Abstract
Aim
To investigate the expression and clinical significance of Holliday junction–recognizing protein (HJURP) in hepatocellular carcinoma (HCC).
Methods
In this study, we detected the expression of HJURP protein in samples of 164 patients with HCC, and based on this, we divided the patients into two cohorts: high expression of HJURP and low expression of HJURP. We analyzed the correlation between HJURP expression and the clinicopathological factors using chi-square test. Survival significance of HJURP was defined by Kaplan–Meier method and log-rank test, and the independent prognostic factors were identified by Cox regression model. Using function assays of HCC cell lines, we investigated the influence of HJURP on the proliferation of HCC cells.
Results
In our study, the proportion of patients with high HJURP expression was 25.6%, which was significantly associated with the tumor size and Barcelona clinic liver cancer stage. Univariate analysis confirmed that high HJURP expression was remarkably associated with poorer overall survival rates (P=0.003), as well as tumor number (P=0.016), tumor differentiation (P=0.047), TNM stage (P=0.005), and Barcelona clinic liver cancer stage (P=0.004). Multivariate analysis confirmed that high HJURP expression (P<0.001) acted as an independent prognostic risk factor of unfavorable prognosis. Real-time polymerase chain reaction analysis revealed that the expression of HJURP was significantly higher in HCC tissues than that in the corresponding normal liver tissues. Moreover, we demonstrated that HJURP overexpression could accelerate HCC cell line proliferation, whereas HJURP knockdown could attenuate the proliferation.
Conclusion
High HJURP expression was an independent prognostic biomarker of HCC, predicting poorer prognosis. HJURP also played an important role in HCC cell proliferation.
Keywords: Holliday junction-recognizing protein, hepatocellular carcinoma, proliferation, prognosis
Introduction
Holliday junction–recognizing protein (HJURP, also known as hFLEG1) has been identified as a chaperon of centromere-specific protein, CENP-A, facilitating the recruitment of CENP-C in centromere, by which it helps to assemble functional kinetochores and mediates chromosome segregation and cell division.1,2 Most cancer cells are characterized by segregation defects and instability of chromosomes at mitotic division.3 Ectopic expression of HJURP has been demonstrated in several kinds of cancers, and its dysfunction suspected in carcinogenesis and cancer progression. HJURP overexpression has been observed in lung, glioblastoma, and breast cancers, which correlated with poor prognosis.4
Hepatocellular carcinoma (HCC) is a deadly threat that causes heavy financial burden worldwide, especially in People’s Republic of China and other Asia-Pacific countries due to its relatively high morbidity. Some of the high risk factors of HCC comprise hepatitis B or C infection, aflatoxin exposure, and hepatic cirrhosis. Recently, several breakthroughs were made with the help of genetic studies on HCC carcinogenesis and its progression. A series of studies along with genome-wide association studies have demonstrated that single nucleotide polymorphisms (SNPs) at 1p36.22, 2q32.2–32.3, 6p21.3, and 21q21.3 are associated with hepatitis B virus (HBV)-related HCC in Chinese patients.5–7 Moreover, Huang et al proved that SNP of HJURP, rs3771333, is associated with higher susceptibility to HCC among Chinese.8 In addition, HJURP expression was significantly higher in HCC tissues compared to paired adjacent non-tumor tissues.8 To date, there are no studies focusing on the correlation between HJURP expression level and HCC progression or prognosis.
In this study, we detected the expression of HJURP in tissue samples obtained from 164 patients with HCC, and based on the cutoff value, we divided the patients into two cohorts: high-expression group and low-expression group. We further analyzed the correlation of HJURP with clinicopathological factors and survival rates. Furthermore, using function assays of HCC cell lines, we investigated the influence of HJURP on the proliferation of HCC cells.
Patients and methods
In this study, a total of 330 patients who were diagnosed with HCC and who underwent R0 resection in Yuhuangding Hospital, Yantai, from 2006 to 2015 were enrolled. Of them, we selected 164 patients who met the following inclusion criteria: (1) availability of tissue samples for immunohistochemistry (IHC), (2) a follow-up time of more than 3 months, and (3) no severe operation-related complications and other tumors except HCC. Moreover, 10 pairs of frozen tissues, including HCC tissues and corresponding normal liver tissues, were collected and stored immediately in liquid nitrogen for mRNA extraction. Our study was approved by the Ethics Committee of Yuhuangding Hospital, and all the tissues were obtained after prior consent of the patients and the approval of Ethics Committee of Yuhuangding Hospital. The tumor TNM stage was identified according to the guideline of seventh American Joint Committee on Cancer/Union for International Cancer Control,9 and the Barcelona clinic liver cancer (BCLC) stage was according to American Association for the Study of Liver Diseases.10
Cells and reagents
HCC cell lines, namely, HuH7, HepG2, and SMMC7721 were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, People’s Republic of China). All the cell lines were cultured in RPMI-1640 medium (HyClone, Chicago, USA) supplemented with 10% fetal bovine serum and 100 U/mL penicillin and 100 μg/mL streptomycin (HyClone) in 5% CO2 resuscitation. Lipofectamine RNAiMAX transfection reagent was purchased from Thermo Fisher Scientific (Waltham, MA, USA). HJURP small interfering RNA (siRNA) and control scrambled RNA were purchased from GeneChem Company (Shanghai, People’s Republic of China). Antibody of HJURP was purchased from Abcam Company (Cambridge, UK), and other antibodies without special instruction were purchased from Santa Cruz Corporation (Austin, TX, USA). All other reagents were purchased from Sigma-Aldrich Corporation without special instruction.
Immunohistochemistry
IHC was used to detect the expression of HJURP with streptavidin peroxidase complex method.11,12 Briefly, samples were deparaffinized with xylene and rehydrated with graded ethanol. A total of 3% hydrogen peroxide was used for endogenous peroxidase inactivation and citrate buffer for better antigen retrieval. Five percent bovine serum albumin (BSA) was used to incubate slides to block nonspecific binding, followed by incubation with primary antibody and subsequently with secondary antibody. Finally, protein visualization was achieved by the application of peroxidase complex reagent and 3,3′-diaminobenzidine solution.
IHC score system
IHC final score was defined as the product of score for positive cells multiplied by score for staining intensity referring to previous studies.13 The scores of positive cell percentage were defined as per the following criteria: 0, <10% positive cells; 1, 10%–30% positive cells; 2, 30%–50% positive cells; and 3, >50% positive cells. The scores of staining intensity were set based on the following criteria: 0, negative staining; 1, weak staining; 2, moderate staining; and 3, strong staining. The total IHC score ranged from 0 to 9. The cutoff of IHC score was generated by receiver operating characteristic curve and set as the point with the highest sum of specialty and sensitivity, which was 3.5 in this study.
RNA extraction and real-time polymerase chain reaction (PCR)
Total RNA of HCC tissues and normal liver tissues was extracted with Trizol reagent according to the manual.14 The cDNA synthesis and quantitative PCR were performed by StepOnePlus real-time PCR system (Thermo Fisher Scientific) and SYBR Green method. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts served as internal controls. The relative amount of mRNA was calculated by 2‒∆∆CT equation. The sequences of primers were based on previous studies and are as follows4: HJURP forward: 5′-GAAGGGATGTACGTGTGACTC-3′; HJURP reverse: 5′-CCATTCTCTGGGAGATGAAGC-3′; GAPDH forward: 5′-TGGAGAATGAGAGGTGGGATG-3′; and GAPDH reverse: 5′-GAGCTTCACGTTCTTGTATCTGT-3′.
Plasmid construction and transfection
HJURP cDNA was purchased from GeneChem (Shanghai, People’s Republic of China) and cloned into pFLAG-CMV2 vector with restriction enzymes (New England BioLabs, MA, USA) after PCR amplification. Empty pFLAG-CMV2 vector was used as the control group in the study. The empty vector or vector with HJURP was transfected with Lipofectamine 2000 according to the manual.
Immunoblotting
The expression of HJURP was detected by Western blotting. Nuclear proteins were extracted from HCC cell lines using Nucleoprotein Extraction Kit (Sangon Company, Shanghai, People’s Republic of China) according to the manual. Each protein sample (20 μg) was boiled with loading buffer, electrophoresed in SDS polyacrylamide gel electrophoresis, and transferred to polyvinylidene fluoride membrane (Pall Corporation, NY, USA). Unspecific antigen binding was blocked with 5% bovine serum albumin. The membrane with protein was incubated in primary antibody (1:1,000) overnight at 4°C and then incubated with corresponding secondary antibody. The proteins were visualized by electro chemiluminescence reagent (EMD Millipore, Billerica, MA, USA).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
HCC cell proliferation was detected by MTT assay.15 Cells in logarithmic phase were first transfected with siRNA, scrambled RNA, empty vector, or Flag-HJURP 24 h before cell passage. Cells were then split into 96-well plates with about 2,000–3,000 cells per well and incubated for 24, 48, and 72 h. At the end, 10 μL MTT (5 mg/mL) was added into each well and incubated for 4 h, and 100 μL DMSO was finally added for dissolving the crystals. Optical density at 490 nm was measured by a spectrophotometer (Molecular Devices Company, CA, USA). Every group had at least six parallel wells. Statistical significance was calculated by Student’s t-test and date were acquired from three independent experiments.
Statistical analysis
All data were analyzed with SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). The correlation between HJURP and clinicopathological factors was evaluated by chi-square test. The survival curves were analyzed by Kaplan–Meier method and the difference was calculated by log-rank test. Independent survival risks were identified by Cox regression model. The difference between the groups compared in HCC cell proliferation test was analyzed by Student’s t-test.
Results
Expression of HJURP in HCC tissues
As a transcription coregulator, HJURP is primarily found in the nucleus of HCC cells (Figure 1). According to the criteria provided in Patients and methods section, all the 164 patients were divided into low-expression group (122/164) and high-expression group (42/164) based on the cutoff value of IHC score. In this study, the percentages of HJURP expressed by low-expression and high-expression groups were 64.4% and 25.6%, respectively.
Figure 1.
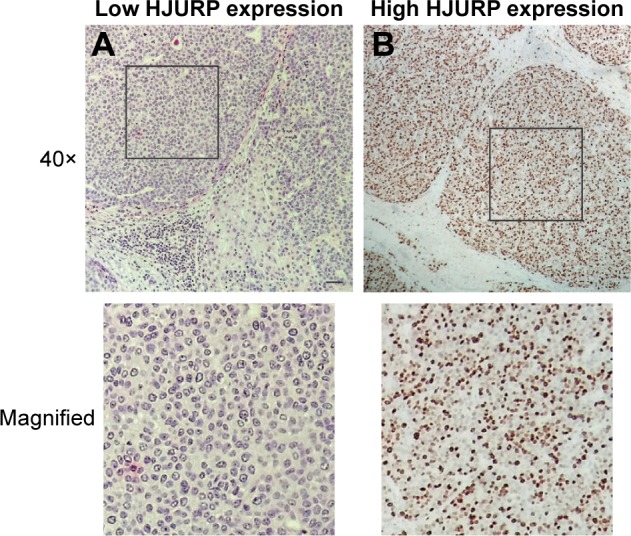
Images of HJURP immunohistochemistry staining. (A) Low HJURP expression and (B) high HJURP expression. Scale bar: 50 μm.
Abbreviation: HJURP, Holliday junction–recognizing protein.
Correlation between HJURP and clinicopathological factors
To investigate HJURP-related clinicopathological factors, the correlation between HJURP and clinicopathological factors was analyzed by chi-square test (Table 1). Interestingly, HJURP overexpression was more likely to exist in tumors >5 cm in our study (P=0.006), indicating that HJURP plays a promoting role in HCC cell proliferation. Moreover, HJURP expression was significantly correlated to BCLC stage, partly because tumor size is an essential index for BCLC stage.
Table 1.
Correlation between HJURP and clinicopatholgical factors
| Characters | Number | HJURP
|
P-value* | |
|---|---|---|---|---|
| Low | High | |||
| Gender | 1.000 | |||
| Female | 43 | 32 | 11 | |
| Male | 121 | 90 | 31 | |
| Age (years) | 0.721 | |||
| <60 | 77 | 56 | 21 | |
| ≥60 | 87 | 66 | 21 | |
| Tumor size (cm) | 0.006 | |||
| ≤5 | 66 | 57 | 9 | |
| >5 | 98 | 65 | 33 | |
| Tumor number | 0.584 | |||
| Single | 103 | 75 | 28 | |
| Multiple | 61 | 47 | 14 | |
| Differentiation | 1.000 | |||
| Well + moderate | 98 | 73 | 25 | |
| Poor | 66 | 49 | 17 | |
| Liver cirrhosis | 0.851 | |||
| Presence | 111 | 83 | 28 | |
| Absence | 53 | 39 | 14 | |
| HBsAg | 0.356 | |||
| Negative | 60 | 42 | 18 | |
| Positive | 104 | 80 | 24 | |
| BCLC stage | 0.002 | |||
| 0&A | 86 | 73 | 13 | |
| B&C | 78 | 49 | 29 | |
| TNM stage | 0.324 | |||
| I–II | 119 | 91 | 28 | |
| III–IV | 45 | 31 | 14 | |
Notes:
Means calculated by chi-square test. Bold represents statistically significant values (P<0.05).
Abbreviations: BCLC, Barcelona clinic liver cancer; HBsAg, hepatitis B surface antigen; HJURP, Holliday junction–recognizing protein.
Prognostic value of HJURP in HCC
All the clinicopathological factors including HJURP were used to analyze their correlation with overall survival rates (Table 2). This study confirmed that multi-tumors (P=0.016), poor differentiation (P=0.047), BCLC stage (P=0.006), and TNM stage (P=0.004) all were significantly associated with unfavorable prognosis of HCC, which was in agreement with the previous reports.16–18 Notably, HJURP expression was also verified to be a prognostic factor. Patients with high HJURP expression seemed to have a poorer survival rate than those with low HJURP expression (P=0.003). Survival curves of low and high HJURP expressions were analyzed using Kaplan–Meier method and statistical significance was analyzed by log-rank test (Figure 2).
Table 2.
Univariate analysis
| Characters | 5-Year survival rate | P-value* |
|---|---|---|
| Gender | 0.302 | |
| Female | 27.9 | |
| Male | 36.2 | |
| Age (years) | 0.850 | |
| <60 | 31.8 | |
| ≥60 | 36.1 | |
| Tumor size (cm) | 0.766 | |
| ≤5 | 33.0 | |
| >5 | 35.7 | |
| Tumor number | 0.016 | |
| Single | 37.3 | |
| Multiple | 28.9 | |
| Differentiation | 0.047 | |
| Well + moderate | 52.2 | |
| Poor | 23.8 | |
| Liver cirrhosis | 0.068 | |
| Presence | 35.1 | |
| Absence | 33.0 | |
| HBsAg | 0.150 | |
| Negative | 41.7 | |
| Positive | 28.1 | |
| BCLC stage | 0.005 | |
| 0&A | 39.8 | |
| B&C | 26.3 | |
| TNM stage | 0.004 | |
| I–II | 37.9 | |
| III–IV | 24.4 | |
| HJURP | 0.003 | |
| Low | 37.8 | |
| High | 24.5 |
Notes:
Means calculated by log-rank test. Bold represents statistically significant values (P<0.05).
Abbreviations: BCLC, Barcelona clinic liver cancer; HBsAg, hepatitis B surface antigen; HJURP, Holliday junction–recognizing protein.
Figure 2.
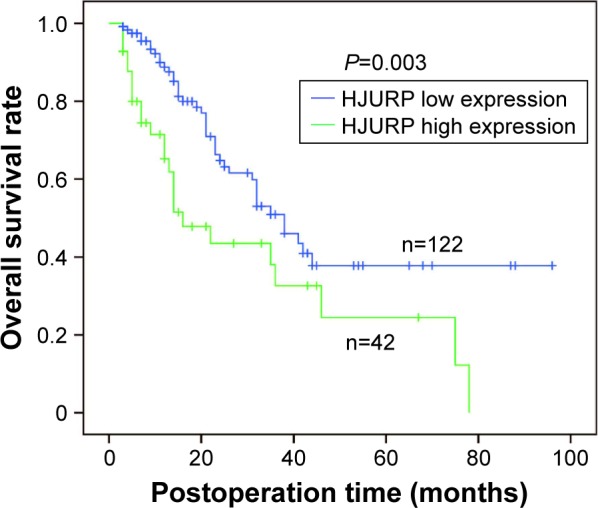
Correlation between HJURP expression and the overall survival rate. Survival curves representing low and high HJURP expressions were displayed by Kaplan–Meier method and statistical significance was analyzed by log-rank test (P=0.003).
Abbreviation: HJURP, Holliday junction–recognizing protein.
Prognostic factors were included into the Cox regression model for the identification of independent risk factors of HCC (Table 3). Patients with liver cirrhosis were also enrolled because its relation with survival rate is also notable without statistical significance (P=0.068). BCLC and TNM stages were excluded because of their interaction with other prognostic factors. In this study, high HJURP expression was identified as an independent prognostic factor (P<0.001, hazard ratio [HR] =2.75, and 95% confidence interval [CI] =1.60–4.73). Moreover, tumor number (P=0.001, HR = 2.50, 95% CI =1.42–4.40), differentiation (P=0.014, HR =2.00, 95% CI =1.15–3.49), and liver cirrhosis (P=0.015, HR =1.95, 95% CI =1.14–3.33) were all confirmed to be the independent risk factors for poorer prognosis.
Table 3.
Multivariate analysis
| Characters | Hazard ratio | 95% CI | P-value* |
|---|---|---|---|
| Tumor number | 0.001 | ||
| Single | 1 | ||
| Multiple | 2.50 | 1.42–4.40 | |
| Differentiation | 0.014 | ||
| Well + moderate | 1 | ||
| Poor | 2.00 | 1.15–3.49 | |
| Liver cirrhosis | 0.015 | ||
| Presence | 1 | ||
| Absence | 1.95 | 1.14–3.33 | |
| HJURP | <0.001 | ||
| Low | 1 | ||
| High | 2.75 | 1.60–4.73 |
Note:
Means calculated by Cox regression model.
Abbreviations: BCLC, Barcelona clinic liver cancer; HJURP, Holliday junction–recognizing protein; CI, confidence interval.
HJURP promotes HCC cell progression
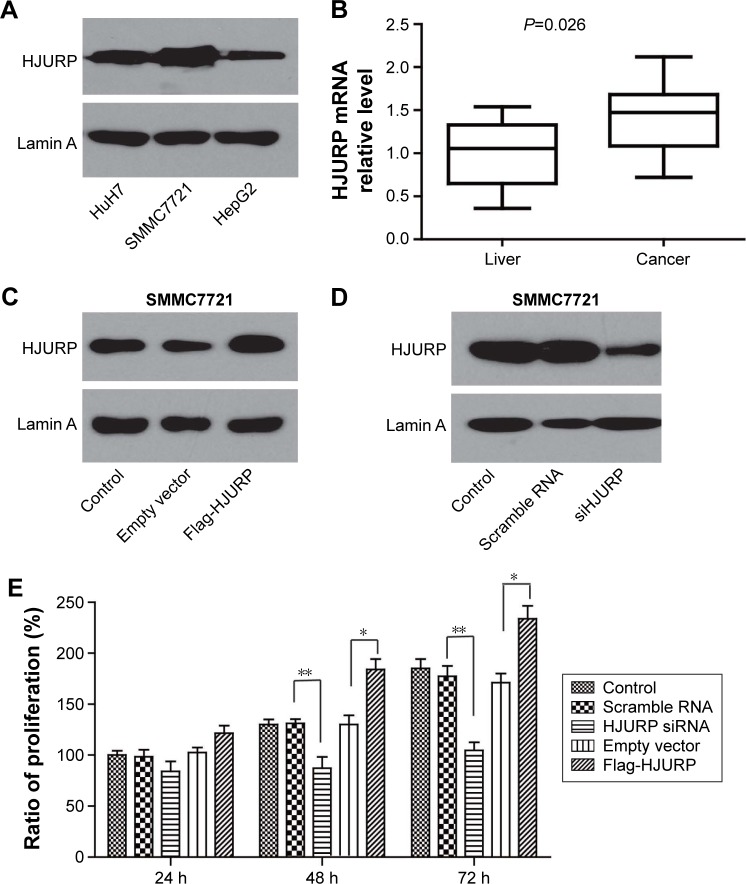
The expression profile in HCC cell lines was first investigated by Western blotting (Figure 3A). In this study, SMMC7721 showed the highest expression of HJURP, whereas HepG2 the lowest among the three cell lines. We speculated that ectopic HJURP expression may promote the progression of HCC cell lines; therefore, we detected mRNA level in HCC tissues and corresponding normal liver tissues of the same patient. Ten patients were enrolled and the average mRNA level of the normal liver was set as baseline, and other mRNA level was standardized to this baseline. The results demonstrated that HJURP in tumor tissues was significantly higher than that in the normal liver tissues (Figure 3B).
Figure 3.
HJURP promotes HCC cell line proliferation. (A) HJURP expression in HCC cell line HuH7, SMMC7721, and HepG2 was detected by Western blotting. (B) HJURP mRNA level in HCC tissues was significantly higher than that of the corresponding normal liver tissues (P=0.026). (C) Successful HJURP overexpression in SMMC7721 was verified by Western blotting. HJURP was cloned into pFLAG-CMV2 by double digestion with restriction enzymes and transfected into SMMC7721. HJURP expression 48 h after transfection was detected by Western blotting. (D) Successful HJURP knockdown in SMMC7721 was verified by Western blotting. HJURP knockdown was performed by siRNA. HJURP expression 48 h after siRNA transfection was detected by Western blotting. (E) Cell proliferation was detected by MTT assay in SMMC7721 cell line after regulating HJURP expression. After 24 h of transfection with scrambled RNA, siRNA, control vector, or Flag-HJURP vector, cells were passaged into 96-well plates and starved for 6 h. Then medium containing 10% FBS was added, which was set as time 0. Cell proliferation at time point 24, 48, or 72 h after time 0 was detected. The proliferation ratio of group without any treatment (control group) at 24 h was set as 100% and other groups were standardized with this baseline. Column graph was displayed with ± SEM. *P<0.05 and **P<0.01.
Abbreviations: FBS, fetal bovine serum; HCC, hepatocellular carcinoma; HJURP, Holliday junction–recognizing protein; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) tetrazolium; SEM, standard error of mean; siRNA, small interfering RNA; siHJURP, siRNA against HJURP.
Analysis of HJURP and clinicopatholgical factors revealed that larger tumors correlated with HJURP overexpression (P=0.006); therefore, we performed proliferation assay after regulating HJURP expression. HepG2 cell line was selected for HJURP overexpression due to its lower basal HJURP level, whereas SMMC7721 was selected as HJURP knockdown cell model. First, successful HJURP overexpression and siRNA knockdown were verified in SMMC7721 by Western blotting (Figure 3C and D). HCC cell proliferation was detected in SMMC7721 cell line by MTT assay at 24, 48, and 72 h (Figure 3E). It appeared that HJURP could accelerate SMMC7721 proliferation and HJURP knockdown could repress the proliferation at 48 and 72 h, demonstrating that HJURP played an important role in promoting HCC cell proliferation.
Discussion
HCC is a highly life-threatening disease worldwide, especially in China because of the extremely high frequency of HBV. Most patients with HCC usually lose their opportunity of surgical resection because of its silent clinical symptoms. New prognostic biomarkers are in urgent need for individual treatment and for improving survival time. In previous studies, Huang et al8 proved that a HJURP non-synonymous SNP was an independent risk factor for HCC. Although no studies have focused on the underlying mechanisms, this report still indicates that HJURP dysfunction may promote susceptibility to HCC among Chinese, inspiring us to investigate HJURP expression and its clinical significance in HCC. In our study, we demonstrated that high HJURP expression was closely associated with larger tumor size, and we further validated that HJURP could promote HCC cell proliferation with experiments in vitro. Furthermore, HJURP has been well acknowledged to regulate cell cycle. However, the regulatory net of HJURP is more complicated than just regulating cell cycle. A number of proteins have been reported to interact with HJURP, including proteins affecting HJURP function and the downstream proteins regulated by HJURP. The most well-known molecule regulated by HJURP is the histone H3 variant, centromere-specific protein (CENP)-A. The cooperation of CENP-A and its chaperon HJURP mediates normal cell cycle, whereas ectopic activation of HJURP is involved in the chromosomal stability and immortality of cancer cells.19 However, the molecules that link HJURP and HCC progression and the precise pathway of HJURP in HCC are still not clear.
In this study, we demonstrated that HJURP overexpression could promote HCC cell proliferation and predict poorer prognosis in HCC patients, suggesting that HJURP function inhibition may decrease HCC cell proliferation and improve prognosis. HJURP may be a potential molecular drug target and even trigger new therapeutic strategy of HCC treatment, which certainly needs new progress in the study of HJURP inhibitors. Unfortunately, no HJURP inhibitors or monoclonal antibodies are currently available to antagonize HJURP function. Effective inhibitors to HJURP or some key downstream proteins may appear in the future. This study has thrown light on the study of HJURP in HCC progression. More studies in the future should focus on the underlying mechanisms of HJURP in HCC progression.
Conclusion
In conclusion, we have demonstrated that HJURP could be considered as an independent prognostic factor in patients with HCC. High expression of HJURP could predict poorer prognosis by itself. HJURP may promote HCC progression by accelerating HCC cell proliferation, indicating the potency of HJURP or its downstream key proteins as potential therapeutic targets in the treatment of HCC.
Acknowledgments
This study was funded by Science and Technology Project of Yantai City (2015WS018, 2016WS006), Youth Research Initiation Foundation of Yuhuangding Hospital (201511), Shandong Province Medical Science and Technology Development Project (2016WS0706).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Tachiwana H, Muller S, Blumer J, Klare K, Musacchio A, Almouzni G. HJURP involvement in de novo CenH3(CENP-A) and CENP-C recruitment. Cell Rep. 2015;11(1):22–32. doi: 10.1016/j.celrep.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Hu Z, Huang G, Sadanandam A, et al. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010;12(2):R18. doi: 10.1186/bcr2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Müller S, Montes de Oca R, Lacoste N, Dingli F, Loew D, Almouzni G. Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3(CENP-A) loading. Cell Rep. 2014;8(1):190–203. doi: 10.1016/j.celrep.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Valente V, Serafim RB, de Oliveira LC, et al. Modulation of HJURP (Holliday junction-recognizing protein) levels is correlated with glioblastoma cells survival. PLoS One. 2013;8(4):e62200. doi: 10.1371/journal.pone.0062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Zhai Y, Hu Z, et al. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet. 2010;42(9):755–758. doi: 10.1038/ng.638. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Qian J, Yang Y, et al. GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet. 2012;8(7):e1002791. doi: 10.1371/journal.pgen.1002791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang DK, Sun J, Cao G, et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet. 2013;45(1):72–75. doi: 10.1038/ng.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang W, Zhang H, Hao Y, et al. A non-synonymous single nucleotide polymorphism in the HJURP gene associated with susceptibility to hepatocellular carcinoma among Chinese. PLoS One. 2016;11(2):e0148618. doi: 10.1371/journal.pone.0148618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minagawa M, Ikai I, Matsuyama Y, Yamaoka Y, Makuuchi M. Staging of hepatocellular carcinoma: assessment of the Japanese TNM and AJCC/UICC TNM systems in a cohort of 13,772 patients in Japan. Ann Surg. 2007;245(6):909–922. doi: 10.1097/01.sla.0000254368.65878.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42(5):1208–1236. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Zhang Q, Li K, et al. Prognostic significance of USP33 in advanced colorectal cancer patients: new insights into β-arrestin-dependent ERK signaling. Oncotarget. 2016;7(49):81223–81240. doi: 10.18632/oncotarget.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu YF, Ge FJ, Han B, et al. High-mobility group box 1 expression and lymph node metastasis in intrahepatic cholangiocarcinoma. World J Gastroenterol. 2015;21(11):3256–3265. doi: 10.3748/wjg.v21.i11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang XQ, Xu YF, Guo S, et al. Clinical significance of nerve growth factor and tropomyosin-receptor-kinase signaling pathway in intrahepatic cholangiocarcinoma. World J Gastroenterol. 2014;20(14):4076–4084. doi: 10.3748/wjg.v20.i14.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Xu Y, Zhang Q, et al. Prognostic significance of TBL1XR1 in predicting liver metastasis for early stage colorectal cancer. Surg Oncol. 2017;26(1):13–20. doi: 10.1016/j.suronc.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Xu YF, Yang XQ, Lu XF, et al. Fibroblast growth factor receptor 4 promotes progression and correlates to poor prognosis in cholangiocarcinoma. Biochem Biophys Res Commun. 2014;446(1):54–60. doi: 10.1016/j.bbrc.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 16.Xiao S, Chang RM, Yang MY, et al. Actin-like 6A predicts poor prognosis of hepatocellular carcinoma and promotes metastasis and epithelial-mesenchymal transition. Hepatology. 2016;63(4):1256–1271. doi: 10.1002/hep.28417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu BS, Xiong SM, Li G, Li JP. Downregulation of SLC5A8 inhibits hepatocellular carcinoma progression through regulation of Wnt/β-catenin signaling. Tumour Biol. 2016;37(10):13445–13453. doi: 10.1007/s13277-016-5170-3. [DOI] [PubMed] [Google Scholar]
- 18.Ji F, Fu S, Guo Z, et al. Prognostic significance of preoperative aspartate aminotransferase to neutrophil ratio index in patients with hepatocellular carcinoma after hepatic resection. Oncotarget. 2016;7(44):72276–72289. doi: 10.18632/oncotarget.10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato T, Sato N, Hayama S, et al. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67(18):8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]