Abstract
It is generally thought that mucosal fluids protect underlying epithelial surfaces against opportunistic infection via their antimicrobial activity. However, our published data show that human tear fluid can protect against the major opportunistic pathogen Pseudomonas aeruginosa independently of bacteriostatic activity. Here, we explored the mechanisms for tear protection, focusing on impacts of tear fluid on bacterial virulence factor expression. Results showed that tear fluid suppressed twitching motility, a type of surface-associated movement conferred by pili. Previously, we showed that twitching is critical for P. aeruginosa traversal of corneal epithelia, exit from epithelial cells after internalization, and corneal virulence. Inhibition of twitching by tear fluid was dose-dependent with dilutions to 6.25% retaining activity. Purified lactoferrin, lysozyme, and contrived tears containing these, and many other, tear components lacked the activity. Systematic protein fractionation, mass spectrometry, and immunoprecipitation identified the glycoprotein DMBT1 (Deleted in Malignant Brain Tumors 1) in tear fluid as required. DMBT1 purified from human saliva also inhibited twitching, as well as P. aeruginosa traversal of human corneal epithelial cells in vitro, and reduced disease pathology in a murine model of corneal infection. DMBT1 did not affect PilA expression, nor bacterial intracellular cyclicAMP levels, and suppressed twitching motility of P. aeruginosa chemotaxis mutants (chpB, pilK), and an adenylate cyclase mutant (cyaB). However, dot-immunoblot assays showed purified DMBT1 binding of pili extracted from PAO1 suggesting that twitching inhibition may involve a direct interaction with pili. The latter could affect extension or retraction of pili, their interactions with biotic or abiotic surfaces, or cause their aggregation. Together, the data suggest that DMBT1 inhibition of twitching motility contributes to the mechanisms by which mucosal fluids protect against P. aeruginosa infection. This study also advances our understanding of how mucosal fluids protect against infection, and suggests directions for novel biocompatible strategies to protect our surface epithelia against a major opportunistic pathogen.
Author summary
Pseudomonas aeruginosa is an opportunistic pathogen that causes life-threatening infections. P. aeruginosa disease is increasing in prevalence while bacteria continue to evolve antibiotic resistance. It is not clear how mucosal fluids usually protect against opportunistic pathogens. Knowing the key ingredients would help us understand susceptibility and develop novel biocompatible therapeutics. Mucosal fluid factors suppressing bacterial virulence may induce less bacterial resistance than traditional antimicrobials. Here we show that DMBT1, an abundant mucosal fluid glycoprotein, enabled tear fluid to inhibit P. aeruginosa twitching motility. We also show DMBT1 directly binds pili, which mediate twitching motility, suggesting a potential mechanism for twitching inhibition. Reflecting the known importance of twitching motility in virulence, purified DMBT1 reduced P. aeruginosa traversal of human cornea epithelial cell layers in vitro, and protected against P. aeruginosa induced disease in vivo, as does whole human tear fluid. These findings contribute to our understanding of mucosal fluid protection against infection, and suggest that DMBT1, or its derivatives, have potential as novel anti-virulence agents that protect against infection.
Introduction
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen ubiquitous in our environment. It is a leading cause of life-threatening infections in debilitated individuals in the hospital setting [1], and of sight-threatening corneal disease in healthy people who wear contact lenses [2, 3]. However, the mechanism(s) by which medical devices at any mucosal surface predispose to infection with P. aeruginosa or other opportunists remains poorly understood [4, 5].
The surface of the eye is normally bathed in tear fluid, which like other mucosal fluids contains many proteins-peptides, lipids, small molecule metabolites, and electrolytes. Indeed, more than 1000 proteins have been identified in healthy human tear fluid [6]. In addition to playing antimicrobial roles, mucosal fluids function to provide lubrication, remove foreign debris, provide homeostatic factors, and repair epithelial damage [7], Our previous studies have confirmed that tear fluid collected from healthy people can protect corneas (of mice) against P. aeruginosa infection in vivo [8].
Of likely relevance to the pathogenesis of contact lens related infections, when a contact lens is worn it dramatically reduces normal tear exchange between the greater tear fluid reservoir and the space between the lens and ocular surface [9, 10]. Suggesting that tear fluid biochemistry is altered under a worn lens, and that this is potentially relevant to the pathogenesis of infection, bacteria inoculated on the back surface of worn lenses grew more efficiently after 8 h of wear compared to 1 h of wear [11]. Candidate antimicrobial tear components in tear fluid that could be impacted by lens wear include; complement, defensins, lactoferrin, lipocalin, lysozyme, secretory phospholipase A2, secretory IgA, soluble mucins (Muc5AC), and/or surfactant proteins (SP-A, SP-D) [12].
However, mechanisms other than antimicrobial activity can also contribute to the protective activity of tear fluid against P. aeruginosa virulence. Indeed, only ~ 50% of P. aeruginosa clinical isolates are susceptible to tear fluid bacteriostatic activity [13], but almost all show reduced virulence in tear fluid. Further, these tear fluid activities are mechanistically separable [13]. Relevant to this, tear fluid can act directly on epithelial cells to enhance their resistance to P. aeruginosa virulence [13, 14] through upregulation of epithelial cell innate defense factors, e.g. RNase7 and ST2 [14, 15], alterations to microRNA expression [14], and the regulation of transcription factors NFκB and AP-1 [14]. Non-bacteriostatic activities of tear fluid also include its capacity to disperse P. aeruginosa biofilms [16], which are thought to be key to the pathogenesis of device-related infections.
A multitude of P. aeruginosa virulence factors can participate in virulence during corneal infection, most playing redundant roles but some required for full virulence [4, 17]. Among the major contributions is twitching motility [18, 19]. Twitching motility is a surface-associated bacterial movement conferred by extension and retraction of type IV pili (T4P) commonly used by Gram-negative bacteria [20]. While assisting bacterial adhesion to surfaces, retraction of pili can bring bacteria into intimate contact with the surface, allowing it to migrate away from the initial point of contact or toward an attractant, to reposition cells with respect to one another (e.g. differentiation within a biofilm), and can also help cells efficiently escape from surfaces when desirable [21]. In P. aeruginosa, the T4P consist of a polymer of the PilA major pilin subunit, while the extension and retraction of pili are controlled by ATPases PilB, PilU and PilT [22, 23]. While twitching motility mutants are able to adhere to, and invade, human corneal epithelial cells grown in vitro, they have a reduced capacity to exit cells after invasion [19]. Twitching mutants are also defective in their ability to traverse multilayers of epithelial cells [19], which may explain their lack of virulence in vivo [18].
Given the protective effect of tear fluid against P. aeruginosa, and the critical role of twitching in corneal pathogenesis, we examined the impact of tear fluid on twitching motility. Systematic fractionation of human tear fluid, combined with mass spectrometry and immuno-precipitation, identified DMBT1 (also known as glycoprotein-340) as required for tear inhibition of twitching motility. DMBT1 purified from saliva was sufficient when used alone for inhibiting twitching, preventing P. aeruginosa traversal of multilayered epithelial in vitro, and for reducing corneal disease severity in a murine model of P. aeruginosa keratitis. These results suggest a novel function for mucosal fluid and specifically DMBT1 in innate defense against infection.
Results
Human tear fluid inhibits P. aeruginosa twitching motility
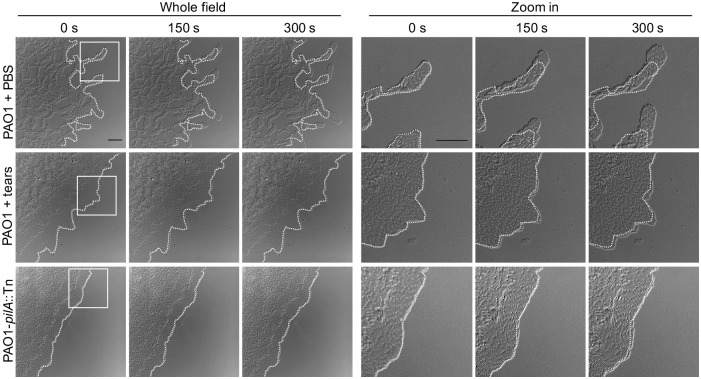
Time-lapse imaging was used to examine the impact of human tear fluid on twitching motility of P. aeruginosa strain PAO1. For this purpose, specialized agar media was used, absorbed with undiluted human tear fluid or PBS (see Methods). For each sample, images were collected with the colony edge positioned half way across the field. To determine speed of bacterial movement, time-lapse imaging was done by repeated 10 second interval image capture over a period of 300 seconds. Velocity of the twitching competent P. aeruginosa strain PAO1 was compared with and without tear fluid added to the media. PAO1-pilA::Tn, lacking twitching motility, was used as a negative control. Fig 1 shows the edge of representative bacterial colonies over a period of 300 seconds (0, 150 and 300 seconds) for PAO1 with and without tear fluid, and compared to the pilA twitching mutant control. Comparison of the colony edge at each time point relative to the dotted white line illustrating the colony edge at the start of the experiment, showed that tear fluid significantly reduced twitching motility compared to PBS (Fig 1, S1, S2 and S3 Videos).
Fig 1. Human tear fluid inhibition of P. aeruginosa twitching motility.
Captured frames of P. aeruginosa PAO1 twitching motility in a 5 min video after 4 h incubation on twitching media absorbed with PBS or undiluted human tear fluid. The twitching mutant PAO1-pilA::Tn served as a negative control. A 5 min time-lapse video was captured at 10 s intervals using a 60 × oil-immersion lens. Dotted lines indicate initial colony edges. Scale bar = 20 μm.
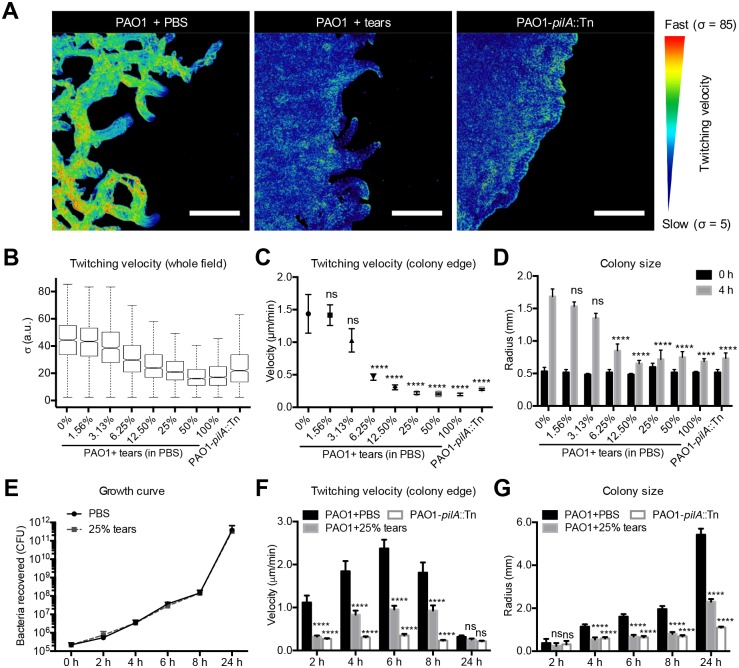
Fig 2 shows quantitative analysis of the impact of tear fluid on reducing twitching motility quantitatively using three methods; i) averaging velocity of all bacteria in the field i.e., bacterial motility in the whole field was corresponded to the standard deviation (σ) of pixel intensity in a 5 min movie (Fig 2A and 2B), ii) examining movement of 10 individual bacteria at the colony edge in a 5 min movie to better represent the concerted movements of twitching [24] (Fig 2C), and iii) quantifying colony size over time for 3 colonies per sample, based on published data that surface-associated twitching movement promotes colony expansion on solid surfaces [21] (Fig 2D). In each instance, tear fluid exposure caused a dose-dependent inhibition of twitching motility (as measured by reduced bacterial velocity and colony size expansion) after 4 h. Tear fluid dilutions up to and including as low as 6.25% retained some degree of inhibition (Fig 2C and 2D). Controls confirmed that tear fluid at 25% dilution, which inhibited twitching motility similarly to undiluted tears, did not inhibit bacterial growth (Fig 2E).
Fig 2. Tear fluid inhibition of P. aeruginosa twitching motility in a dose-dependent manner.
(A) PAO1 motility was measured as the standard deviation (σ) of pixel intensity in a 5 min video. Larger standard deviations correspond to greater pixel intensity modulation resulting from higher bacterial motility. Blue represents slower movement. Scale bar = 50 μm. (B) Effect of tear fluid concentration on PAO1 twitching motility quantified using the above method. (C) Effect of tear fluid concentration on PAO1 twitching quantified by displacement of bacteria in the leading edge of the colony from the first to last slide of the same experiment shown in panel B. Ten single bacteria in the leading edge in each video were tracked. (D) Effect of tear fluid concentration on PAO1 colony size at time zero and 4 h incubation times on twitching media in the same experiment shown in panel B. (E) P. aeruginosa PAO1 growth on twitching media in PBS or 25% tear fluid. (F) PAO1 twitching velocity measured in 5 min videos on twitching media with PBS or 25% human tear fluid after different incubation times. (G) PAO1 colony size expansion on twitching media with PBS or 25% human tear fluid after different incubation times. In each panel, data are expressed as the mean ± SEM per sample from at least three independent experiments. Significance was determined by one-way ANOVA with Tukey's post-hoc analysis for twitching velocity and growth, and two-way ANOVA with Tukey's post-hoc analysis for colony size. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
Since we had previously shown that P. aeruginosa could overcome the cytoprotective activities of tear fluid with prolonged exposure (8 h or more) [13], we also examined the impact of tear fluid (25%) on PAO1 twitching motility for up to 24 h. Tear fluid (25%) maintained inhibition of twitching for up to 8 h (Fig 2F and 2G). At 24 h, twitching velocity was very slow in PBS, and not significantly different from tear fluid-treated PAO1 or the pilA mutant (Fig 2F). However, tear fluid retained significant inhibition of colony size expansion at 24 h (Fig 2G). While that result likely reflected the cumulative effects of tear-mediated twitching inhibition, it suggested that the bacteria do not readily adapt to tear inhibition of twitching, or that the tear factor(s) involved are not readily compromised.
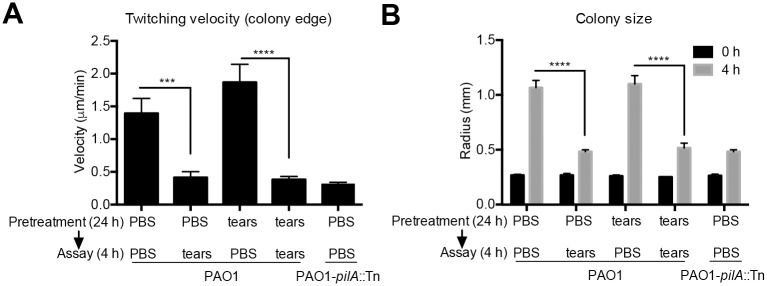
The above hypotheses were supported by experiments in which P. aeruginosa was pretreated with 25% tear fluid on twitching media for 24 h, before transfer to fresh media (PBS or 25% tear fluid) for 4 h. Results showed that 24 h tear pretreatment did not affect bacterial susceptibility to inhibition of twitching as measured by twitching velocity (Fig 3A) or colony size expansion (Fig 3B). The data also showed that tear-exposed P. aeruginosa recovered twitching once placed in PBS indicating reversible inhibition.
Fig 3. P. aeruginosa does not adapt to tear fluid inhibition of twitching motility.
PAO1 was pretreated with PBS or 25% human tear fluid for 24 h on twitching media, then transferred to new twitching media with PBS or 25% human tear fluid for 4 h. Bacterial twitching velocity in 5 min videos (A) and colony size expansion (B) were measured. Data are shown as the mean ± SEM per sample from three independent experiments. Significance was determined using two-way ANOVA with Tukey's post-hoc analysis. ****, P < 0.0001; ***, P < 0.001; ns, not significant.
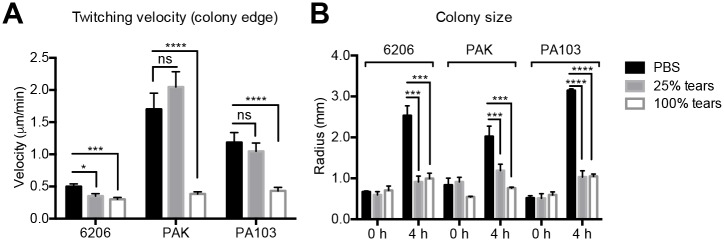
Several other P. aeruginosa strains were also tested for susceptibility to tear inhibition of twitching motility (Fig 4). Strains 6206, PAK, and PA103 were each susceptible to tear inhibition as measured by reduced velocity (Fig 4A) or reduced colony size expansion (Fig 4B). Some variability was noted between these strains and PAO1. For example, undiluted tear fluid was required for reducing twitching velocity of strains PAK and PA103. Nevertheless, these data show that tear fluid inhibition of twitching motility is not restricted to one strain.
Fig 4. Human tear fluid inhibition of twitching motility on multiple P. aeruginosa strains.
(A) Twitching velocity of P. aeruginosa strains 6206, PAK, and PA103 under the same experimental conditions used for PAO1. (B) Colony size on twitching media for P. aeruginosa strains 6206, PAK, and PA103 at 0 and 4 h incubation times. In each panel, data are expressed as the mean ± SEM per sample from at least three independent experiments. Significance was determined by one-way ANOVA with Tukey's post-hoc analysis for twitching velocity and growth, and two-way ANOVA with Tukey's post-hoc analysis for colony size. ****, P < 0.0001; ***, P < 0.001; *, P < 0.05; ns, not significant.
Identification of DMBT1 as a tear fluid factor inhibiting twitching motility
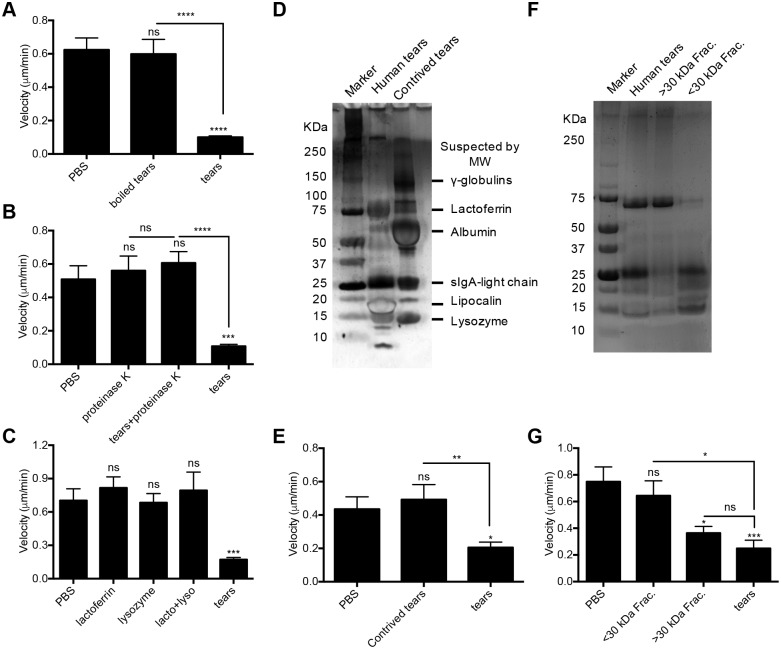
To begin to identify the tear factor(s) involved in twitching inhibition, human tear fluid was boiled or treated with proteinase K prior to bacterial exposure. In each instance, tears lost their inhibitory effects on twitching motility (Fig 5A and 5B) suggesting a heat-labile protein(s) was involved. Lysozyme and lactoferrin (alone or combined) at concentrations found in human tears [25] had no effect on twitching motility (Fig 5C). Commercially available contrived human tears containing lysozyme, lipocalin, albumin, lactoferrin, and gamma-globulins, and resembling tear fluid by SDS-PAGE (Fig 5D), also had no effect (Fig 5E). However, separation of human tear fluid into molecular weight fractions greater or less than ~30 kDa (Fig 5F), revealed that > ~30 kDa fractions inhibited twitching motility, while < ~30 kDa fractions did not (P < 0.05, ANOVA) (Fig 5G).
Fig 5. Heat-sensitive protein(s) in human tear fluid > 30 kDa inhibit P. aeruginosa twitching motility.
P. aeruginosa PAO1 twitching velocity on twitching media was measured after 4 h. (A) Human tear fluid (25%) heated at 95°C for 10 min (boiled) then centrifuged to remove precipitated proteins lost inhibitory activity against twitching. (B) Human tear fluid (25%) treated by proteinase K (100 μg/mL) at 42°C for 2 h lost inhibitory activity. (C) Purified lysozyme (2 mg/mL), lactoferrin (2 mg/mL) or a combination cocktail do not inhibit P. aeruginosa PAO1 twitching motility. (D) Protein analysis of human tears and contrived tears by SDS-PAGE. (E) Contrived tears do not inhibit P. aeruginosa PAO1 twitching motility. (F) Protein analysis of human tear fluid fractions by SDS-PAGE after separation using a ~30 kDa cut-off column. (G) Tear fractions > 30 kDa inhibited PAO1 twitching motility. Data are shown as the mean ± SEM per sample from three independent experiments. Significance was determined using one-way ANOVA with Tukey's post-hoc analysis. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
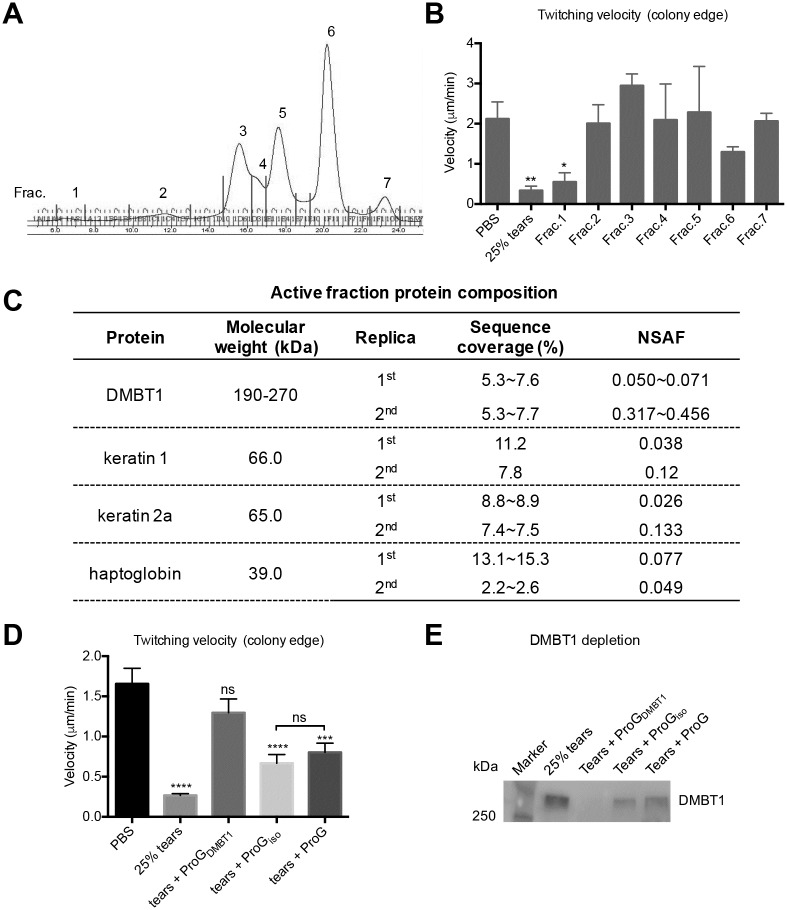
Human tear fluid was separated into 7 fractions by size exclusion chromatography (Fig 6A & S2A Fig), and each fraction tested for inhibition of twitching motility in duplicate experiments. Consistent with our previous results (Fig 5F), only the high molecular weight fractions (Fraction 1 in Fig 6A, and Fraction 2 in S2A Fig) significantly inhibited P. aeruginosa PAO1 twitching motility (Fig 6B & S2B Fig). Since the active fraction in the first experiment (S2A & S2B Fig) was of high molecular weight, a different column material (Superose 6) was used for the second fractionation to obtain better protein separation. Proteins in the active fractions (Fraction 1 in Fig 6A, and Fraction 2 in S2A Fig) were analyzed by mass spectrometry. Results revealed the presence of only 4 proteins; DMBT1 (Deleted in Malignant Brain Tumors 1), keratin 1, keratin 2a and haptoglobin, were present in active fractions of both experiments (Fig 6C & S1 Table).
Fig 6. Identification of DMBT1 as the tear fluid inhibitor of P. aeruginosa twitching motility.
(A) Human tear fluid was separated into 7 fractions using size exclusion chromatography. (B) Effect of tear fractions on twitching velocity of PAO1 reveals a high Mw fraction retains inhibitory activity. (C) Mass spectrometric analysis of high Mw tear fractions from two size-exclusion experiments reveal 4 proteins common to fractions inhibiting twitching motility. (D) DMBT1-depleted human tear fluid does not inhibit twitching motility of PAO1. (E) Western blot analysis of samples used in (D) shows depletion of DMBT-1 from tear fluid, and partial depletion by isotype control and protein G only beads control. Data shown in panels B and D as mean ± SEM from three independent experiments. Significance was determined using a one-way ANOVA with Tukey's post-hoc analysis. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
It has been shown that spectrum counting and mass spectrometry chromatograms correlate with quantitative changes in protein amount [26]. Considering that large proteins tend to contribute more peptide/spectra than small ones, the NSAF (normalized spectral abundance factor) was used to account for the effect of protein length on spectral counts, which allowed a comparison of individual protein abundance in multi-protein complexes [27]. Based on NSAF values, DMBT1 showed the highest relative abundance in active fractions for both experiments, and was therefore considered the most likely candidate for tear inhibition of P. aeruginosa twitching motility.
To directly evaluate DMBT1 involvement in tear inhibition of twitching motility, DMBT1 was immunoprecipitated from human tear fluid. Human tear fluid-depleted of DMBT1 lost inhibition of P. aeruginosa twitching motility compared to 25% tears, with no significant difference in twitching velocity found between PBS and tear-fluid depleted of DMBT1 (Fig 6D). It was noted that tear fluid treated with isotype control antibody and protein G only beads partially inhibited twitching motility (Fig 6D). Western immunoblot (Fig 6E) confirmed that DMBT1 was efficiently removed from tear fluid. Fig 6E also showed that the isotype control and protein G only beads partially depleted DMBT1 suggesting a degree of non-specific binding, but consistent with observed partial effects on twitching velocity (Fig 6D). Together, these data suggested that DMBT1 was required for human tear fluid inhibition of P. aeruginosa twitching motility.
DMBT1 purified from saliva also inhibits P. aeruginosa twitching motility
DMBT1 is expressed in multiple tissues and body fluids and can undergo modifications that could affect its function at specific sites [28, 29]. Since DMBT1 is abundant in saliva, we tested if human saliva could inhibit P. aeruginosa twitching motility. Results confirmed a significant reduction in P. aeruginosa twitching velocity by human saliva treatment from a mean (± SEM) of 1.16 (± 0.14) μm/min in PBS controls to 0.27 (± 0.02) μm/min with human saliva treatment (P < 0.0001, one-way ANOVA, Tukey's post-hoc analysis). The latter velocity was not significantly different from the reduction achieved by 25% tears 0.48 (± 0.06) μm/min (P = 0.2316). Thus, DMBT1 was purified from human saliva, and tested for inhibition of twitching motility. DMBT1 purification was achieved by exploiting DMBT1 binding to/aggregation of Streptococcus pyogenes, and the bound DMBT1 then released from the aggregated S. pyogenes with EDTA treatment [30, 31]. Results (S3 Fig & S2 Table) showed that after purification using S. pyogenes, DMBT1 was the only protein common in two independent fractions (two experimental replicates), and the major protein in the purified fractions based on NSAF. This fraction was referred to as "purified DMBT1" in subsequent experiments.
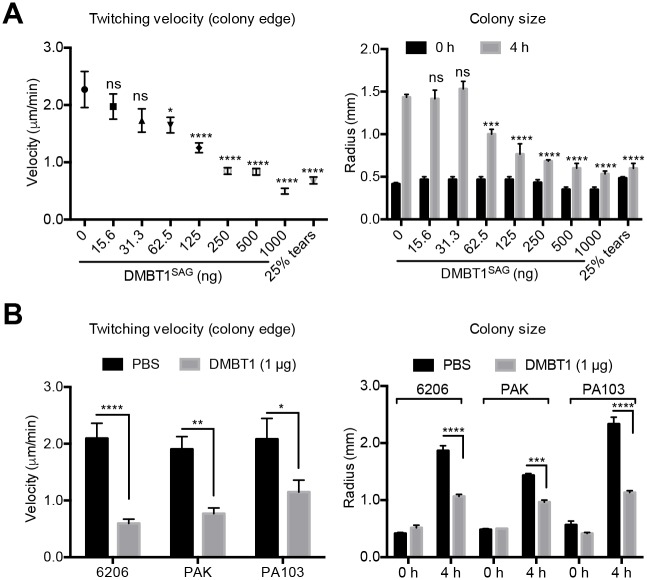
The purified DMBT1 fraction from saliva caused a dose-dependent inhibition of PAO1 twitching velocity and colony size expansion (Fig 7A). In each instance, a significant inhibition was achieved with concentrations of DMBT1 equal to or greater than 12.5 ng/μl (5 μl drop placed onto twitching media) (Fig 7A). DMBT1 at 1 μg in PBS (placed on twitching media) also inhibited twitching motility of all of three other P. aeruginosa strains (Fig 7B).
Fig 7. Saliva-purified DMBT1 inhibits twitching of multiple P. aeruginosa strains.
DMBT1 solutions (5 μl) at different concentrations up to 200 ng/μl DMBT1 in PBS were dropped onto twitching media then inoculated with bacteria and incubated for 4 h at 37°C. Twitching velocity and colony size were quantified. (A) Purified DMBT1 from saliva inhibited P. aeruginosa PAO1 twitching velocity and colony size expansion in a dose-dependent manner. (B) Purified DMBT1 from saliva (1 μg) inhibited twitching velocity and colony size expansion in three other P. aeruginosa strains. In each panel, data are expressed as mean ± SEM per sample from three independent experiments. Significance was determined by one-way ANOVA with Tukey's post-hoc analysis for twitching velocity, and two-way ANOVA with Tukey's post-hoc analysis for colony size. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
Mass spectrometry analysis of high Mw fractions that inhibit P. aeruginosa twitching motility, from human tear fluid (S1 Table) or saliva (S2 Table) showed that only DMBT1 was present in all samples, and that it was the most abundant protein further supporting the hypothesis that DMBT1 is responsible for inhibition of twitching motility in tear fluid and saliva.
Saliva-purified DMBT1 inhibits P. aeruginosa traversal of multilayered human corneal epithelial cells in vitro
Our previous studies have shown that twitching motility contributes to P. aeruginosa traversal of corneal epithelial cells [19], and that tear fluid protects against P. aeruginosa traversal [8]. Thus, we hypothesized that purified DMBT1 would also inhibit P. aeruginosa traversal of multilayered human corneal epithelial cells.
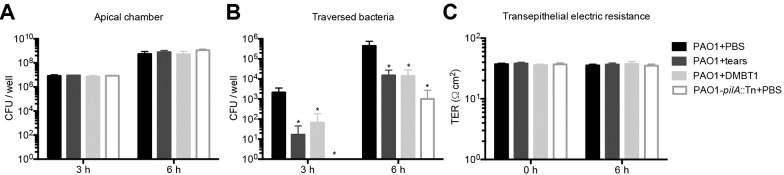
Human corneal epithelial cells (hTCEpi) were grown on Transwell filters (3 μm pore-size) and airlifted for 7 days to form multilayers. P. aeruginosa PAO1 was added to the apical surface with DMBT1 solution, PBS or human tear fluid. After 3 and 6 h, viable bacteria from the apical and basal chambers were counted. Human tear fluid (50%) or DMBT1 (100 ng/μl) had no effect on bacterial growth in the apical chamber (Fig 8A), but significantly inhibited P. aeruginosa traversal at 3 h and 6 h (Fig 8B). As expected, the pilA mutant showed significantly reduced traversal compared to wild-type PAO1. Transepithelial resistance (TER) was unaffected in each sample over the 6 h incubation (Fig 8C) consistent with our published data for wild-type P. aeruginosa [19].
Fig 8. DMBT1 purified from saliva inhibits P. aeruginosa traversal of multilayered human corneal epithelial (hTCEpi) cells in vitro.
Traversal of P. aeruginosa PAO1 or its pilA mutant across cultured airlifted human corneal epithelial cells in vitro. PAO1 was incubated in PBS, treated with 50% human tear fluid, or with 100 ng/μl of DMBT1. The pilA mutant was added in PBS. (A) Viable bacterial counts (means ± SD) in the apical chamber were determined at 3 and 6 h to evaluate bacterial growth. (B) Viable bacterial counts (means ± SD) in the basal chamber were determined at 3 and 6 h to estimate traversed bacteria. In each instance significance was determined using one-way ANOVA with Tukey’s post-hoc analysis. *, P < 0.05. (C) TER (Ω·cm2) across the human corneal epithelial cells over 6 h. A Transwell filter without cells was used as a control. TER values shown represent TER(sample)—TER(bank).
DMBT1 purified from saliva protects against P. aeruginosa corneal infection in vivo
Our previous studies showed that twitching motility was important for P. aeruginosa virulence in a murine scarification model of corneal infection [18], and that human tear fluid can protect against P. aeruginosa corneal infection in both scarification and healing injury models [8]. Thus, we explored if DMBT1 could protect against P aeruginosa infection in vivo using a mouse model.
After scarification injury and 6 h healing, mouse corneas were inoculated with P. aeruginosa PAO1 in PBS or DMBT1 (see Methods). Representative images (Fig 9A) show that corneas inoculated with PAO1 in PBS presented with clear signs of infection after day 1 that progressed further by day 2. DMBT1 treated corneas showed reduced disease pathology at day 1, and greatly reduced pathology at day 2. Quantification of disease severity using a grading system that accounted for area of infection, density of opacity, and surface irregularity (Fig 9B) [32] showed DMBT1 treated corneas had a significant reduction in area of infection at day 1 and day 2 (Fig 9C), and in corneal opacity at day 2 (Fig 9D). Surface irregularity was relatively unaffected (Fig 9E). Overall disease severity was significantly reduced in DMBT1 treated eye at both time points (Fig 9F) showing that DMBT1 protected corneas from P. aeruginosa keratitis.
Fig 9. DMBT1 purified from saliva protects against P. aeruginosa corneal infection.

(A) Representative images of C57BL/6 murine corneas at 24 and 48 h and post-infection with P. aeruginosa PAO1 in PBS or DMBT1 (150 ng/μL). (B) Schematic for grading disease severity of infected murine corneas. Effect of DMBT1 on corneal infection disease severity scores at 24 and 48 h comparing (C) area of infection, (D) density of opacity, (E) corneal surface irregularity, and (F) total disease severity, the sum of values shown in (C), (D), and (E). Data are reported as the mean ± SEM per group over three independent experiments (6 mice per group in total). Significance of differences between groups was determined using the Mann-Whitney U test. **, P < 0.01; *, P < 0.05; ns, not significant.
DMBT1 does not affect P. aeruginosa PilA expression or cyclicAMP but does bind pili
Salivary DMBT1 is a well recognized agglutinin for Gram-positive and Gram-negative bacteria except P. aeruginosa [31, 33–36]. Tear fluid DMBT1 was also shown to bind Staphylococcus aureus, but not P. aeruginosa, using anti-DMBT1 antibody in a dot-immunoblot assay [29]. We confirmed that DMBT1 purified from saliva aggregates S. pyogenes [30], but does not aggregate P. aeruginosa (S4 and S5 Videos), suggesting a novel mechanism for inhibiting P. aeruginosa twitching motility.
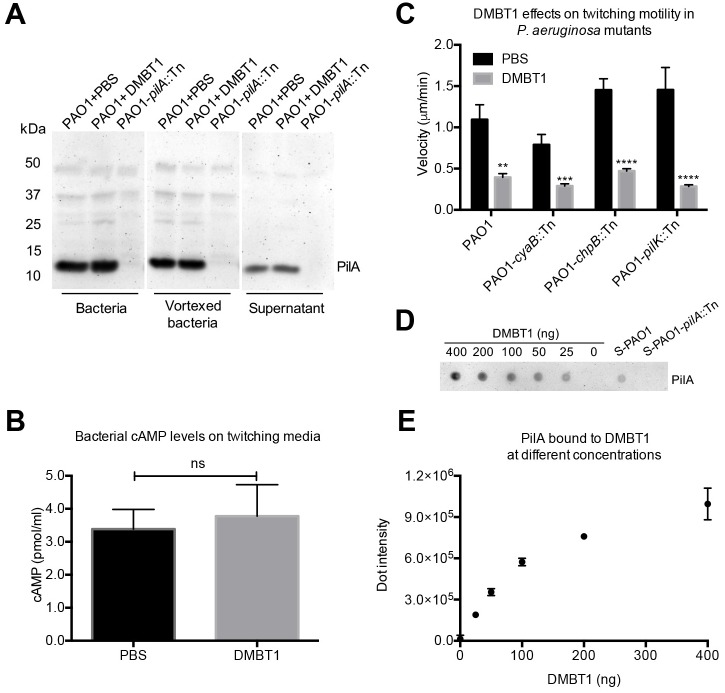
Type IV pilus (T4P) production and twitching motility in P. aeruginosa is controlled by the Pil-Chp pathway (encoded by gene cluster IV) containing pilG/H/I/J/K and chpA/B/C/D/E genes [37]. The Chp system controls T4P production by modulation of cyclicAMP; twitching motility is cyclicAMP-independent [38]. To begin to explore the mechanism for DMBT1 inhibition of twitching motility, we examined some key elements of the pathways involved. Western immunoblots showed no difference in PilA expression by P. aeruginosa PAO1 after DMBT1 exposure for 4 h (Fig 10A) suggesting that pilin production was unaffected. Purified DMBT1 also had no effect on cyclicAMP levels in P. aeruginosa PAO1 collected from twitching media after 4 h (Fig 10B). Since many chemotaxis mutants of P. aeruginosa lose twitching motility, it is difficult to determine if DMBT1 inhibition of twitching motility involves chemotaxis genes [38]. However, the latter study identified three mutants of P. aeruginosa strain PAK that retained twitching motility; mutants in cyaB (encoding an adenylate cyclase to control cAMP synthesis), chpB (encoding a methylesterase) which can adjust the methylation status of the sensor module in the pili-mediated chemotaxis system [37], and pilK (encoding a methyltransferase) [38]. In the present study, cyaB, chpB, and pilK mutants of strain PAO1 also retained twitching motility under control conditions (Fig 10C). DMBT1 inhibited twitching of all three mutants (Fig 10C), suggesting that those genes are not needed for DMBT1-mediated twitching inhibition. Dot-immunoblot assays, however, showed that purified DMBT1 could bind pili extracted from P. aeruginosa strain PAO1 (Fig 10D and 10E), suggesting that the mechanism for inhibition of twitching motility involves a direct interaction with P. aeruginosa pili.
Fig 10. DMBT1 does not affect P. aeruginosa PilA expression or cyclicAMP levels, but inhibits twitching of PAO1 mutants in cyaB, chpB, and pilK.
(A) Western immunoblot of PilA expression in P. aeruginosa PAO1 after DMBT1 (100 ng/μl) treatment on twitching media for 4 h. PilA protein levels were unaffected by DMBT1 exposure in all samples (bacteria, vortexed bacteria, and supernatant, see Methods). (B) CyclicAMP levels of P. aeruginosa PAO1 treated with DMBT1 (100 ng/μl) on twitching media for 4 h. (C) Effects of DMBT1 (100 ng/μl) on twitching velocity of PAO1 mutants in cyaB, chpB and pilK after 4 h. Data are shown as the mean ± SEM per sample from three independent experiments. Significance was determined using one-way ANOVA with Tukey's post-hoc analysis. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant. (D) Dot-immunoblot assay using anti-PilA antibody to show the binding of PAO1 pili to DMBT1 after 40 min incubation with a pili-containing extract from PAO1. Diluted extracts from PAO1 (S-PAO1) or its pilA mutant (S-PAO1-pilA::Tn) (see Methods) were used as controls. A representative experiment of two independent experiments is shown. (E) Quantification of dot-intensity from the dot-immunoblot assay shown in D. Data are shown as the mean ± SEM of triplicate measurements from each sample.
Discussion
Previously, we showed that tear fluid could protect human corneal epithelial cells and mouse corneas against P. aeruginosa infection [8, 19], a protective activity mechanistically separable from bacteriostatic activity. Here, we explored the mechanisms involved and found that human tear fluid can inhibit surface-associated twitching motility by P. aeruginosa, dependent on the glycoprotein DMBT1. DMBT1, also known as GP340, is abundant in various mucosal fluids. Used alone, DMBT1 purified from saliva inhibited twitching motility, suggesting it is both necessary and sufficient for the protective activity of tear fluid. DMBT1 did not suppress P. aeruginosa viability, or aggregate bacteria, and its inhibition of twitching was not associated with changes in pilin expression or bacterial cyclicAMP. However, DMBT1 bound pili extracted from P. aeruginosa suggesting a direct interaction with pili is involved in twitching inhibition. Reflecting the known importance of twitching in virulence, DMBT1 protected multilayers of human corneal epithelial cells against P. aeruginosa traversal, and reduced disease severity in an in vivo animal model. Thus, this glycoprotein, or an active derivative, may have potential as a biocompatible therapeutic intervention against P. aeruginosa that acts without suppressing bacterial viability or bacterial aggregation. Although this study was focused on tear fluid and the cornea, the findings might be broadly applicable to other mucosal surfaces that P. aeruginosa infects given that DMBT1 is present in human saliva, the small intestine, and airways [28, 29].
DMBT1 was first isolated from saliva using affinity adsorption to Streptococcus mutans, and identified as a ~300–400 kDa glycoprotein [28]. It belongs to the scavenger receptor cysteine-rich (SRCR) protein superfamily, which is highly conserved down to sponges as secreted or membrane-bound proteins [39]. The human chromosome contains one copy of the gene encoding DMBT1 located on chromosome 10q26.13. However, there are different human DMBT1 alleles within the population and different isoforms in different tissues governed by alternative splicing and post-translation modification, e.g. DMBT1 variations include the number of SRCR domains or patterns of glycosylation [28, 39, 40]. Approximately 25% of the molecular mass of salivary-derived DMBT1SAG (Salivary Agglutinin) is due to glycosylation (~10% for N-glycosylation, and ~15% for O-glycosylation) [41, 42]. Differences in DMBT1 glycosylation were reported between saliva-derived DMBT1SAG and lung-derived DMBT1GP340, the latter lacking Lewis (Le) antigens [43]. Moreover, two isoforms of DMBT1 derived from human tear fluid expresses sialyl-Lea antigens [44], not sialyl-Lex antigens expressed by DMBT1SAG. In our studies, DMBT1 depletion from human tear fluid removed inhibition of twitching motility, while DMBT1 purified from saliva inhibited twitching motility. Thus, reported differences in sialyl-Le antigens do not affect this function suggesting that tear fluid and salivary DMBT1 isoforms share a common domain(s) to fulfill twitching inhibition. Detailed structure function studies to identify domains inhibiting twitching will require further investigation.
The SRCR domains play a key role in the function of DMBT1 in mucosal immunity as a bacterial agglutinin that binds many pathogens including Gram-positive and Gram-negative bacteria, and viruses [31, 33–36]. However, DMBT1 does not aggregate P. aeruginosa strain PAO1 [29], a finding confirmed in the present study (S4 and S5 Videos).
DMBT1 is known to bind other mucosal fluid antimicrobial and immune defense proteins including; SP-D [45], SP-A [46], lactoferrin [47] and secretory IgA [48]. Thus, some of its apparent activities can depend on its binding partners. However, no other known defense factors were found in the mass spectrometry analysis of the high Mw fractions of tear fluid or DMBT1 purified from saliva, which both inhibited twitching motility. While the active tear fraction containing DMBT1 did contain three other proteins in replicate experiments; keratin 1, keratin 2a, and haptoglobin (Fig 4C), none of them were present in the saliva-purified fractions containing DMBT1 that inhibited twitching.
That neither tear fluid nor purified DMBT1 inhibited the growth of P. aeruginosa strain PAO1 (Figs 2E and 8A) is consistent with our previously published data showing a lack of tear fluid bacteriostatic activity against many P. aeruginosa isolates [13].
Thus, the mechanism by which DMBT1 inhibits P. aeruginosa twitching motility appears to be independent of bacterial aggregation, known DMBT1 binding partners, other proteins present in the active fractions with DMBT1, and bacteriostatic activity. Our data also indicated that pilin expression, and bacterial cyclicAMP levels were unaffected by DMBT1 exposure, and DMBT1 could inhibit twitching motility of P. aeruginosa mutants in pilK (encoding a methyltransferase), chpB (encoding a methylesterase), or cyaB (encoding an adenylate cyclase), suggesting that none of these factors were involved.
Purified DMBT1 did, however, bind pili extracted from P. aeruginosa PAO1 suggesting that twitching motility inhibition involves direct interaction with pili. Such interactions could affect numerous aspects of pilus function including; their extension or retraction, their interactions with surfaces (biotic and abiotic), or cause their aggregation. However, twitching motility could also be compromised by DMBT1 at other levels including an alteration of gene expression in the Pil-Chp pathway (or its regulation), or interfering with small molecule regulation, e.g. cyclic-di-GMP [21, 22]. Targets for future study could include pilus extension or retraction motor proteins, e.g. ATPases PilB or PilT respectively [49, 50], or the chemosensory protein PilJ which directly interacts with PilA [23], and also controls pilus extension [51].
It is possible that DMBT1 interacts with other P. aeruginosa surface antigens or structures, in addition to pili, that affect twitching motility and/or other bacterial functions. While DMBT1 did not affect bacterial swimming motility (S5 Video), the full spectrum of DMBT1-P. aeruginosa interactions, and their consequences, will require further investigation.
The protective mechanism of DMBT1 in our in vitro and in vivo infection models is likely to involve suppression of twitching motility, given that twitching is critical to pathogenesis in the cornea [18]. While enabling bacteria to traverse surface epithelial cells, twitching may be used for trafficking along/through the basal lamina [52], and/or for disseminating within the underlying corneal stroma [53]. Twitching is also important for biofilm formation [54], a key virulence determinant when infection is device-related [55, 56].
However, activities of DMBT1 other than inhibition of twitching might have contributed to its protective activities in our in vivo experiments, for example through binding and association with other tear defense proteins. Surfactant protein D (SP-D) which readily binds DMBT1 can protect corneal epithelial cells against P. aeruginosa invasion [57], and it contributes to clearing P. aeruginosa from the ocular surface [58]. IgA, another binding partner, can prevent P. aeruginosa binding to mouse corneas and reduces severity of P. aeruginosa keratitis [59]. Both factors can also function as opsonins facilitating phagocytosis and clearance of P. aeruginosa [60, 61]. Also possible, is that DMBT1 influences pathogenesis via direct effects on resident or infiltrating host cells. Indeed, it can stimulate a dose-dependent chemokinesis (random migration) of alveolar macrophages, suggesting role(s) in respiratory inflammatory and immune responses [46]. It is also able to activate classical and lectin pathways of the complement system [62, 63]. Teasing apart the relative contributions of different DMBT1 activities in protecting the cornea against P. aeruginosa infection in vivo will not be straightforward.
Mice also express a homolog of DMBT1 [64]. The ability of P. aeruginosa to infect control eyes in our study likely reflects characteristics of the infection model, in which murine corneas were washed with PBS prior to bacterial inoculation, and mice sustained under anesthesia for 4 h after inoculation. This methodology would remove murine tear fluid, and reduce tear flow, likely compromising the ability of murine DMBT1 to exert protective effects.
In sum, the results of this study suggest that DMBT1 inhibition of twitching motility contributes to mechanisms by which mucosal fluids protect against P. aeruginosa infection, and is likely accomplished by direct binding to pili. Twitching motility, important to P. aeruginosa virulence both in vitro and in vivo, is also key to biofilm formation. Thus, discovery that DMBT1 modulates bacterial virulence factor expression adds to our understanding of how mucosal fluids defend tissue surfaces against infection. Further, DMBT1 or its derivatives may hold promise for development of biocompatible strategies for preventing P. aeruginosa infection that act by altering expression of virulence genes rather than agglutinating bacteria, or suppressing their viability. Whether wearing a contact lens or other device at mucosal surfaces impacts the quantity, location, or integrity of DMBT1, and if any changes relate to pathogenesis of infection, remains to be determined.
Materials and methods
Ethics statement
Human tear fluid and saliva were collected from healthy volunteers under a protocol approved by the Committee for the Protection of Human Subjects, University of California Berkeley. Informed, written consent was obtained from all participants. All procedures involving mice were carried out in accordance with standards established by the Association for the Research in Vision and Ophthalmology, under the protocol AUP-2016-08-9021 approved by the Animal Care and Use Committee, University of California Berkeley, an AAALAC accredited institution. The protocol adheres to PHS policy on the humane care and use of laboratory animals, and the guide for the care and use of laboratory animals. Anesthesia was achieved by intraperitoneal injection of an anesthetic cocktail containing ketamine (80 mg/Kg) and xylazine (10 mg/Kg), or ketamine (50 mg/Kg) and medetomidine (0.75 mg/Kg) for sustained anesthesia. Euthanasia was performed using carbon dioxide inhalation.
Bacterial strains and culture conditions
P. aeruginosa strains PAO1, PAK, PA103 and 6206 were used. Bacteria were grown on tryptic soy agar (TSA) plates at 37°C for 16 h to obtain lawn cultures. P. aeruginosa PAO1 transposon insertion mutant pilA::Tn (PW8621) [65] lacking twitching motility, was grown on TSA with 60 μg/mL tetracycline, and used as a negative control. P. aeruginosa PAO1 transposon insertion mutants cyaB::Tn (PW6387), chpB::Tn (PW1760) and pilK::Tn (PW1757) [65] were also grown on TSA with 60 μg/mL tetracycline. Each of the transposon mutants was verified by PCR (S1 Fig) using previously reported primers [65] or primers designed for pilK. For twitching motility assays, bacteria were grown on twitching motility Gellan Gum media (TMGG, 0.8 g Gellan gum, 0.4 g tryptone, 0.2 g yeast extract, 0.2 g NaCl, 0.1 g MgSO4·7H2O, in 100 mL H2O) at 37°C in a humidified chamber for different times. Streptococcus pyogenes (ATCC19615) was grown in Brain and Heart Infusion (BHI) broth at 37°C overnight and used for purification of DMBT1.
Reagents
Tear fluid was collected using a 30 μl volume capillary tube. Subjects were non-contact lens wearers, males and females, between 18 and 45 years of age, and with no ocular infection or inflammation at the time of collection. Approximately 5% of the tear fluid from each subject was plated on TSA to control for bacterial contamination; sterile tear fluid was pooled (from 6 to 8 subjects) and stored at -80°C until used. In different experiments, human tear fluid (25%) was boiled at 95°C for 10 min to denature heat-liable components, treated with proteinase k (Sigma-Aldrich, 100 μg/mL) at 42°C for 2 h, or fractionated using sterile water pre-rinsed Microcon centrifugal filter devices with membrane cutoffs of ~30 kDa (Millipore). Saliva was obtained from healthy volunteers, clarified by centrifugation at 3,800 x g for 10 min then used for testing its effect on twitching motility, and for DMBT1 purification as described below. Purified lactoferrin and lysozyme from human milk (2 mg/mL in PBS) were purchased from Sigma-Aldrich. Contrived tear fluid containing lysozyme, albumin, and γ-globulins was purchased from Ursa BioScience (MD, USA).
Twitching motility assays
Twitching motility was measured using a method modified from the microscope slide assay described previously [66]. Bacteria were grown on TSA plates (supplemented with tetracycline if needed) at 37°C for 16 h. Twitching motility Gellan Gum media was dried for 20 min in a sterile airflow (BSL2 Biosafety Cabinet) before use and then 5 μL of tear fluid, PBS or other solution was dropped onto the twitching media until completely absorbed. Bacteria grown on TSA were collected, and mixed using a plastic inoculation loop and subsequently inoculated onto the twitching media using a sterile toothpick to achieve a tip-sized inoculum. A glass coverslip was gently placed onto the twitching media to create an interstitial space. The slides were then incubated at 37°C for 4 h unless otherwise stated. After indicated incubation times, 5 min time-lapse videos were captured at 10 s intervals via differential interference contrast (DIC) microscopy using a Nikon ECLIPS Ti microscope with a 60× oil-immersion objective at 37°C.
Quantification of twitching motility
Twitching motility was quantified with three different methods. Firstly, individual bacterial twitching motility was quantified as the degree to which they modulated light in a DIC image. To normalize the contrast of each image and eliminate contrast bias in areas of high focus/illumination, a band pass filter (2–40 pixels) was used in ImageJ. The degree of modulation was measured as the standard deviation of intensity of each pixel during the length of the movie. The standard deviation map was then thresholded so that only regions containing bacteria were analyzed. A histogram of the standard deviation map was then used to measure the distribution of bacterial motility in each sample. A notched boxplot was used to represent each result. In a notched box plot, if the notches do not overlap then the distributions are significantly different. Secondly, twitching velocity was measured as the twitching distance of the colony leading edge divided by time. The bacterial distance traveled in a 5 min video was measured from location in the first slide to location in the last slide using ImageJ. Different treatment groups for bacteria were done in triplicate and ten bacteria were tracked in each video. Thirdly, bacterial colony size was measured soon after inoculation onto twitching media (time zero) and after different incubation times.
Bacterial growth in the presence of tear fluid
P. aeruginosa PAO1 bacteria were grown on TSA media overnight and then diluted to OD600 0.03 by use of TSB (tryptic soy broth) media. One microliter of diluted bacteria was dropped onto twitching media absorbed with PBS or 25% human tear fluid and incubated at 37°C for up to 24 h. After collection and serial dilution in PBS, samples were plated onto TSA agar and incubated at 37°C overnight to determine numbers of viable bacteria expressed as Colony Forming Units (CFU). Experiments were repeated five times.
Size-exclusion chromatography of human tear fluid
Size-exclusion chromatography was performed on an AKTAmicro system using a Superdex 200 10/300 GL column (GE Healthcare) in the first separation, or a Superose 6 10/300 GL column (GE Healthcare) in the second separation, equilibrated in PBS (pH 7.4). To minimize peak broadening, short lengths of 0.15 mm i. d. tubing were used between the injection valve and the fraction collector. Human tear fluid was injected onto the column, and fractions of 250 μl were collected. Protein was detected by UV absorbance at 280 nm. Eluted fractions were pooled according to protein peaks and concentrated using a ~3 kDa cut-off filter (Millipore). The activity of eluted fractions against P. aeruginosa twitching motility was then assessed as described above.
Mass spectrometry
Mass spectrometry (MS) was performed at the Proteomics/Mass Spectrometry Laboratory, University of California, Berkeley. A nano LC column was packed in a 100 μm inner diameter glass capillary with an emitter tip. The column consisted of 10 cm of Polaris C18 5 μm packing material (Varian, Agilent, CA), followed by 4 cm of Partisphere 5 SCX (Whatman, Sigma-Aldrich, MO). The column was loaded by use of a pressure bomb and washed extensively with buffer A (see below). The column was then directly coupled to an electrospray ionization source mounted on a Thermo-Fisher LTQ XL linear ion trap mass spectrometer. An Agilent 1200 HPLC equipped with a split line to deliver a flow rate of 300 nl/min was used for chromatography. Peptides were eluted using a 4-step MudPIT procedure [67]. Buffer A was 5% acetonitrile/ 0.02% heptaflurobutyric acid (HFBA); buffer B was 80% acetonitrile/ 0.02% HFBA. Buffer C was 250 mM ammonium acetate/ 5% acetonitrile/ 0.02% HFBA; buffer D was same as buffer C, but with 500 mM ammonium acetate.
Protein identification and quantification were done with Integrated Proteomics Pipeline (IP2, Integrated Proteomics Applications, Inc. San Diego, CA) using ProLuCID/Sequest, DTASelect2 and Census [68–70]. Tandem mass spectra were extracted into ms1 and ms2 files from raw files using RawExtractor [71], and searched against the human protein database plus sequences of common contaminants, concatenated to a decoy database in which the sequence for each entry in the original database was reversed [72].
Immunoprecipitation of DMBT1
Immunoprecipitation was performed using mouse monoclonal anti-DMBT1 antibody (ab17779, Abcam, MA), or a mouse IgG1 isotype control (Thermo Fisher, NY), and protein G-magnetic beads (New England BioLabs, MA) according to manufacturer’s protocols. After coating protein G magnetic beads with either DMBT1 antibody or isotype control in a 20 μl reaction, the complex was incubated with 25 μl of 25% tears for 60 min at 4°C. Non-coated protein G magnetic beads were also used as a negative control. The supernatant was then collected for assessing its activity on twitching motility, and its DMBT1 protein content by Western immunoblotting (anti-DMBT1 antibody was diluted 1:1000). The beads were washed 3 times with PBS and then eluted with 20 μl of 0.1 M glycine (pH 2.5) for 3 min at room temperature twice. The supernatant was collected and neutralized with 2 M tris (pH 9.0). The proteins bound to beads were analyzed by MS as described previously.
Purification of DMBT1 from human saliva
DMBT1 was purified from human saliva rather than tear fluid because saliva is more abundant and easier to collect. Purification of DMBT1 was performed as described previously [31, 73]. Briefly, clarified saliva was diluted 50% with PBS. Streptococcus pyogenes was incubated in BHI broth overnight at 37°C, collected by centrifugation at 3,800 x g for 5 min, and washed three times with PBS. Bacterial concentration was adjusted to ~5 x109 CFU/mL. Equal volumes of bacterial suspension and diluted saliva were then mixed and incubated at 37°C for 60 min. Bacterial cells were collected again by centrifugation at 3,800 x g for 5 min, and washed three times with PBS. PBS (1.5 mL) containing 5 mM EDTA was then used to release bound protein at room temperature for 5 min. The bacterial culture was centrifuged at 15,000 x g for 5 min, the supernatant filtered using a 0.22 μm filter, and then dialyzed (Slide-A-Lyzer dialysis cassettes, Thermo Fisher, NY) against PBS at 4°C overnight. Dialyzed eluate was subjected to gel filtration chromatography on a Superose 6 10/300 GL column (GE Healthcare, CA) equilibrated in PBS (pH 7.4). The eluate at void volume was collected and used as purified DMBT1 from saliva. The presence and purity of DMBT1 was verified by mass spectrometry as described above. DMBT1 concentration was measured using a micro BCA protein assay kit (Thermo Scientific, IL, USA).
Bacterial traversal assay
Telomerase-immortalized human corneal epithelial cells (~ 6 × 104 cells) were seeded onto 24-well polyester tissue culture treated Transwell™ inserts (3 μm pore size, Corning Costar, NY) in KGM-2 medium containing 1.15 mM CaCl2 for 7 days, then airlifted for 7 days as previously described [74]. P. aeruginosa strain PAO1, or its pilA mutant (~1.3 × 106 CFU) was inoculated on the apical surface of the cells in PBS, human tear fluid (50% in PBS) or DMBT1 (100 ng/μl in PBS) for 6 h at 37°C (5% CO2). Transepithelial resistance (TER) (Ω·cm2) was measured using an Epithelial Voltohmeter (World Precision Instruments, Inc., FL) before inoculating the bacteria, and after 6 h incubation. Transwell™ inserts without corneal cells served as negative controls. After 3 and 6 h, bacterial viable counts in the apical and basal chambers were determined to measure bacterial growth and epithelial traversal.
Murine corneal infection
All procedures were approved by the University of California, Berkeley Animal Care and Use Committee. The scarification with healing murine model of corneal infection was used as previously described [8] with minor modification. Briefly, C57BL/6 mice (6 to 12 weeks old) were anesthetized by intraperitoneal injection of an anesthetic cocktail containing ketamine (80 mg/Kg) and xylazine (10 mg/Kg). Eyes were checked for corneal clarity using a stereomicroscope prior to the initiation of experiments. Three parallel scratches were made on the right cornea of each anesthetized animal using a sterile 25 5/8-gauge needle. Mice were checked every 15 min until they woke up. After 6 h of epithelial healing, mice were anesthetized with a cocktail containing ketamine (50 mg/Kg) and medetomidine (0.75 mg/Kg), 70 μl per 25 g of body weight. Healing corneas were then washed with PBS (500 μl), then inoculated with 5 μl of a P. aeruginosa PAO1 suspension containing ~2 ×103 CFU bacteria in DMEM mixed with either PBS or 200 ng/μl of DMBT1 at a ratio of 1:3. After 4 h infection under sustained anesthesia, an anesthesia reversal agent, atipamezole (3.75 mg/Kg), 50 μl per 25 g of body weight, was used to wake the mice. Mice were observed daily and ocular images were captured at 24 and 48 h post-inoculation using 2–3% isoflurane in oxygen inhalation for anesthesia. Corneal disease severity was graded by a masked observer using a previously described scoring system [32], which assesses area of infection, density of opacity, surface regularity and overall disease severity.
Immunoblot assays
To study the effects of DMBT1 on pilin expression, PilA was measured by Western immunoblot using sample preparation methods based on previous studies [75]. PAO1 or its pilA mutant were treated with PBS or DMBT1 (100 ng/μl) as described above in the twitching motility assays section. After 4 h incubation, bacteria were washed from the twitching media and cover-slip with 50 mM Na2CO3 (pH 9.6). Bacterial OD600 was adjusted to 0.6, and 300 μl of bacterial culture centrifuged at 13,000 x g for 5 min, and re-suspended into SDS-PAGE sample buffer ("bacteria" sample). Another 300 μl of bacterial culture was extensively vortexed for 3 min to remove pili, centrifuged at 15,000 x g for 20 min, and the pellet dissolved in SDS-PAGE sample buffer ("vortexed bacteria" sample). The supernatant was placed at 4°C overnight, after adding MgCl2 to a concentration of 100 mM, and the next day centrifuged at 15,000 x g for 20 min. The pellet was dissolved in same volume of SDS-PAGE sample buffer ("supernatant" sample). All samples were heated at 95°C for 10 min, separated by SDS-PAGE (20% gel), and probed with antibody to PilA (1:5000) (a kind gift from Dr. Joanne Engel, University of California, San Francisco), then goat anti rabbit-HRP antibody (1:5000, Abcam, MA).
Dot-immunoblotting was used to test if DMBT1 could bind P. aeruginosa pili. Briefly, to prepare an extract of pili, a suspension of P. aeruginosa PAO1 in PBS was prepared to an OD600 of ~10. The suspension was vortexed for 3 min, centrifuged at 15,000 x g for 20 min, and the supernatant collected. MgCl2 solution (1 M) was added to the supernatant to a final concentration of 100 mM, and the supernatant placed at 4°C overnight. After centrifugation at 15,000 x g for 20 min, the pellet was resuspended in PBS (500 μl) to form a pili-containing extract. The same method was used to prepare a negative control extract of PAO1-pilA::Tn. For dot-immunoblot assays, 2 μl of DMBT1 in PBS (400 ng/μl and serial dilutions in PBS) were spotted onto a nitrocellulose membrane (0.2 μm pore-size, BioRad), along with a PBS control, the pili-containing extract from PAO1 (positive control), and the extract from the pilA mutant (negative control). The extracts were diluted 1 in 500 in PBS for use as controls. After the membrane was dry, it was blocked with 5% BSA for 1 h at room temperature, then washed with PBS for 5 min. The membrane was then incubated with the original (undiluted) pili-containing extract of PAO1 for 40 min at room temperature, then washed 5 times with PBS. Membranes were then probed with anti-PilA primary antibody (1:5000) and Goat anti-Rabbit HRP-conjugated secondary antibody (1:5000). Dot intensity was measured using AlphaView FluoChem HD2 software.
CyclicAMP assay
Intracellular cyclicAMP of P. aeruginosa was measured as described previously [38] with minor modification. PAO1 was incubated on twitching media with DMBT1 (100 ng/μl) or PBS at 37°C for 4 h as described above in twitching motility assays. Bacteria were washed from twitching media with 0.9 M NaCl at 4°C, made with superpure water from Cayman Chemical (Ann Arbor, MI) and kept on ice. Bacterial suspensions were adjusted to the same OD600 value of ~1.0. Two equal volumes of each suspension were centrifuged at 13,000 x g for 2 min at 4°C, and the bacterial pellets washed twice with 1 mL of cold 0.9 M NaCl (final OD600 ~ 2.5). Bacterial pellets were suspended in 200 μl of 0.1 N HCl (made with superpure water) and incubated on ice for 10 min with occasional vortexing to lyse the bacteria. Lysates were centrifuged at 13,000 x g for 5 min at 4°C to remove cellular material, and the supernatant was assayed for cAMP concentration using a Cyclic AMP EIA Kit (Cayman Chemical) following the manufacturer’s protocol for sample acetylation.
Statistical analysis
Data were expressed as a mean ± standard error of mean (SEM) unless otherwise stated. The significance of differences between groups was assessed by one or two-way ANOVA with Tukey's post-hoc analysis, or using the Mann-Whitney U test for in vivo experiments. P values of less than 0.05 were considered significant.
Supporting information
Represents a 5 min time-lapse movie of P. aeruginosa twitching motility captured at an interval of 10 s with a 60 × oil-immersion lens. Scale bar = 20 μm. Frame rate = 10 fps.
(AVI)
Represents a 5 min time-lapse movie of P. aeruginosa twitching motility captured at an interval of 10 s with a 60 × oil-immersion lens. Scale bar = 20 μm. Frame rate = 10 fps.
(AVI)
Represents a 5 min time-lapse movie of P. aeruginosa twitching motility captured at an interval of 10 s with a 60 × oil-immersion lens. Scale bar = 20 μm. Frame rate = 10 fps.
(AVI)
Represents a 10 s time-lapse movie of P. aeruginosa movement captured at no delay with a 40 × lens. Frame rate = 10 fps.
(AVI)
Represents a 10 s time-lapse movie of P. aeruginosa movement captured at no delay with a 40 × lens. Frame rate = 10 fps.
(AVI)
PAO1-pilA::Tn (PW8621), PAO1-cyaB::Tn (PW6387), and PAO1-chpB::Tn (PW1760) were verified by PCR with primers provided by the insertion mutant library database. PAO1-pilK::Tn (PW1757) was verified by PCR with the following primers; pilK flanking primers pilK-F (5'-AGATGCGCAACTCGGTATCC-3') and pilK-R (5'-TTCAGGGTTTCGGCGATCTC-3'). The red square was used to label target products.
(TIF)
(A) Fractions of human tear fluid separated by size exclusion chromatography (first experiment). (B) Effect of tear fractions on P. aeruginosa PAO1 twitching velocity measured in 5 min videos of each sample. Data are expressed as the mean ± SEM per sample from three independent experiments. Significance was determined using one-way ANOVA with Tukey's post-hoc analysis. **** P < 0.0001, ***P < 0.001.
(TIF)
(A) SDS-PAGE with silver stain (left panel) suggested DMBT1 was present after S. pyogenes treatment, and was confirmed by Western immunoblot (right panel) using anti-DMBT1 antibody. (B) and (C) Two independent experiments each showing that size-exclusion chromatography after DMBT1 purification from human saliva using S. pyogenes generated a high Mw fraction (fraction 1). Proteins were separated from aggregated S. pyogenes in human saliva using EDTA (5 mM). (D) Mass spectrometric analysis of fraction 1 after DMBT1 purification from saliva revealed the presence of DMBT1 in two independent experiments.
(TIF)
Results shown for two independent fractionations of human tear fluid using size-exclusion chromatography.
(TIF)
Each fraction inhibited twitching motility of P. aeruginosa PAO1. Results represent two independent experiments.
(TIF)
Acknowledgments
Our thanks to Dr. Joanne Engel (UCSF, San Francisco, CA) for providing the antibody to PilA, and to our colleagues from UC Berkeley, Berkeley, CA: Dr. Chris Jeans, for separating proteins from human tears and saliva; Drs. Lori Kohlstaedt and Vincent Coates, for mass spectrometry analysis; Dr. Benjamin Smith, for image analysis, and Ms. Hart Horneman, for scoring corneal pathology in the murine model. P. aeruginosa PAO1 and its mutants were obtained from the P. aeruginosa mutant collection, University of Washington, Seattle WA.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Institutes of Health (R01 EY024060, SF), and by the China Postdoctoral Council (20140085, JL). The P. aeruginosa mutant collection was supported by the National Institutes of Health (P30 DK089507, University of Washington, Seattle, WA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.MacVane SH. Antimicrobial resistance in the intensive care unit: A focus on Gram-negative bacterial infections. J Intensive Care Med. 2017;32(1):25–37. 10.1177/0885066615619895 [DOI] [PubMed] [Google Scholar]
- 2.Yildiz EH, Airiani S, Hammersmith KM, Rapuano CJ, Laibson PR, Virdi AS, et al. Trends in contact lens-related corneal ulcers at a tertiary referral center. Cornea. 2012;31(10):1097–102. 10.1097/ICO.0b013e318221cee0 [DOI] [PubMed] [Google Scholar]
- 3.Ng AL, To KK, Choi CC, Yuen LH, Yim SM, Chan KS, et al. Predisposing factors, microbial characteristics, and clinical outcome of microbial keratitis in a tertiary centre in Hong Kong: A 10-year experience. J Ophthalmol. 2015;2015:769436 10.1155/2015/769436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans DJ, Fleiszig SM. Why does the healthy cornea resist Pseudomonas aeruginosa infection? Am J Ophthalmol. 2013;155(6):961–70.e2. 10.1016/j.ajo.2013.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans DJ, Fleiszig SM. Microbial keratitis: could contact lens material affect disease pathogenesis? Eye Contact Lens. 2013;39(1):73–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou L, Zhao SZ, Koh SK, Chen L, Vaz C, Tanavde V, et al. In-depth analysis of the human tear proteome. J Proteomics. 2012;75(13):3877–85. 10.1016/j.jprot.2012.04.053 [DOI] [PubMed] [Google Scholar]
- 7.Ohashi Y, Dogru M, Tsubota K. Laboratory findings in tear fluid analysis. Clin Chim Acta. 2006;369(1):17–28. 10.1016/j.cca.2005.12.035 [DOI] [PubMed] [Google Scholar]
- 8.Kwong MS, Evans DJ, Ni M, Cowell BA, Fleiszig SM. Human tear fluid protects against Pseudomonas aeruginosa keratitis in a murine experimental model. Infect Immun. 2007;75(5):2325–32. 10.1128/IAI.01404-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNamara NA, Fleiszig SM. Human tear film components bind Pseudomonas aeruginosa. Adv Exp Med Biol. 1998;438:653–8. [DOI] [PubMed] [Google Scholar]
- 10.McNamara NA, Polse KA, Brand RJ, Graham AD, Chan JS, McKenney CD. Tear mixing under a soft contact lens: effects of lens diameter. Am J Ophthalmol. 1999;127(6):659–65. [DOI] [PubMed] [Google Scholar]
- 11.Wu YT, Zhu LS, Tam KP, Evans DJ, Fleiszig SM. Pseudomonas aeruginosa survival at posterior contact lens surfaces after daily wear. Optom Vis Sci. 2015;92(6):659–64. 10.1097/OPX.0000000000000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDermott AM. Antimicrobial compounds in tears. Exp Eye Res. 2013;117:53–61. 10.1016/j.exer.2013.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleiszig SM, Kwong MS, Evans DJ. Modification of Pseudomonas aeruginosa interactions with corneal epithelial cells by human tear fluid. Infect Immun. 2003;71(7):3866–74. 10.1128/IAI.71.7.3866-3874.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mun JJ, Tam C, Evans DJ, Fleiszig SM. Modulation of epithelial immunity by mucosal fluid. Sci Rep. 2011;1:8 10.1038/srep00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mun J, Tam C, Chan G, Kim JH, Evans D, Fleiszig S. MicroRNA-762 is upregulated in human corneal epithelial cells in response to tear fluid and Pseudomonas aeruginosa antigens and negatively regulates the expression of host defense genes encoding RNase7 and ST2. PLoS One. 2013;8(2):e57850 10.1371/journal.pone.0057850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu YT, Tam C, Zhu LS, Evans DJ, Fleiszig SM. Human tear fluid reduces culturability of contact lens associated Pseudomonas aeruginosa biofilms but induces expression of the virulence associated type III secretion system. Ocul Surf. 2017; 15(1): 88–96. 10.1016/j.jtos.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willcox MD. Pseudomonas aeruginosa infection and inflammation during contact lens wear: a review. Optom Vis Sci. 2007;84(4):273–8. 10.1097/OPX.0b013e3180439c3e [DOI] [PubMed] [Google Scholar]
- 18.Zolfaghar I, Evans DJ, Fleiszig SM. Twitching motility contributes to the role of pili in corneal infection caused by Pseudomonas aeruginosa. Infect Immun. 2003;71(9):5389–93. 10.1128/IAI.71.9.5389-5393.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alarcon I, Evans DJ, Fleiszig SM. The role of twitching motility in Pseudomonas aeruginosa exit from and translocation of corneal epithelial cells. Invest Ophthalmol Vis Sci. 2009;50(5):2237–44. 10.1167/iovs.08-2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. 10.1146/annurev.micro.56.012302.160938 [DOI] [PubMed] [Google Scholar]
- 21.Burrows LL. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol. 2012;66:493–520. 10.1146/annurev-micro-092611-150055 [DOI] [PubMed] [Google Scholar]
- 22.Leighton TL, Buensuceso RN, Howell PL, Burrows LL. Biogenesis of Pseudomonas aeruginosa type IV pili and regulation of their function. Environ Microbiol. 2015;17(11):4148–63. 10.1111/1462-2920.12849 [DOI] [PubMed] [Google Scholar]
- 23.Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2015;112(24):7563–8. 10.1073/pnas.1502025112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Semmler AB, Whitchurch CB, Mattick JS. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology. 1999;145 (Pt 10):2863–73. [DOI] [PubMed] [Google Scholar]
- 25.Tiffany JM. The normal tear film. Dev Ophthalmol. 2008;41:1–20. 10.1159/000131066 [DOI] [PubMed] [Google Scholar]
- 26.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem. 2005;77:6218–24. 10.1021/ac050846r [DOI] [PubMed] [Google Scholar]
- 27.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, et al. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A. 2006;103(50):18928–33. 10.1073/pnas.0606379103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madsen J, Mollenhauer J, Holmskov U. Gp-340/DMBT1 in mucosal innate immunity. Innate Immun. 2010;16(3):160–7. 10.1177/1753425910368447 [DOI] [PubMed] [Google Scholar]
- 29.Jumblatt MM, Imbert Y, Young WW Jr., Foulks GN, Steele PS, Demuth DR. Glycoprotein 340 in normal human ocular surface tissues and tear film. Infect Immun. 2006;74(7):4058–63. 10.1128/IAI.01951-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loimaranta V, Hytonen J, Pulliainen AT, Sharma A, Tenovuo J, Stromberg N, et al. Leucine-rich repeats of bacterial surface proteins serve as common pattern recognition motifs of human scavenger receptor gp340. J Biol Chem. 2009;284(28):18614–23. 10.1074/jbc.M900581200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kukita K, Kawada-Matsuo M, Oho T, Nagatomo M, Oogai Y, Hashimoto M, et al. Staphylococcus aureus SasA is responsible for binding to the salivary agglutinin gp340, derived from human saliva. Infect Immun. 2013;81(6):1870–9. 10.1128/IAI.00011-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowell BA, Wu C, Fleiszig SM. Use of an animal model in studies of bacterial corneal infection. ILAR J. 1999;40(2):43–50. [DOI] [PubMed] [Google Scholar]
- 33.Ligtenberg AJ, Veerman EC, Nieuw Amerongen AV, Mollenhauer J. Salivary agglutinin/glycoprotein-340/DMBT1: a single molecule with variable composition and with different functions in infection, inflammation and cancer. Biol Chem. 2007;388(12):1275–89. 10.1515/BC.2007.158 [DOI] [PubMed] [Google Scholar]
- 34.Brittan JL, Nobbs AH. Group B Streptococcus pili mediate adherence to salivary glycoproteins. Microbes Infect. 2015;17(5):360–8. 10.1016/j.micinf.2014.12.013 [DOI] [PubMed] [Google Scholar]
- 35.Prakobphol A, Xu F, Hoang VM, Larsson T, Bergstrom J, Johansson I, et al. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J Biol Chem. 2000;275(51):39860–6. 10.1074/jbc.M006928200 [DOI] [PubMed] [Google Scholar]
- 36.Hartshorn KL, Ligtenberg A, White MR, Van Eijk M, Hartshorn M, Pemberton L, et al. Salivary agglutinin and lung scavenger receptor cysteine-rich glycoprotein 340 have broad anti-influenza activities and interactions with surfactant protein D that vary according to donor source and sialylation. Biochem J. 2006;393(Pt 2):545–53. 10.1042/BJ20050695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sampedro I, Parales RE, Krell T, Hill JE. Pseudomonas chemotaxis. FEMS Microbiol Rev. 2015;39(1):17–46. 10.1111/1574-6976.12081 [DOI] [PubMed] [Google Scholar]
- 38.Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol. 2010;76(4):889–904. 10.1111/j.1365-2958.2010.07135.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ligtenberg AJ, Karlsson NG, Veerman EC. Deleted in malignant brain tumors-1 protein (DMBT1): a pattern recognition receptor with multiple binding sites. Int J Mol Sci. 2010;11(12):5212–33. 10.3390/ijms1112521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polley S, Louzada S, Forni D, Sironi M, Balaskas T, Hains DS, et al. Evolution of the rapidly mutating human salivary agglutinin gene (DMBT1) and population subsistence strategy. Proc Natl Acad Sci U S A. 2015;112(16):5105–10. 10.1073/pnas.1416531112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oho T, Yu H, Yamashita Y, Koga T. Binding of salivary glycoprotein-secretory immunoglobulin A complex to the surface protein antigen of Streptococcus mutans. Infect Immun. 1998;66(1):115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holmskov U, Lawson P, Teisner B, Tornoe I, Willis AC, Morgan C, et al. Isolation and characterization of a new member of the scavenger receptor superfamily, glycoprotein-340 (gp-340), as a lung surfactant protein-D binding molecule. J Biol Chem. 1997;272(21):13743–9. [DOI] [PubMed] [Google Scholar]
- 43.Eriksson C, Frangsmyr L, Danielsson Niemi L, Loimaranta V, Holmskov U, Bergman T, et al. Variant size- and glycoforms of the scavenger receptor cysteine-rich protein gp-340 with differential bacterial aggregation. Glycoconj J. 2007;24(2–3):131–42. 10.1007/s10719-006-9020-1 [DOI] [PubMed] [Google Scholar]
- 44.Schulz BL, Oxley D, Packer NH, Karlsson NG. Identification of two highly sialylated human tear-fluid DMBT1 isoforms: the major high-molecular-mass glycoproteins in human tears. Biochem J. 2002;366(Pt 2):511–20. 10.1042/BJ20011876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madsen J, Tornoe I, Nielsen O, Lausen M, Krebs I, Mollenhauer J, et al. CRP-ductin, the mouse homologue of gp-340/deleted in malignant brain tumors 1 (DMBT1), binds gram-positive and gram-negative bacteria and interacts with lung surfactant protein D. Eur J Immunol. 2003;33(8):2327–36. 10.1002/eji.200323972 [DOI] [PubMed] [Google Scholar]
- 46.Tino MJ, Wright JR. Glycoprotein-340 binds surfactant protein-A (SP-A) and stimulates alveolar macrophage migration in an SP-A-independent manner. Am J Respir Cell Mol Biol. 1999;20(4):759–68. 10.1165/ajrcmb.20.4.3439 [DOI] [PubMed] [Google Scholar]
- 47.Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417(6888):552–5. 10.1038/417552a [DOI] [PubMed] [Google Scholar]
- 48.Reichhardt MP, Jarva H, de Been M, Rodriguez JM, Jimenez Quintana E, Loimaranta V, et al. The salivary scavenger and agglutinin in early life: diverse roles in amniotic fluid and in the infant intestine. J Immunol. 2014;193(10):5240–8. 10.4049/jimmunol.1401631 [DOI] [PubMed] [Google Scholar]
- 49.Chiang P, Sampaleanu LM, Ayers M, Pahuta M, Howell PL, Burrows LL. Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology. 2008;154(Pt 1):114–26. 10.1099/mic.0.2007/011320-0 [DOI] [PubMed] [Google Scholar]
- 50.Bertrand JJ, West JT, Engel JN. Genetic analysis of the regulation of type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa. J Bacteriol. 2010;192(4):994–1010. 10.1128/JB.01390-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeLange PA, Collins TL, Pierce GE, Robinson JB. PilJ localizes to cell poles and is required for type IV pilus extension in Pseudomonas aeruginosa. Curr Microbiol. 2007;55(5):389–95. 10.1007/s00284-007-9008-5 [DOI] [PubMed] [Google Scholar]
- 52.Alarcon I, Kwan L, Yu C, Evans DJ, Fleiszig SM. Role of the corneal epithelial basement membrane in ocular defense against Pseudomonas aeruginosa. Infect Immun. 2009;77(8):3264–71. 10.1128/IAI.00111-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tam C, LeDue J, Mun JJ, Herzmark P, Robey EA, Evans DJ, et al. 3D quantitative imaging of unprocessed live tissue reveals epithelial defense against bacterial adhesion and subsequent traversal requires MyD88. PLoS One. 2011;6(8):e24008 10.1371/journal.pone.0024008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30(2):295–304. [DOI] [PubMed] [Google Scholar]
- 55.Mack D, Rohde H, Harris LG, Davies AP, Horstkotte MA, Knobloch JK. Biofilm formation in medical device-related infection. Int J Artif Organs. 2006;29(4):343–59. [DOI] [PubMed] [Google Scholar]
- 56.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–22. [DOI] [PubMed] [Google Scholar]
- 57.Ni M, Evans DJ, Hawgood S, Anders EM, Sack RA, Fleiszig SM. Surfactant protein D is present in human tear fluid and the cornea and inhibits epithelial cell invasion by Pseudomonas aeruginosa. Infect Immun. 2005;73(4):2147–56. 10.1128/IAI.73.4.2147-2156.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mun JJ, Tam C, Kowbel D, Hawgood S, Barnett MJ, Evans DJ, et al. Clearance of Pseudomonas aeruginosa from a healthy ocular surface involves surfactant protein D and is compromised by bacterial elastase in a murine null-infection model. Infect Immun. 2009;77(6):2392–8. 10.1128/IAI.00173-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masinick SA, Montgomery CP, Montgomery PC, Hazlett LD. Secretory IgA inhibits Pseudomonas aeruginosa binding to cornea and protects against keratitis. Invest Ophthalmol Vis Sci. 1997;38(5):910–8. [PubMed] [Google Scholar]
- 60.Giannoni E, Sawa T, Allen L, Wiener-Kronish J, Hawgood S. Surfactant proteins A and D enhance pulmonary clearance of Pseudomonas aeruginosa. Am J Respir Cell Mol Biol. 2006;34(6):704–10. 10.1165/rcmb.2005-0461OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Restrepo CI, Dong Q, Savov J, Mariencheck WI, Wright JR. Surfactant protein D stimulates phagocytosis of Pseudomonas aeruginosa by alveolar macrophages. Am J Respir Cell Mol Biol. 1999;21(5):576–85. 10.1165/ajrcmb.21.5.3334 [DOI] [PubMed] [Google Scholar]
- 62.Boackle RJ, Connor MH, Vesely J. High molecular weight non-immunoglobulin salivary agglutinins (NIA) bind C1Q globular heads and have the potential to activate the first complement component. Mol Immunol. 1993;30(3):309–19. [DOI] [PubMed] [Google Scholar]
- 63.Leito JT, Ligtenberg AJ, van Houdt M, van den Berg TK, Wouters D. The bacteria binding glycoprotein salivary agglutinin (SAG/gp340) activates complement via the lectin pathway. Mol Immunol. 2011;49(1–2):185–90. 10.1016/j.molimm.2011.08.010 [DOI] [PubMed] [Google Scholar]
- 64.Blackburn AC, Hill LZ, Roberts AL, Wang J, Aud D, Jung J, et al. Genetic mapping in mice identifies DMBT1 as a candidate modifier of mammary tumors and breast cancer risk. American Journal of Pathology. 2007;170(6):2030–41. 10.2353/ajpath.2007.060512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100(24):14339–44. 10.1073/pnas.2036282100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turnbull L, Whitchurch CB. Motility assay: twitching motility. Methods Mol Biol. 2014;1149:73–86. 10.1007/978-1-4939-0473-0_9 [DOI] [PubMed] [Google Scholar]
- 67.Washburn MP, Wolters D, Yates JR 3rd. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. 10.1038/85686 [DOI] [PubMed] [Google Scholar]
- 68.Xu T, Venable JD, Park SK, Cociorva D, Lu B, Liao L, et al. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol Cell Proteomics. 2006;5(10):S174–S. [Google Scholar]
- 69.Tabb DL, McDonald WH, Yates JR 3rd. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1(1):21–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park SK, Venable JD, Xu T, Yates JR 3rd. A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008;5(4):319–22. 10.1038/nmeth.1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, et al. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom. 2004;18(18):2162–8. 10.1002/rcm.1603 [DOI] [PubMed] [Google Scholar]
- 72.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2(1):43–50. [DOI] [PubMed] [Google Scholar]
- 73.Loimaranta V, Jakubovics NS, Hytonen J, Finne J, Jenkinson HF, Stromberg N. Fluid- or surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties. Infect Immun. 2005;73(4):2245–52. 10.1128/IAI.73.4.2245-2252.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robertson DM, Li L, Fisher S, Pearce VP, Shay JW, Wright WE, et al. Characterization of growth and differentiation in a telomerase-immortalized human corneal epithelial cell line. Invest Ophthalmol Vis Sci. 2005;46(2):470–8. 10.1167/iovs.04-0528 [DOI] [PubMed] [Google Scholar]
- 75.Whitchurch CB, Leech AJ, Young MD, Kennedy D, Sargent JL, Bertrand JJ, et al. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol. 2004;52(3):873–93. 10.1111/j.1365-2958.2004.04026.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Represents a 5 min time-lapse movie of P. aeruginosa twitching motility captured at an interval of 10 s with a 60 × oil-immersion lens. Scale bar = 20 μm. Frame rate = 10 fps.
(AVI)
Represents a 5 min time-lapse movie of P. aeruginosa twitching motility captured at an interval of 10 s with a 60 × oil-immersion lens. Scale bar = 20 μm. Frame rate = 10 fps.
(AVI)
Represents a 5 min time-lapse movie of P. aeruginosa twitching motility captured at an interval of 10 s with a 60 × oil-immersion lens. Scale bar = 20 μm. Frame rate = 10 fps.
(AVI)
Represents a 10 s time-lapse movie of P. aeruginosa movement captured at no delay with a 40 × lens. Frame rate = 10 fps.
(AVI)
Represents a 10 s time-lapse movie of P. aeruginosa movement captured at no delay with a 40 × lens. Frame rate = 10 fps.
(AVI)
PAO1-pilA::Tn (PW8621), PAO1-cyaB::Tn (PW6387), and PAO1-chpB::Tn (PW1760) were verified by PCR with primers provided by the insertion mutant library database. PAO1-pilK::Tn (PW1757) was verified by PCR with the following primers; pilK flanking primers pilK-F (5'-AGATGCGCAACTCGGTATCC-3') and pilK-R (5'-TTCAGGGTTTCGGCGATCTC-3'). The red square was used to label target products.
(TIF)
(A) Fractions of human tear fluid separated by size exclusion chromatography (first experiment). (B) Effect of tear fractions on P. aeruginosa PAO1 twitching velocity measured in 5 min videos of each sample. Data are expressed as the mean ± SEM per sample from three independent experiments. Significance was determined using one-way ANOVA with Tukey's post-hoc analysis. **** P < 0.0001, ***P < 0.001.
(TIF)
(A) SDS-PAGE with silver stain (left panel) suggested DMBT1 was present after S. pyogenes treatment, and was confirmed by Western immunoblot (right panel) using anti-DMBT1 antibody. (B) and (C) Two independent experiments each showing that size-exclusion chromatography after DMBT1 purification from human saliva using S. pyogenes generated a high Mw fraction (fraction 1). Proteins were separated from aggregated S. pyogenes in human saliva using EDTA (5 mM). (D) Mass spectrometric analysis of fraction 1 after DMBT1 purification from saliva revealed the presence of DMBT1 in two independent experiments.
(TIF)
Results shown for two independent fractionations of human tear fluid using size-exclusion chromatography.
(TIF)
Each fraction inhibited twitching motility of P. aeruginosa PAO1. Results represent two independent experiments.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.