Abstract
Cancer stem cells represent the putative tumor-driving subpopulation thought to account for drug resistance, relapse, and metastatic spread of epithelial and other cancer types. Accordingly, cell surface markers for therapeutic delivery to cancer stem cells are subject of intense research. Somatostatin receptor 2 and nucleolin are known to be overexpressed by various cancer types, which have elicited comprehensive efforts to explore their therapeutic utilization. Here, we evaluated somatostatin receptor 2 targeting and nucleolin targeting for therapeutic delivery to cancer stem cells from lung cancer. Nucleolin is expressed highly but not selectively, while somatostatin receptor 2 is expressed selectively but not highly by cancer cells. The non-small cell lung cancer cell lines A549 and H1299, displayed average levels of both surface molecules as judged based on analysis of a larger cell line panel. H1299 compared to A549 cells showed significantly elevated sphere-forming capacity, indicating higher cancer stem cell content, thus qualifying as suitable test system. Nucleolin-targeting 57Co-DOTA-AS1411 aptamer showed efficient internalization by cancer cells and, remarkably, at even higher efficiency by cancer stem cells. In contrast, somatostatin receptor 2 expression levels were not sufficiently high in H1299 cells to confer efficient uptake by either non-cancer stem cells or cancer stem cells. The data provides indication that the nucleolin-targeting AS1411 aptamer might be used for therapeutic delivery to non-small cell lung cancer stem cells.
Introduction
Lung cancer is the most common cause of cancer death in industrialized countries, with non-small cell lung cancer (NSCLC) as the most common form accounting for about 80% of the cases [1, 2]. NSCLC is often diagnosed after the occurrence of metastases. At this stage, a curative therapy is no longer an option and a rapid disease progression results in five year survival rates below 15% [2].
Cancer stem cells (CSCs) represent a small subpopulation of the cancer cells with stem-like properties such as ability for self-renewal and asymmetric division, that enables them to restore heterogeneous tumors [3–5]. After their initial discovery in breast cancer, CSCs were subsequently also found in various other solid cancer types, including NSCLC [4–7]. Of importance, CSCs display high tumorigenicity, elevated drug resistance and propensity for metastatic spread, and therefore are thought to be responsible for relapse of drug resistant metastatic cancer [4–7]. This has elicited intense searches for biomarkers for, and therapeutic strategies against CSCs in general, and NSCLC-CSCs in particular. Different cell surface proteins are presently discussed to identify NSCLC-CSCs including, CD133, EpCAM, CXCR4, and ABCG2 [6, 7]. A common property across cancer types is the ability to form tumor spheres under non-adherent culture conditions, in the presence of defined growth factors. This has advanced to a standard assay for determining the CSC numbers [5–7].
Small molecule drugs including chemotherapeutics, have the advantage of rapid uptake by cancer cells, but the disadvantage of rapid extrusion by CSCs, via multiple drug resistance proteins, such as ABCG2 [6, 7]. Macromolecular drugs, such as nucleic acids mediating RNA-interference, would have the advantage to escape these extrusion mechanisms [8–11].
However, these drugs are typically entering cells at low efficiency, thus requiring special delivery mechanisms [8–11]. Somatostatin receptor 2 (SSTR2) and nucleolin (NCL) are under intense investigation, based on their overexpression at the surface of cancer cells [12–18]. SSTR2 is a cell surface receptor overexpressed in neuroendocrine tumors [12–14] and peptide-based SSTR2-targeting, for example by radiolabeled DOTATATE, is already used for diagnostic imaging. Furthermore, the potential of DOTATATE for delivery of therapeutic agents has been explored in various studies [12–14, 18]. While SSTR2 is a classical cell surface receptor, NCL was discovered by chance, tracing back to the identification of a G-quadruplex forming aptamer, later on referred to as AS1411, with anti-cancer activity [15–17, 19]. NCL commonly locates to the nucleus, but AS1411 was shown to bind to NCL at the surface of cancer cells, where the protein is also located for yet unclear reasons [17]. Development of AS1411 reached clinical phase trial 2 in renal cancer, where, however, it failed to show efficacy [19]. Investigations are presently ongoing to evaluate, whether AS1411 can be used for drug delivery, including proof-of-concept that the aptamer may qualify for the delivery of nucleic acid-based therapeutics to cancer cells [10, 11].
Here, we set out to explore, whether SSTR2 or NCL can be utilized for efficient delivery of radionuclides to NSCLC-CSCs. We established H1299-derived spheres as CSC model system. We report that, although NCL does not display strong tendency towards selective overexpression by cancer cells, efficient uptake into NSCLC-CSCs was conferred by the NCL-targeting AS1411 aptamer. This is in contrast to SSTR2, which was more selectively overexpressed by cancer cells, but SSTR2-targeting DOTATATE was inferior for delivery purposes, as can be explained by low expression of the receptor.
Materials and methods
Cell culture
Cell lines were grown in a humidified incubator (at 37°C and 5% CO2). For passaging, cells were washed with phosphate buffered saline, and detached with TrypLE Express (Invitrogen, Karlsruhe, Germany). A549 (ATCC, CLL-185) was grown in Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich, Copenhagen, Denmark), and H1299 (ATCC, CLR-5803) was grown in Roswell Park Memorial Institute 1640 medium with L-glutamine (Sigma-Aldrich). Both media were supplemented with 10% fetal bovine serum (Sigma-Aldrich), and 1% Penicillin-Streptomycin (Sigma-Aldrich). All experiments were performed with 80% confluent cells. All pictures of cells were taken using the IX71® Inverted Microscope (Olympus Ballerup, Denmark), with a 10X/0.3phC lens.
In order to create the comparative RNA and protein panels, the following cell lines were used; 184A1 (CRL-8798), MCF-10A (CRL-10317), MCF-12A (CRL-10782), ZR-75-1 (CRL-1500), MCF7 (HTB-22), T47-D (HTB-133), MDA-MB-361 (HTB-27), BT-474 (HTB-20), HCC70 (CRL-2315), BT-20 (HTB-19), MDA-MB-231 (CRM-HTB-26), HCC1500 (CRL-2329), HCC1569 (CRL-2330), BT-549 (HTB-122), MDA-MB-436 (HBT-130), NCI-H69 (HTB-119), PC-3 (CRL-1435) and U-251 MG (formerly known as U-373 MG) (ECACC 09063001). All were obtained from ATCC (Wesel, Germany) except for U-251 MG that was obtained at the ECACC (Porton Down, UK). Cell lines were maintained following the guidelines of the providers.
Quantitative RT-PCR
Total RNA was harvested from adherent cells using the RNeasy Mini Kit (Qiagen, Copenhagen, Denmark) according to the instructions of the supplier. The RNA concentration was measured on the NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Hvidovre, Denmark), and stored at -80°C until use. RNA was reverse transcribed using the RevertAid Minus First strand cDNA synthesis kit (Life Technologies, Nærum, Denmark) using 1 μg total RNA and oligo(dT) primers, following the instructions of the manufacturer. qRT-PCR was performed with Maxima® Probe/ROX qPCR Master Mix (Thermo Fisher Scientific) and taqman assays for NCL (Life Technologies, Hs01066668-m1) and SSTR2 (Life Technologies, Hs00265624_s1). 10 ng cDNA was used per reaction, as recommended by the providers’ protocols. ACTB (Life Technologies, Hs99999903-m1) and GAPDH (Life Technologies, Hs99999905-m1) were used as references for normalization. The qRT-PCR was performed in a ABI Prism 7300 (Thermo Fisher Scientific), for 2 min at 50°C, followed by 15 min at 95°C and 40 cycles at 95°C for 15 sec and 60°C for 1 min. Expression levels were analyzed using the Biogazelle qBaseplus software (www.qbaseplus.com), and normalized to ACTB and GAPDH. Moreover, the expression levels were referred to a virtual common reference (REF), representing the mean expression level of the TP53 and the CDKN1A mRNA in MCF7 cells.
Western blot analysis
Adherent cells were harvested in proteolytic RIPA buffer, mixed with EDTA-Free protease inhibitor (Roche, Hvidovre, Denmark). The protein concentration was measured by the Pierce™ BCA protein assay kit (Thermo Fisher Scientific). Gel electrophoresis was performed under reducing conditions with 10% SDS-polyacrylamide gels. Proteins were blotted onto a polyvinylidene difluoride (PVDF) membrane and the membrane incubated with a monoclonal mouse anti-NCL IgG1 antibody (1:5000, 4E2, Abcam, Cambridge, UK) [20]. After rinsing, the membrane was incubated with HRP conjugated polyclonal rabbit-anti-mouse secondary antibody (1:5000, Jackson Immunoresearch, Suffolk, UK) and visualized on X-ray film using Super-Signal West Dura Extended Duration substrate kit according to the manufacturer’s instructions (Thermo Fisher Scientific).
After stripping, the process was repeated on the same membrane with a monoclonal mouse GAPDH-specific IgM antibody (1:5000, 3C2, Sigma-Aldrich). Developed films were digitalized, and the protein expression levels were quantified with the Image Studio™ Litesoftware (www.licor.com). NCL expression levels were adjusted to the individual GAPDH levels and, to compare, normalized to the mean value of the normal epithelial breast cell lines 184A1 and MCF-10A.
Sphere formation assays
For the determination of sphere forming activity, adherently growing NSCLC cell lines H1299 and A549 were harvested and re-suspended in sphere-forming medium, containing 0.4% bovine serum albumin (Sigma-Aldrich), 10 ng/ml Epidermal Growth Factor (Sigma-Aldrich), 20 ng/ml Basic Fibroblast Growth Factor (VWR, Søborg, Denmark) and, in the case of H1299, 0.25 μg/ml human insulin (Sigma-Aldrich). To ensure a single cell suspension, the cells were filtered through 20 μm Steriflip filters (Millipore, Hellerup, Denmark), before being counted on a Cedex XS cell counter (Roche). The cells were seeded at densities of 250, 500, 1000, 2000, 4000 or 8000 cells/well in 150 μl medium in Corning™ Ultra-Low Attachment 96-Well Plates (Sigma-Aldrich). At day 6, spheres were counted under the microscope. Three pictures were taken per well, and the diameter of spheres was determined with the Cell^F program (Olympus Europe). For the sphere-based viability assay, 20μl CellTiterBlue™ (Promega, Nacka, Sweden) was added per well, and the plate incubated for 23 hours prior to viability readout with a Wallac Victor3 1420 Multilabel Counter (Perkin Elmer, Skovlunde, Denmark) at 560ex/590em nm. Experiments were performed with 10 wells per setting, and repeated three times.
Radiochemistry and subcellular distribution
DOTA-conjugated AS1411 (Trilink Biotechnologies, CA, USA) and DOTATATE (ABX, Radeberg, Germany) were labeled with 57Co. AS1411 was mixed with potassium acetate (0.1 M, pH 4.5) and 57CoCl2 (Perkin Elmer). The mixture was heated for 3 min at 850 W in a domestic microwave. The mixture was allowed to cool to room temperature, and potassium phosphate buffer 0.1 M, pH 7.55 was added. The aptamer solution was annealed for 1 min in a domestic microwave at 850 W. The radiochemical purity was analyzed using size exclusion HPLC employing a Biosep S2000 (Phenomenex, Værløse, Denmark) eluted with phosphate buffered saline. For labeling of DOTATATE, see [20]. H1299 cells were seeded as adherent cells and spheres in 24-well conventional and Ultra-Low Attachment plates, respectively using a uniform seeding density of 10,000 cells per well corresponding to 20,000 cells per ml. Three days after seeding, cells were incubated in serum-free medium containing the radioactive labeled conjugates, at a final concentration of 20 kBq/ml. The subcellular distribution of 57Co-DOTA-AS1411 (specific activity 49.4 MBq/nmol) and 57Co-DOTATATE (specific activity 4 MBq/nmol) was analyzed at the indicated time points. Non-labeled DOTA-AS1411 or DOTATATE in 1000-fold excess, were added to a separate set of wells for blocking of specific uptake (“block”) and evaluated at the late time points as previously described [20]. The radioactivity in the different cell fractions was measured in a 2470 Wizard Automatic Gamma Counter (Perkin Elmer). A separate series of cells was seeded in parallel and counted at the respective time points in order to normalize to cell number. The experiment was performed three times in triplicate.
Statistical analyses
The Student’s t test was uniformly used to evaluate statistical significance. p-values < 0.05 were considered statistically significant.
Results
Expression of SSTR2 and NCL in cancer versus normal cells
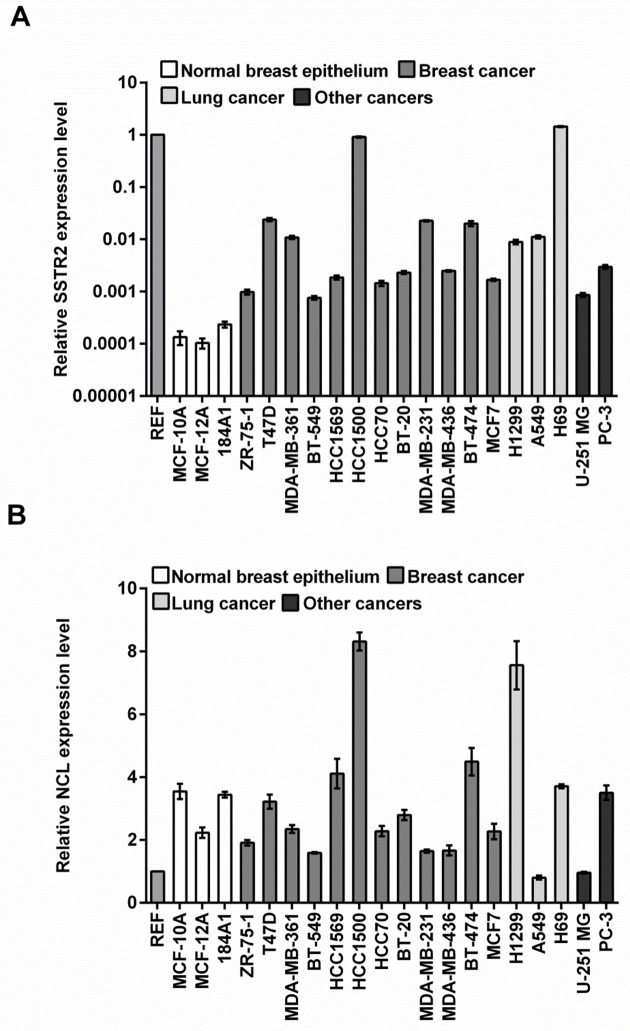
In order to evaluate if SSTR2 and NCL can potentially be used for therapeutic delivery in NSCLC cells, we initially determined the relative mRNA expression levels by real-time PCR in the two NSCLC cell lines A549 and H1299 and in a panel of other cancer cell lines (Fig 1). For comparison, we used three normal, i.e. non-tumorigenic, mammary gland epithelial cell lines (MCF-10A, MCF-12A, and 184A1). SSTR2 was higher expressed in all cancer cell lines versus the normal cell lines up to about 10,000-fold. The majority of the cancer cell lines displayed about 10- to 100-fold upregulation (56- and 70-fold respectively for H1299 and A549). However, the expression only caused elevation from a very low to a low or moderate level, since the majority of the cancer cell lines showed 100- to 1,000-fold reduced levels compared to the common reference value (Fig 1A). NCL showed a remarkably different pattern (Fig 1B). Only four cancer cell lines had elevated mRNA levels compared to normal epithelial cells, including the breast cancer cell line HCC1500 and the NSCLC cell line H1299 with about 2- to 3-fold increase. However, when comparing to the common reference value, NCL was in general expressed at substantially higher levels than SSTR2.
Fig 1. Real-Time PCR quantification of mRNA.
Real-time PCR showing relative expression levels of SSTR2 (A) and NCL (B) in a panel of human cell lines displayed on a logarithmic and a non-logarithmic scale, respectively. Expression levels were normalized to GAPDH and ACTB as housekeeping genes. We defined a common reference value (REF; see Materials and methods), to allow for comparison of expression levels between the two genes. The average level and SEM for each cell line were calculated from three wells.
Next, we evaluated the protein levels of NCL in a subset of the cell lines from the panel and quantified the expression relative to the expression levels of GAPDH (Fig 2). Albeit that protein levels seemed to be generally elevated in cancer versus normal cells, the increase was moderate.
Fig 2. Western blot detection of NCL.
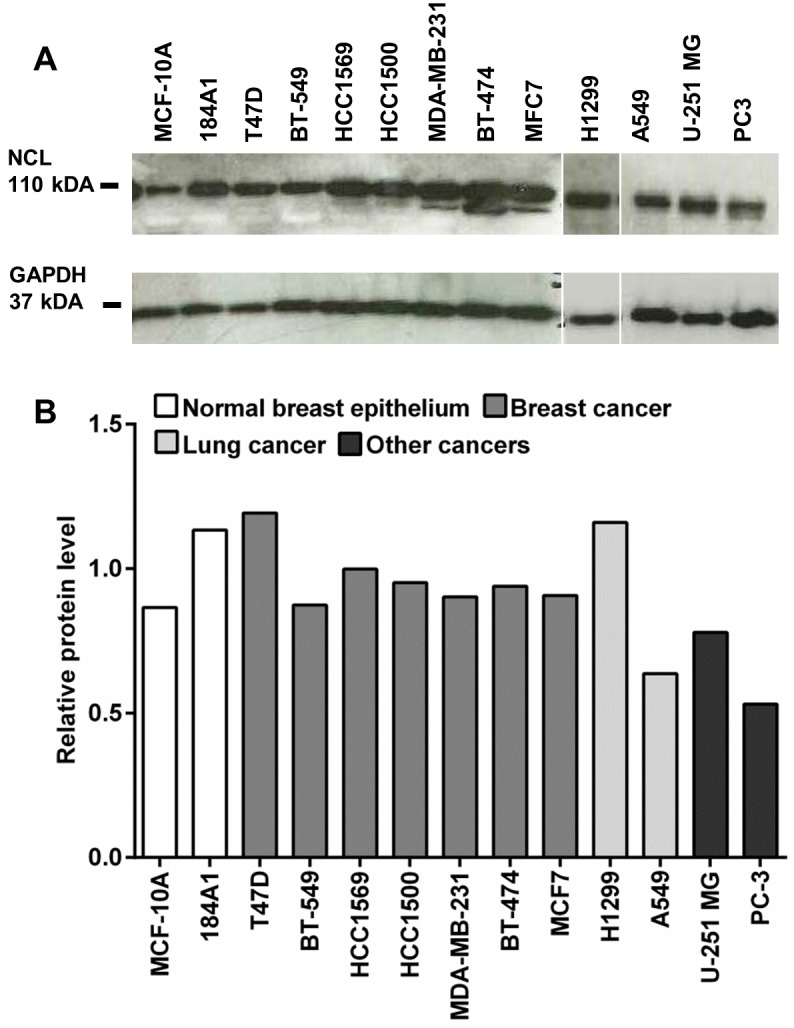
(A) Western blot of extracts from different cell lines were first probed with anti-NCL antibody and afterwards reprobed with anti-GAPDH antibody for equal loading control. (B) Quantification of NCL expression relative to the expression of GAPDH and normalized to the mean value of the normal epithelial breast cell lines MCF-10A and 184A1.
These results suggest that SSTR2 was selectively overexpressed in cancer cells, but at comparably low levels, while NCL was expressed at high overall levels but only displayed a moderate increase of protein levels compared to normal cells. Furthermore, both H1299 and A549 cells expressed about the same elevated levels of SSTR2 compared to normal cells. In contrast, NCL was higher expressed in H1299 cells compared to A549 cells both at the mRNA and protein level.
Differential sphere formation capacity
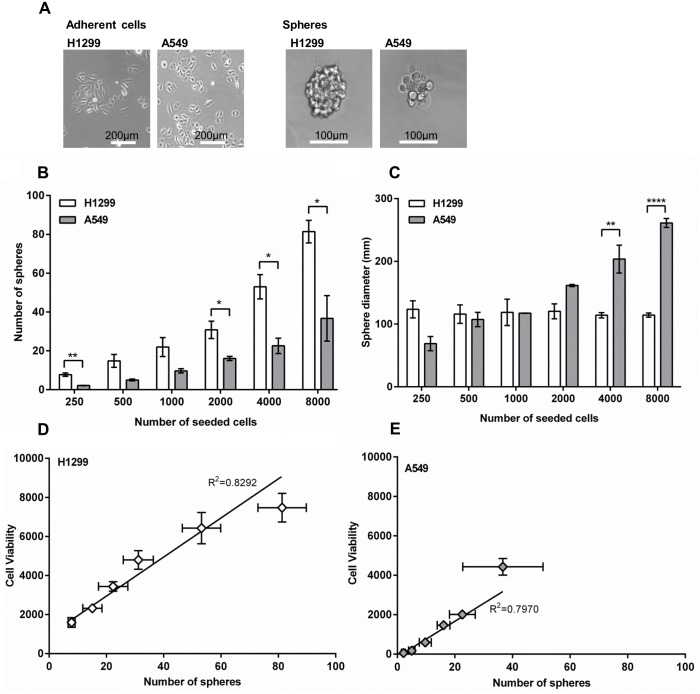
To establish a suitable test system for in vitro targeting studies of CSCs, we compared the sphere-forming abilities of A549 and H1299 cells. In non-adherent, serum-free culture conditions, non-CSCs undergo cell death by anoikis but CSCs survive to give rise to tumor spheres [21] so that, conceptually, the number of spheres is equivalent to the number of CSCs in the sample. In terms of general morphology, A549 cells formed poorly connected spheres, while H1299 cells formed more defined spheres (Fig 3A). Determination of sphere numbers at different seeding densities primarily suggested a good correlation for both NSCLC cell lines (Fig 3B). However, H1299 cells were about twice as potent as A549 cells in sphere formation. Regardless of seeding density, H1299 cells formed spheres of a uniform size of about 120 μm. By contrast, the size of A549 spheres increased from about 70 μm at the lowest seeding density to about 250 μm at the highest seeding density (Fig 3C). This strongly indicates that cell aggregation contributes to, or is responsible for A549 spheres, in agreement with the morphological appearance. Cell viability assays, used as alternative readout for spheroid formation, displayed good correlation with sphere numbers for both cell lines (Fig 3D and 3E). Overall, the sphere-forming ability of H1299 cells was significantly higher than the one of A549 cells, in which cell aggregation may add confounding effects. These results indicated H1299 cells as suitable model system for testing delivery by SSTR2 and NCL targeting.
Fig 3. Sphere formation capacity.
(A) The two NSCLC cell lines H1299 and A549 were grown as adherent cells or spheres for six days. (B) The number of lung cancer spheres after six days at different number of seeded cells. Mean and SEM from 3–4 plates. (C) Diameter of A549 and H1299 lung cancer spheres. Mean and SEM of 3–4 plates. The number of H1299 spheres (D) and A549 spheres (E) correlated with cell viability. SEM for sphere count and cell viability from 3–4 plates plotted as vertical and horizontal error lines, respectively. T-test depicts difference between A549 and H1299 at identical number og seeded cells p-value <0.05; *, <0.01; **; <0.0001; ****.
The NCL-targeting AS1411 aptamer shows efficient uptake by NSCLC CSCs
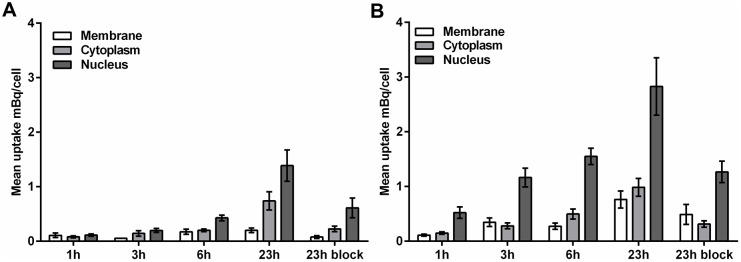
Next, we explored SSTR2 and NCL targeting for potential delivery to NSCLC-CSCs, by performing uptake studies with radioactive labeled SSTR2-targeting peptide (57Co-DOTATATE) and NCL-targeting AS1411 aptamer (57Co-DOTA-AS1411) in both H1299 adherent cells and spheres. In agreement with the low expression levels of SSTR2, there was no uptake of 57Co-DOTATATE in H1299 non-CSCs or CSCs. In contrast, there was a time-dependent uptake of 57Co-DOTA-AS1411 in both H1299 non-CSCs (Fig 4A) and CSCs (Fig 4B). Interestingly, the total cell-associated 57Co-DOTA-AS1411 uptake was higher in CSCs compared to non-CSCs at all time-points. Most of the radioactivity was found in the nuclear fraction in both cell-types and up until the latest time point investigated (23 hours), uptake of 57Co-DOTA-AS1411 continued to increase, providing an about 2-fold increase in both total and nuclear uptake in CSCs versus non-CSCs (Fig 4A and 4B). The competitive blocking experiment indicated that most of the uptake was specific, although internalization was not completely blocked. The results show that the NCL-targeting AS1411 was efficiently internalized by non-CSCs and even more efficiently by CSCs from NSCLC and could be used for therapeutic delivery.
Fig 4. Uptake of 57Co labeled DOTATATE in the NSCLC cell line H1299.
The subcellular distribution of 57Co-DOTA-AS1411 in H1299 adherent cells (A) and in H1299 spheres (B) as a function of increasing incubation time (1–23 h). In each case a blocking experiment was performed with 1000-fold excess AS1411 (23h block). Mean uptake (mBq/cell) is expressed as mean ± SEM from triplicate experiments.
Discussion
Evidence has accumulated that NSCLC, as suggested for other solid cancer types, is driven by CSCs [6, 7]. One particular hallmark of CSCs is drug resistance, caused by rapid extrusion of chemotherapeutics and other small molecule drugs, via multidrug resistance transporter systems such as ABCG2, which has been found on CD133-positive putative NSCLC-CSCs [6]. Opposed to this, macromolecular drugs, such as nucleic acids causing RNA-interference, and peptides would not be subjected to these drug resistance mechanisms [8–11, 22]. However, especially nucleic acids bear the problem of efficient delivery to cancer cells [9–11]. We here evaluated two broadly discussed cell surface targets, namely SSTR2 and NCL, for their ability to confer uptake in NSCLC-CSCs in vitro.
Both SSTR2 and NCL have previously been reported to be overexpressed by cancer cells [13–17]. Our present data confirms that SSTR2 is upregulated in cancer versus normal cell lines by a factor of 10- to 10,000-fold, however the SSTR2 expression levels in cancer cells were very low compared to the reference, also in the two NSCLC cell lines (Fig 1). By contrast, most cancer cell lines displayed NCL expression levels lower or equal to those observed in normal epithelial cell lines, and a moderate 2- to 3-fold increase in a smaller subset of the cancer cells at the mRNA level. These findings are largely recapitulated at the protein level, albeit NCL levels were equal to or higher in cancer cells versus normal epithelial cells. However, with about 2-fold, the increase was moderate (Fig 2). For yet unknown reasons, NCL is also present at the cell surface [17]. As the western blot analysis reflects the total protein, we cannot rule out that cancer cells may expose more dramatically elevated NCL levels at the surface than normal cells as has been found in other studies [23–25]. On the other hand, our uptake studies indicate that cell surface NCL is trafficked to the nucleus, which represents the predominant localization of the protein.
While the data argued that SSTR2 opposed to NCL would represent a cancer-specific molecule for targeting, we also included a common reference to gain an estimate of the overall intensity of expression of SSTR2 and NCL. This methodology neither takes into consideration that the qRT-PCR assays for SSTR2 and NCL may have different performance, nor the different levels of post-transcriptional and -translational regulation. Thus, differences are not necessarily expected to reflect true numerical differences. However, they may point to the correct order of magnitude.
Different markers and their combinations are currently discussed for NSCLC-CSCs [6, 7]. Expanding on the characterization of NSCLC-CSC markers was not the focus of our present study. Instead, in order to identify a suitable test system for uptake by NSCLC-CSCs, we approached the problem phenomenologically, by utilizing a marker-independent sphere formation assay. H1299 cells formed spheres more efficiently than A549 cells, for which cell aggregation may partly contribute to the rather irregularly shaped spheres. This data is in accordance with recent reports, indicating decreased sphere formation potential and in vivo tumorigenicity of A549 versus H1299 cells [26] and in turn may suggest a higher CSC content for H1299 than for A549 cells (Fig 3B and 3C). To this end, we identified it as suitable and robust test system for studying uptake in NSCLC-CSCs. We verified that the cells were CSCs by performing qPCR assays to analyze the expression of the three known markers oct4, sox2 and nanog (S1 Fig). All three markers were upregulated in cancer stem cells compared to bulk cancer cells supporting the notion that the cells grow under sphere conditions were truly CSCs.
Utilizing this model system, there was no uptake of 57Co-DOTATATE by H1299 cells. This was also supported by the mRNA analysis, which showed low levels of SSTR2 expression in the NSCLC cell lines. 57Co-DOTA-AS1411 on the other hand displayed efficient uptake and 4.5-fold higher in CSCs versus non-CSCs after 3 hours. At later time points, CSCs internalized about 2-fold the amount of the 57Co-DOTA-AS1411 internalized by non-CSCs. These results are in line with a recent study, in which Fonseca et al. reported efficient internalization of NCL-targeting liposomes by both breast cancer non-CSCs and CSCs [27]. Fonseca et al. associated increased NCL mRNA levels in stem cells with improved uptake, but increased accessibility or faster trafficking in CSCs could also represent conceivable reasons. We also cannot rule out that the absence of serum from the medium used for sphere cultivation contributes to the increased uptake by CSCs, albeit 0.4% bovine serum albumin was contained. It was recently reported that prolonged exposure in vitro to AS1411, causes a switch from NCL-dependent internalization to an NCL-independent macropinocytosis, which is triggered by the aptamer itself [28]. The competitive blocking experiments indicated that at 23 h, the NCL-dependent uptake still dominated (Fig 4A and 4B). Albeit, AS1411 previously has been shown the have anti-cancer activity in malignant cells, we saw no effect on proliferation and stemness (S2 Fig). This is most likely due owing to the fact that we only use 0.2 nM AS1411 (corresponding to 20 kBq/ml 57Co-AS1411), which is far below the 1–10 uM GI50 (50% growth inhibition) found for most malignant cell lines [29].
In conclusion, our data indicate that NCL-targeting via AS1411 in principle can be utilized for efficient delivery of radionuclides to NSCLC-CSCs. This might be accompanied by a trade-off that is to be made between selectivity for cancer cells and efficient uptake.
Supporting information
The expression levels of oct4, sox2 and nanog were normalized to actin. Data and error bars are presented as mean ± SEM. ***p<0.001, ****p<0.001. The experiments were repeated three times.
(TIF)
H1299 spheres and bulk cells were incubated with 0.2 nM AS1411 for 24h. (A) Proliferation was measured by the WST assay. Relative expression levels of oct4, sox2 and nanog were analyzed by qPCR in spheres (A) or in bulk cancer cells (B). Expression levels were normalized to actin and control cells (no AS1411). Data and error bars are presented as mean ± SEM. All the experiments were repeated three times.
(TIF)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by: Danish Council for Independent Research; Medical Sciences (DFF-4004-00268) http://ufm.dk/forskning-og-innovation/rad-og-udvalg/det-frie-forskningsrad, Receiver: JM; Lundbeck Foundation grant to the Nanomedicine Research Center for Cancer Stem Cell-Targeting Therapeutics (NanoCAN) http://www.lundbeckfonden.com/, Receiver: JM.
References
- 1.World Health Organization. International agency for cancer research, Population fact sheets 2012. http://globocan.iarc.fr/Pages/fact_sheets_population.aspx.
- 2.Reck M, Popat S, Reinmuth N, De Ruysscher D, Kerr KM, Peters S, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25 Suppl 3:iii27–39. 10.1093/annonc/mdu199 . [DOI] [PubMed] [Google Scholar]
- 3.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3(12):895–902. 10.1038/nrc1232 . [DOI] [PubMed] [Google Scholar]
- 4.Moghbeli M, Moghbeli F, Forghanifard MM, Abbaszadegan MR. Cancer stem cell detection and isolation. Med Oncol. 2014;31(9):69 10.1007/s12032-014-0069-6 . [DOI] [PubMed] [Google Scholar]
- 5.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–8. 10.1073/pnas.0530291100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106(38):16281–6. 10.1073/pnas.0905653106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15(3):504–14. 10.1038/sj.cdd.4402283 . [DOI] [PubMed] [Google Scholar]
- 8.Malhi S, Gu X. Nanocarrier-mediated drugs targeting cancer stem cells: an emerging delivery approach. Expert Opin Drug Deliv. 2015;12(7):1177–201. 10.1517/17425247.2015.998648 . [DOI] [PubMed] [Google Scholar]
- 9.Wu Y, Crawford M, Yu B, Mao Y, Nana-Sinkam SP, Lee LJ. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol Pharm. 2011;8(4):1381–9. 10.1021/mp2002076 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pofahl M, Wengel J, Mayer G. Multifunctional nucleic acids for tumor cell treatment. Nucleic Acid Ther. 2014;24(2):171–7. 10.1089/nat.2013.0472 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong Y, Meng F, Deng C, Zhong Z. Ligand-directed active tumor-targeting polymeric nanoparticles for cancer chemotherapy. Biomacromolecules. 2014;15(6):1955–69. 10.1021/bm5003009 . [DOI] [PubMed] [Google Scholar]
- 12.Bodei L, Ferone D, Grana CM, Cremonesi M, Signore A, Dierckx RA, et al. Peptide receptor therapies in neuroendocrine tumors. J Endocrinol Invest. 2009;32(4):360–9. 10.1007/BF03345728 . [DOI] [PubMed] [Google Scholar]
- 13.Zou Y, Xiao X, Li Y, Zhou T. Somatostatin analogues inhibit cancer cell proliferation in an SSTR2-dependent manner via both cytostatic and cytotoxic pathways. Oncol Rep. 2009;21(2):379–86. . [PubMed] [Google Scholar]
- 14.Xu C, Zhang H. Somatostatin receptor based imaging and radionuclide therapy. Biomed Res Int. 2015;2015:917968 10.1155/2015/917968 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J, Zhao Z, Liu Q, Ye M, Hu B, Wang J, et al. Study of the Function of G-Rich Aptamers Selected for Lung Adenocarcinoma. Chem Asian J. 2015;10(7):1519–25. 10.1002/asia.201500187 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koutsioumpa M, Papadimitriou E. Cell surface nucleolin as a target for anti-cancer therapies. Recent Pat Anticancer Drug Discov. 2014;9(2):137–52. . [DOI] [PubMed] [Google Scholar]
- 17.Reyes-Reyes EM, Salipur FR, Shams M, Forsthoefel MK, Bates PJ. Mechanistic studies of anticancer aptamer AS1411 reveal a novel role for nucleolin in regulating Rac1 activation. Mol Oncol. 2015;9(7):1392–405. 10.1016/j.molonc.2015.03.012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delpassand ES, Samarghandi A, Zamanian S, Wolin EM, Hamiditabar M, Espenan GD, et al. Peptide receptor radionuclide therapy with 177Lu-DOTATATE for patients with somatostatin receptor-expressing neuroendocrine tumors: the first US phase 2 experience. Pancreas. 2014;43(4):518–25. 10.1097/MPA.0000000000000113 . [DOI] [PubMed] [Google Scholar]
- 19.Rosenberg JE, Bambury RM, Van Allen EM, Drabkin HA, Lara PN Jr., Harzstark AL, et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest New Drugs. 2014;32(1):178–87. 10.1007/s10637-013-0045-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thisgaard H, Olsen BB, Dam JH, Bollen P, Mollenhauer J, Hoilund-Carlsen PF. Evaluation of cobalt-labeled octreotide analogs for molecular imaging and auger electron-based radionuclide therapy. J Nucl Med. 2014;55(8):1311–6. 10.2967/jnumed.114.137182 . [DOI] [PubMed] [Google Scholar]
- 21.Charpentier M, Martin S. Interplay of Stem Cell Characteristics, EMT, and Microtentacles in Circulating Breast Tumor Cells. Cancers (Basel). 2013;5(4):1545–65. 10.3390/cancers5041545 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Craik DJ, Fairlie DP, Liras S, Price D. The future of peptide-based drugs. Chem Biol Drug Des. 2013;81(1):136–47. 10.1111/cbdd.12055 . [DOI] [PubMed] [Google Scholar]
- 23.Subramanian N, Srimany A, Kanwar JR, Kanwar RK, Akilandeswari B, Rishi P, et al. Nucleolin-aptamer therapy in retinoblastoma: molecular changes and mass spectrometry-based imaging. Mol Ther Nucleic Acids. 2016;5(8):e358 10.1038/mtna.2016.70 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding Y, Song N, Liu C, He T, Zhuo W, He X, et al. Heat shock cognate 70 regulates the translocation and angiogenic function of nucleolin. Arterioscler Thromb Vasc Biol. 2012;32(9):e126–34. 10.1161/ATVBAHA.112.247502 . [DOI] [PubMed] [Google Scholar]
- 25.El Khoury D, Destouches D, Lengagne R, Krust B, Hamma-Kourbali Y, Garcette M, et al. Targeting surface nucleolin with a multivalent pseudopeptide delays development of spontaneous melanoma in RET transgenic mice. BMC Cancer. 2010;10:325 10.1186/1471-2407-10-325 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung LY, Tang SJ, Wu YC, Sun GH, Liu HY, Sun KH. Galectin-3 augments tumor initiating property and tumorigenicity of lung cancer through interaction with beta-catenin. Oncotarget. 2015;6(7):4936–52. 10.18632/oncotarget.3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fonseca NA, Rodrigues AS, Rodrigues-Santos P, Alves V, Gregorio AC, Valerio-Fernandes A, et al. Nucleolin overexpression in breast cancer cell sub-populations with different stem-like phenotype enables targeted intracellular delivery of synergistic drug combination. Biomaterials. 2015;69:76–88. 10.1016/j.biomaterials.2015.08.007 . [DOI] [PubMed] [Google Scholar]
- 28.Reyes-Reyes EM, Teng Y, Bates PJ. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010;70(21):8617–29. 10.1158/0008-5472.CAN-10-0920 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bates PJ, Reyes-Reyes EM, Malik MT, Murphy EM, O’Toole MG, Trent JO. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms, Biochim Biophys Acta. 2016, 10.1016/j.bbagen.2016.12.015 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expression levels of oct4, sox2 and nanog were normalized to actin. Data and error bars are presented as mean ± SEM. ***p<0.001, ****p<0.001. The experiments were repeated three times.
(TIF)
H1299 spheres and bulk cells were incubated with 0.2 nM AS1411 for 24h. (A) Proliferation was measured by the WST assay. Relative expression levels of oct4, sox2 and nanog were analyzed by qPCR in spheres (A) or in bulk cancer cells (B). Expression levels were normalized to actin and control cells (no AS1411). Data and error bars are presented as mean ± SEM. All the experiments were repeated three times.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.