and C. S. Newlon. 1996. DNA replication fork pause sites dependent on transcription. Science 272:1030-1033. [DOI] [PubMed] [Google Scholar]
- 12.Elías-Arnanz, M., and M. Salas. 1997. Bacteriophage φ29 DNA replication arrest caused by codirectional collisions with the transcription machinery. EMBO J. 16:5775-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elías-Arnanz, M., and M. Salas. 1999. Resolution of head-on collisions between the transcription machinery and bacteriophage φ29 DNA polymerase is dependent on RNA polymerase translocation. EMBO J. 18:5675-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French, S. 1992. Consequences of replication fork movement trough transcription units in vivo. Science 258:1362-1365. [DOI] [PubMed] [Google Scholar]
- 15.Gotta, S. L., O. L. J. Miller, and S. French. 1991. rRNA transcription rate in Escherichia coli. J. Bacteriol. 173:6647-6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirose, S., S. Hiraga, and T. Okazaki. 1983. Initiation site of deoxyribonucleotide polymerization at the replication origin of the Escherichia coli chromosome. Mol. Gen. Genet. 189:422-431. [DOI] [PubMed] [Google Scholar]
- 17.Ivessa, A. S., B. A. Lenzmeier, J. B. Bessler, L. K. Goudsouzian, S. L. Schnakenberg, and V. A. Zakian. 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 12:1525-1536. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, T., and T. Horiuchi. 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1:465-474. [DOI] [PubMed] [Google Scholar]
- 19.Kornberg, A., and T. Baker. 1992. DNA replication, 2nd ed., p. 641-647. W. H. Freeman and Co., New York, N.Y.
- 20.Krasilnikov, A. S., A. Podtelezhnikov, A. Vologodskii, and S. M. Mirkin. 1999. Large-scale effects of transcriptional DNA supercoiling in vivo. J. Mol. Biol. 292:1149-1160. [DOI] [PubMed] [Google Scholar]
- 21.Krasilnikova, M. M., G. M. Samadashwily, A. S. Krasilnikov, and S. M. Mirkin. 1998. Transcription through a simple DNA repeat blocks replication elongation. EMBO J. 17:5095-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krasilnikova, M. M., E. V. Smirnova, A. S. Krasilnikov, and S. M. Mirkin. 2001. A new trick for an old dog: TraY binding to a homopurine-homopyrimidine run attenuates DNA replication. J. Mol. Biol. 313:271-282. [DOI] [PubMed] [Google Scholar]
- 23.Lee, J., and A. Goldfarb. 1991. lac repressor acts by modifying the initial transcribing complex so that it cannot leave the promoter. Cell 66:793-798. [DOI] [PubMed] [Google Scholar]
- 24.Linskens, M. H., and J. A. Huberman. 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:4927-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, B., and B. M. Alberts. 1995. Head-on collision between a DNA replication apparatus and RNA. Science 267:1131-1137. [DOI] [PubMed] [Google Scholar]
- 26.Liu, B., M. L. Wong, R. L. Tinker, E. P. Geiduschek, and B. M. Alberts. 1993. The DNA replication fork can pass RNA polymerase without displacing the nascent transcript. Nature 366:33-39. [DOI] [PubMed] [Google Scholar]
- 27.Liu, L. F., and J. C. Wang. 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 84:7024-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López-Estraño, C., J. B. Schvartzman, D. B. Krimer, and P. Hernández. 1999. Characterization of the pea rDNA replication fork barrier: putative cis-acting and trans-acting factors. Plant Mol. Biol. 40:99-110. [DOI] [PubMed] [Google Scholar]
- 29.López-Estraño, C., J. B. Schvartzman, D. B. Krimer, and P. Hernández. 1998. Co-localization of polar replication fork barriers and rRNA transcription terminators in mouse rDNA. J. Mol. Biol. 277:249-256. [DOI] [PubMed] [Google Scholar]
- 30.Marians, K. J. 2000. PriA-directed replication fork restart in Escherichia coli. Trends Biochem. 25:185-189. [DOI] [PubMed] [Google Scholar]
- 31.Marians, K. J. 1992. Prokaryotic DNA replication. Annu. Rev. Biochem. 61:673-719. [DOI] [PubMed] [Google Scholar]
- 32.Marians, K. J., W. Soeller, and S. L. Zipursky. 1982. Maximal limits of the Escherichia coli replication factor Y effector site sequences in pBR322 DNA. J. Biol. Chem. 257:5656-5662. [PubMed] [Google Scholar]
- 33.Maric, C., B. Levacher, and O. Hyrien. 1999. Developmental regulation of replication fork pausing in Xenopus laevis ribosomal RNA genes. J. Mol. Biol. 291:775-788. [DOI] [PubMed] [Google Scholar]
- 34.Martin-Parras, L., P. Hernández, M. Martinez-Robles, and J. B. Schvartzman. 1991. Unidirectional replication as visualised by two-dimensional agarose gel electrophoresis. J. Mol. Biol. 220:843-855. [DOI] [PubMed] [Google Scholar]
- 35.McGlynn, P., and R. G. Lloyd. 2002. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 18:413-419. [DOI] [PubMed] [Google Scholar]
- 36.McLean, M. J., K. H. Wolfe, and K. M. Devine. 1998. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol. 47:691-696. [DOI] [PubMed] [Google Scholar]
- 37.Nakajima, N., H. Ozeki, and Y. Shimura. 1981. Organization and structure of an E. coli tRNA operon containing seven tRNA genes. Cell 23:239-249. [DOI] [PubMed] [Google Scholar]
- 38.Nomura, M., and E. A. Morgan. 1977. Genetics of bacterial ribosomes. Annu. Rev. Genet. 11:297-347. [DOI] [PubMed] [Google Scholar]
- 39.Olavarrieta, L., P. Hernández, D. B. Krimer, and J. B. Schvartzman. 2002. DNA knotting caused by head-on collision of transcription and replication. J. Mol. Biol. 322:1-6. [DOI] [PubMed] [Google Scholar]
- 40.Peter, B. J., C. Ullsperger, H. Hiasa, K. J. Marians, and N. R. Cozzarelli. 1998. The structure of supercoiled intermediates in DNA replication. Cell 94:819-827. [DOI] [PubMed] [Google Scholar]
- 41.Putter, V., and F. Grummt. 2002. Transcription termination factor TTF-I exhibits contrahelicase activity during DNA replication. EMBO Rep. 3:147-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rocha, E. P., and A. Danchin. 2003. Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat. Genet. 34:377-378. [DOI] [PubMed] [Google Scholar]
- 43.Samadashwily, G. M., G. Raca, and S. M. Mirkin. 1997. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 17:298-304. [DOI] [PubMed] [Google Scholar]
- 44.Sánchez-Gorostiaga, A., C. López-Estraño, D. B. Krimer, J. B. Schvartzman, and P. Hernández. 2004. Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol. Cell. Biol. 24:398-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staudenbauer, W. L. 1977. Replication of the ampicillin resistance plasmid RSF1030 in extracts of Escherichia coli: separation of the replication cycle into early and late stages. Mol. Gen. Genet. 156:27-34. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi, Y., T. Horiuchi, and T. Kobayashi. 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 17:1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, H.-Y., S. Shyy, J. C. Wang, and L. F. Liu. 1988. Transcription generates positively and negatively supercoiled domains in the template. Cell 53:433-440. [DOI] [PubMed] [Google Scholar]
- 48.Yanish-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, Z., D. M. Macalpine, and G. M. Kapler. 1997. Developmental regulation of DNA replication: replication fork barriers and programmed gene amplification in Tetrahymena thermophila. Mol. Cell. Biol. 17:6147-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]