Abstract
Chromosomal rearrangements associated with acute myeloid leukemia (AML) include fusions of the genes encoding the acetyltransferase MOZ or MORF with genes encoding the nuclear receptor coactivator TIF2, p300, or CBP. Here we show that MOZ-TIF2 acts as a dominant inhibitor of the transcriptional activities of CBP-dependent activators such as nuclear receptors and p53. The dominant negative property of MOZ-TIF2 requires the CBP-binding domain (activation domain 1 [AD1]), and coimmunoprecipitation and fluorescent resonance energy transfer experiments show that MOZ-TIF2 interacts with CBP directly in vivo. The CBP-binding domain is also required for the ability of MOZ-TIF2 to extend the proliferative potential of murine bone marrow lineage-negative cells in vitro. We show that MOZ-TIF2 displays an aberrant nuclear distribution and that cells expressing this protein have reduced levels of cellular CBP, leading to depletion of CBP from PML bodies. In summary, our results indicate that disruption of the normal function of CBP and CBP-dependent activators is an important feature of MOZ-TIF2 action in AML.
CREB binding protein (CBP) is a histone acetyltransferase that is required as a cofactor by many DNA-binding transcription factors and as such plays a central role in the regulation of gene expression and the integration of signaling pathways (22). In vivo, CBP associates with subnuclear structures called PML bodies, which contain the structural component PML and at least 10 other proteins such as Sp100 (66). Both CBP and its homolog p300 function in cell proliferation, differentiation, and apoptosis (7, 21). Studies with heterozygous null mice have demonstrated a requirement for CBP in normal hematopoiesis. CBP-null embryos were found to have defective hematopoiesis (44, 65), whereas mice heterozygous for the CBP gene developed a range of hematopoietic diseases (33) and showed reduced self-renewal of hematopoietic stem cells (51). Moreover, genetic abnormalities involving the CBP/p300 genes in acute myeloid leukemia (AML) provide further evidence for their role in the regulation of hematopoietic development.
Reciprocal translocations fusing genes encoding monocytic leukemia zinc finger protein (MOZ/MYST3) or MOZ-related factor (MORF/MYST4) to CBP (8, 48), p300 (10) or transcriptional intermediary factor TIF2 (1, 6, 9, 15, 36, 42, 47) are recurrent in the M4 and M5 subtypes of AML. MOZ is a member of the MOZ/YBF2/SAS2/TIP60 homology domain (MYST) family of protein acetyltransferases, which contain a C2HC zinc finger and acetyltransferase domain that together comprise the MYST domain (64). MOZ and MORF acetylate histones in vitro but share little sequence homology with other MYST proteins outside the histone acetyltransferase (HAT) domain, suggesting diverse cellular functions for this family (56, 64). MOZ was reported to modestly enhance reporter activation by Runx domain proteins such as AML1, although this activity was not dependent on its histone acetyltransferase activity (11, 12, 32). Thus, while it is likely to have a role in chromatin modification, the function of MOZ in vivo remains to be established.
TIF2/GRIP1 (24, 62) is a member of the p160 family of coactivators, which also includes SRC1 (28, 46) and ACTR/pCIP/AIB1 (4, 13, 60). These proteins function as coactivators for nuclear receptors (NRs) and associate tightly with NRs via LXXLL motifs (23, 60). The C termini of p160 proteins contain the AD1 and AD2 domains, which recruit CBP/p300 and CARM-1, respectively, to NR target promoters (13, 24, 28, 31, 54, 62). A leucine-rich α-helix in AD-1 (17, 54) is conserved in Ets-2, IRF3, p53, and E1A proteins, which compete with p160s for binding to the C terminus of CBP (39).
The t(8:16)(p11;p13) translocation produces a fusion protein containing the N terminus of MOZ (amino acids 1 to 1547) and almost the entire sequence of CBP (amino acids 244 to 2441) (8). MOZ-TIF2 translocations derive from inversions of chromosome 8, inv8(p11q13) (42), and give rise to fusion proteins containing the N terminus of MOZ, including the plant homeodomain and MYST domains (amino acids 1 to 1117) and the C terminus of TIF2 (amino acids 869 to 1713 or 939 to 1713), including AD1 and AD2. The acquisition of MOZ-CBP and MOZ-TIF2 translocations is likely to affect cell function in a number of ways, including (i) a reduction of the normal cellular complement of parental proteins and (ii) aberrant function of the resultant fusion proteins, including both dominant negative and gain-of-function effects. MOZ-TIF2 expression was recently shown to immortalize lineage-deficient (Lin−) murine bone marrow cells and to induce AML in mice (16). While the molecular mechanisms involved remain unclear, it was demonstrated that the CBP-binding function was necessary for inducing myeloproliferative disease in these studies (16). Due to the function of TIF2, CBP, and possibly MOZ in transcriptional regulation and chromatin modification, it is likely that MOZ-CBP and MOZ-TIF2 fusion proteins affect gene expression networks that are important in cell differentiation and proliferation. Moreover, it has been suggested that MOZ-TIF2 may mimic MOZ-CBP action by interaction with CBP in vivo (9).
In this study we investigated the molecular mechanisms underlying MOZ-TIF2 action in vivo. We show that MOZ-TIF2 has an aberrant subcellular localization, which is distinct from that of wild-type MOZ and other known nuclear compartments. We demonstrate that MOZ-TIF2 interacts with CBP via the AD1 domain and that as a consequence of this, the transcriptional activities of CBP-dependent activators such as NRs and p53 are compromised. Finally, we show that expression of MOZ-TIF2 correlates with a depletion of CBP from PML bodies and reduced cellular CBP levels. This ability to interact with CBP and impair CBP function is critical to the ability of MOZ-TIF2 to transform lineage-deficient bone marrow cells in vitro.
MATERIALS AND METHODS
Plasmid construction.
pcDNA3.0 plasmids containing MOZ and MOZ-TIF2 cDNAs were gifts from M. Carapeti and D. G. Gilliland. To remove the MOZ 3′ untranslated region (UTR) sequence, a PCR fragment was generated from an internal CelII restriction site to the termination codon, with BglII and XbaI sites inserted at the 3′ end (primers 5′-GGACTGGCTCAGCTGGCTCC-3′ and 5′-AAAATCTAGAAGATCTTCATCTTCTCATGTAAGGTCC-3′). The modified vector pcDNA3.0PT was created by insertion of the sequence 5′-AGCTGCCGCGGTTACTCGAGTGTGGGCCCTATAGCTCAGCACTTTCTAGAAGGCC-3′ between the HindIII and ApaI sites. The CelII-XbaI PCR product was inserted into pcDNA3.0PT, and the rest of the MOZ coding sequence was inserted as an ApaI-CelII fragment. Approximately 1 kb of MOZ-TIF2 3′ UTR sequence was removed through digestion of the full-length cDNA with ApaI and XbaI and insertion into pcDNA3.0PT.
The 5′ UTRs were removed, and an N-terminal FLAG tag was added to both MOZ and MOZ-TIF2 via PCR (forward primer, FLAG-5′-AAAAGGGCCCCACCATGGACTACAAGGACGACGACGATAAGGGGATGGTAAAACTCGCAAAC-3′; reverse primer encompassing a natural EcoRV restriction site in the MOZ cDNA sequence, 5′-GTTCTTGGATATCACG-3′).
MOZ-N was created by digestion of pcDNA3.0PT FLAG-MOZ with EcoRI and XbaI and insertion of a termination codon and restriction enzyme sites at the 3′ end (primers 5′-AATTTGGGCGGCCGCGAATTCGCTAGCGGATCCTGAAGATCTT-3′ and 5′-CTAGAAGATCTTCAGGATCCGCTAGCGAATTCGCGGCCGCCCA-3′).
(i) MOZ-TIF2ΔAD1.
PCRs were carried out with BstEII and Delta-reverse primers and the Delta-forward and SspBI primers, and the products were combined by recombinant PCR. The resulting fragment was digested with BstEII and SspBI restriction enzymes and cloned into the MOZ-TIF2 vectors (BstEII, 5′-GAGCCTGGTGACCAG CCTGGC-3′; Delta-reverse, 5′-CCATGGCCTGAAAGCATCGAACAGTTCTCAAGT CAG-3′; Delta-forward, 5′-CCATGGCCTGAAAGCATCGAACAGTTCTCAAGTC AG-3′; SspBI, 5′-GTTGTTACTGTACATGCTGGTG-3′).
(ii) pSG5, cyan fluorescent protein (CFP), and yellow fluorescent protein (YFP) constructs.
Full-length mouse CBP cDNA was removed from pRSV-CBP-HA (a gift from R. Goodman) as a BamHI fragment (resulting in the loss of the hemagglutinin [HA] tag) and cloned into the pSG5 and pECFP-C1 vectors. Tagged MOZ, MOZ-TIF2, and MOZ-TIF2ΔAD1 were digested with ApaI and XbaI and inserted into digested pEYFP-C1 (Clontech).
(iii) pMIE-MCS, pMIE-MCS FLAG-MOZ-TIF2, and pMIE-MCS FLAG-MOZ-TIF2 ΔAD1.
A multiple cloning site was introduced into pMIE (35) via the EcoRI and XhoI site by using primers 5′-AATTCTGTCCGCGGTTGTAGGGATCCTTAAGATCTAAC-3′ and 5′-TCGAGTTAGATCTTAAGGATCCCTACAACCGCGGACAG-3′. Full-length FLAG-MOZ-TIF2 and FLAG-MOZ-TIF2ΔAD1 cDNAs were removed from pEYFP-FLAG-MOZ-TIF2 and pEYFP-FLAG-MOZ-TIF2ΔAD1 by using KspI and BglII and cloned into pMIE-MCS vectors.
GST pull-down assays.
Glutathione S-transferase (GST) pull-down assays were carried out as described previously, using pGEX-DMH as the control plasmid (54). The fusion constructs GST-SRC1/p160 interaction domain (SID) (54) and GST-estrogen receptor α (ERα)-AF2 (23) were as previously described. GST-retinoid X receptor α (RXRα)-AF2 was a gift from K. Chatterjee, and full-length GST-CBP was a gift from A. Harel-Bellan. NR ligand-binding domain (LBD) pull-down assays were carried out in the presence of 10−7 M ligand (GST-ERα, 17β-estradiol; GST-RXRα, 9-cis-retinoic acid) or vehicle.
Antibodies.
For Western blotting, coimmunoprecipitations, and immunofluorescence, the following antibodies were used: anti-CBP A22 and C20, anti-MOZ N19, anti-PML A20, anti-rabbit immunoglobulin G (IgG)-peroxidase, anti-mouse IgG-peroxidase (Autogen Bioclear), anti-α-tubulin, anti-FLAG M2 and anti-FLAG M2-agarose affinity gel, anti-goat IgG-fluorescein isothiocyanate or -tetramethyl rhodamine isocyanate, anti-mouse IgG-fluorescein isothiocyanate, anti-rabbit IgG-tetramethyl rhodamine isocyanate (Sigma), anti-PML 83-2 (a gift from A. Dejean), anti-SP100 (a gift from T. G. Hofmann), and antivinculin (a gift from J. Norman).
Cell culture and transient transfections.
Constructs pGAL4-RXRα, p(Gal4)5-E1bΔ-Luc (54), pSP6-MOR, and pSG5-FLAG-SRC1e (28) have been described previously. The following plasmids were obtained as gifts: p(PPRE)3-tk-Luc (S. Ali); pSG5-TIF2 (C. Bevan); pCH110, p(ERE)3-tk-Luc, pEF-BOS, pGAL4-ERα, and pGAL4-RARα (M. Parker); pSG5-HA-GRIP1 (M. Stallcup); pRep4-RARE-luc (A. Zelent); IRGC-E4-Luc and pcDNA3-p53 (X. Liu); and pBax-Luc, pGADD45-Luc, and pMDM2-Luc (C. J. Di Como).
COS-1 and HEK293 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum and 2 mM glutamine at 5% CO2. U937 cells were maintained in phenol red-free RPMI 1640 containing 5% dextran-charcoal-stripped fetal calf serum (DCSS) at 5% CO2.
For reporter assays, COS-1 cells were seeded 24 h prior to transfection in six-well plates at 1 ×105 to 3 ×105 cells/well in 2 ml of phenol red-free Dulbecco's modified Eagle medium supplemented with 5% DCSS. Cells were transiently transfected with 500 ng of β-galactosidase reporter (pCH110), 500 ng of luciferase reporter [p(Gal4)5-E1bΔ-Luc or p(ERE)3-tk-Luc], expression vectors (100 ng unless otherwise stated), and empty vector to standardize the quantity of DNA in each well. Transfections were carried out with the CalPhos mammalian transfection kit (Clontech) according to the manufacturer's protocol. Fresh medium containing 10−7 M ligand (17β-estradiol, 9-cis-retinoic acid, or all-trans-retinoic acid) or vehicle was added at 24 h posttransfection, and the cells incubated for an additional 24 h before harvesting. Cells were resuspended in 100 μl of lysis buffer (Dual Light System kit; Applied Biosystems), 5 μl of extract was assayed (in duplicate) for luciferase activity, and the results normalized by using β-galactosidase activity.
For p53 reporter assays, SaOS-2 cells were seeded 6 h prior to transfection in six-well plates, and the cells were transiently transfected with 100 ng of β-galactosidase reporter pCH110, 500 ng of p53-responsive luciferase reporters (IRGC-E4-Luc, pBax-Luc, pGADD45-Luc, and pMDM2-Luc), 100 ng of pcDNA3-p53 expression vector, and 750 ng of MOZ-TIF2, along with empty vector to standardize the quantity of DNA in each well. Cells were harvested after 48 h and assayed as described above.
For U937 transient transfections, 2 × 107 cells per electroporation were harvested by centrifugation (2,000 × g, 5 min), washed with phosphate-buffered saline (PBS), and resuspended in 200 μl of RPMI 1640 supplemented with 0.1% glucose. Cells were electroporated (Bio-Rad Gene Pulser II; 300 V, 950 μF, maximum capacitance) with 1 μg of pEF-BOS, 4 μg of pRep4-RARE-luc or p(PPRE)3-tk-luc, or MOZ-TIF2 or MOZ-TIF2ΔAD1 DNA (as indicated in Fig. 3D), and pcDNA3.0PT to standardize the total amount of DNA to 10 μg. After electroporation, cells were recovered by addition of 800 μl RPMI 1640 containing 10% DCSS. After 3 h another 1 ml of 10% DCSS-RPMI 1640 containing ligand [pRep4-RARE-luc, all-trans-retinoic acid and 9-cis-retinoic acid; p(PPRE)3-tk-luc, rosiglitazone and 9-cis-retinoic acid; final concentrations, 10−7 M) or the equivalent volume of vehicle was added. Cells were harvested after a further 21 h, and extracts were assayed as described for COS-1 transfections.
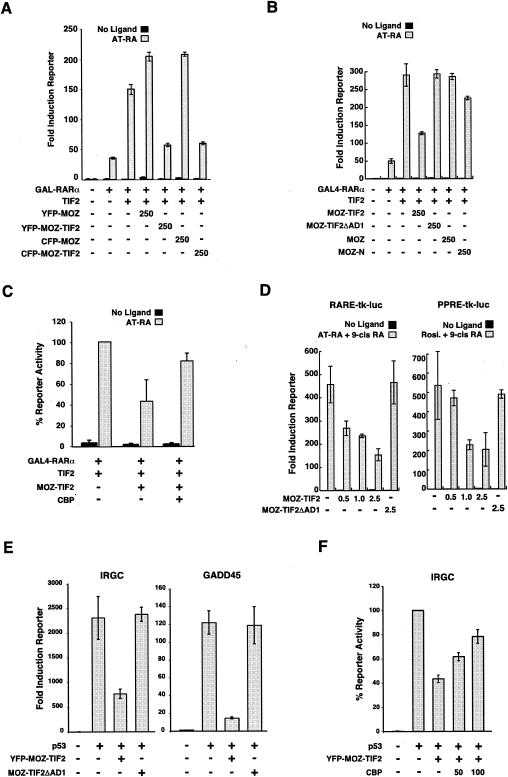
FIG. 3.
Role of CBP in the inhibition of NR-mediated and p53-mediated transcription activation by MOZ-TIF2. (A) GAL4-RARα/TIF2-mediated reporter assays in COS-1 cells expressing MOZ and MOZ-TIF2 fusion proteins tagged with YFP or CFP. (B) GAL4-RARα/TIF2-dependent activation of a (Gal4)5-1bΔ-luc reporter gene in COS-1 cells in the presence of various MOZ-containing proteins. (C) Rescue experiment showing the effect of overexpressed CBP on MOZ-TIF2-mediated inhibition of GAL4-RARα/TIF2 reporter activation in COS-1 cells. (D) Dose-dependent inhibition of endogenous PPAR- and RAR-mediated transcription activation by MOZ-TIF2 is dependent on the AD1 domain in U937 cells. (E) p53-dependent activation of the IRGC-luc and the GADD45-luc reporter genes in SaOs-2 cells in the presence of various MOZ-containing proteins. (F) Rescue experiment showing the effect of overexpressed CBP on MOZ-TIF2-mediated inhibition of p53-dependent reporter activation of the IRGC-luc reporter gene in SaOs-2 cells. Error bars indicate standard errors of the means.
Live cell imaging.
Cells were seeded at 5 × 104 cells on 22-mm-diameter circular coverslips, and 24 h later they were transfected with 1 μg of pEYFP-HA-MOZ-TIF2 by using TransFast (Promega). At 18 h posttransfection the coverslip was mounted in a steel coverslip holder in CO2-independent medium (Invitrogen) supplemented with l-glutamine and then overlaid with mineral oil (Sigma). The coverslip holder was inserted in a Patch Slice MicroIncubator, which was kept at a constant temperature of 37°C by a TC-202A temperature controller (Digitimer; Harvard Apparatus Medical Systems Research Products. Hertfordshire, United Kingdom). After the MicroIncubator was mounted on the microscope stage, images were taken at 5-min intervals with an ORCA ER charge-coupled-device camera (Hamamatsu, Hamamatsu City, Japan) attached to a Nikon TE300 inverted microscope. Five optical sections (z sections) with 0.1-μm intervals, obtained with a high-speed Piezo focus drive (Orbit II) fixed to a 60×, 1.4-numerical-aperture objective, were captured at the individual time points by using Openlab 3.09 software (Improvision, Coventry, United Kingdom). The individual sections were merged by using the Openlab software, processed with Adobe Photoshop, and saved as a quick-time movie. Stills were made with Volocity software (Improvision).
Immunoprecipitation and immunofluorescence.
For coimmunoprecipitations, HEK293 cells were seeded in 10-cm-diameter dishes at 1.5 × 106 cells/dish and transfected with 10 μg of FLAG construct and 10 μg of pSG5-CBP. Immunocomplexes were isolated from whole-cell extracts after 48 h by using anti-FLAG M2-agarose affinity gel. Bound proteins were washed, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed by immunoblotting with anti-CBP A22 and C20 antibodies and a secondary antibody-horseradish peroxidase conjugate.
For immunofluorescence, cells were plated onto coverslips in six-well plates at 1 × 105 to 3 × 105 cells/well and, where appropriate, transfected 24 h later by using TransFast (Promega) according to the manufacturer's protocol. After 48 h the cells were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. The cells were washed and, after blocking in 3% bovine serum albumin-PBS, incubated with primary antibodies (1:50 dilution for overexpressed proteins, 1:10 for CBP, 1:30 for PML, and 1:200 for SP100). After 1 h of incubation, the cells were washed and incubated with the secondary antibody (1:100 dilution). DNA was stained with 0.5 μg of Hoechst 33258 per ml or 0.1 μg of DAPI (4′,6′-diamidino-2-phenylindole) per ml. The samples were viewed with a fluorescence microscope (Zeiss Axiovert 200 M) or a confocal laser scanning microscope (Zeiss LSM510).
FRET analysis.
Fluorescence emission spectra were acquired with a confocal laser scanning microscope (Zeiss LSM 510 META). Images were taken with a 40×, 1.3-numerical aperture objective. The emission spectra (fingerprints) of donor and acceptor fluorophores alone were saved as references to allow separation of fluorescence signals from YFP and CFP. Fluorescent resonance energy transfer (FRET) was determined by selective photobleaching of the acceptor fluorophore. Three successive prebleach (CFP and YFP) images were taken by scanning with 453- and 514-nm light, acquiring 10.7-nm-bandwidth images in the range of 463 to 634.9 nm. Intranuclear regions of interest were selected, and YFP was bleached by repeated scanning (300 iterations) with 100% power of the 514-nm laser. Postphotobleach images were acquired with 453- and 514-nm lasers. These images were spectrally unmixed by using the YFP and CFP fingerprints. CFP and YFP fluorescence intensities were recorded. The increase or dequenching of CFP emission (Em) is a direct measure of FRET efficiency (E%) and was calculated as follows: E% = [1 − (CFP Em before YFP photobleach/CFP Em after YFP photobleach)] × 100 (55).
Fluorescence-activated cell sorting (FACS) for Western blotting.
A total of 1.5 × 105 HEK293 cells were plated per 100-mm-diameter dish, and 24 h later they were transfected with 20 μg of expression plasmid encoding either YFP, YFP-FLAG-MOZ-TIF2, or YFP-HA-MOZ-TIF2-ΔAD1. At 24 h posttransfection, cells were incubated in the absence or presence of 10 μM lactacystin for a further 24 h and then washed with PBS, taken off by incubation with 5 mM EDTA, and resuspended in 3 ml of PBS. Cells were sorted with a Becton Dickinson (Franklin Lakes, N.J.) FACS Vantage instrument at an excitation wavelength of 488 nm. A total of 105 cells were collected per condition and analyzed by Western blotting with anti-CBP A22 (1:500 dilution), anti-α-tubulin (1:1,000), and antivinculin (1:1,000) antibodies with the respective peroxidase conjugates (1:5,000).
Purification and infection of Lin− cells (hematopoietic progenitors).
Lin− cells were purified from the bone marrow of 8- to 10-week-old 129SvEv mice. After centrifugation through a Ficoll gradient, mononucleate cells were enriched in progenitors by depletion of cells presenting myeloid, erythroid, and lymphoid differentiation markers, using commercially available reagents (Stem Cell Technologies, Vancouver, British Columbia, Canada). Purified Lin− cells were grown for 36 h in medium containing interleukin 3 (IL-3), IL-6, and stem cell factor (SCF). The cells were then plated on retronectin-coated (Takara-Shuzo, Shiga, Japan), non-tissue culture-treated plates and exposed to the supernatant of packaging, ecotropic Phoenix cells transiently transfected with the indicated retroviral vectors. Transduced cells were sorted with a FACS Vantage instrument.
Serial replating assays.
Transduced, sorted Lin− cells were plated in methylcellulose medium supplemented with 100 ng of SCF per ml and 20 ng each of IL-3 and IL-6 per ml. After 10 days, single-cell suspensions were prepared from pooled colonies, and 10,000 cells were plated in a secondary methylcellulose culture. The cells were serially replated until the control cells were unable to give colonies (usually after three to five platings). The values shown are the means from one independent experiment performed in triplicate.
RESULTS
MOZ-TIF2 interacts with CBP in vitro and in vivo.
To explore the mechanisms underlying its leukemogenic action, we investigated MOZ-TIF2 interactions with potential target proteins such as NRs and CBP. In vitro-translated TIF2, MOZ, and MOZ-TIF2 proteins (shown schematically in Fig. 1A) were assessed for binding to GST-NR-LBD fusion proteins (Fig. 1B). TIF2 exhibited strong ligand-dependent binding to ERα and RXRα as expected, whereas MOZ-TIF2 did not bind either receptor in the presence or absence of ligand. This is consistent with the absence of the TIF2 NR interaction domain in the MOZ-TIF2 fusion protein, and it rules out NR interaction with the C terminus of TIF2 or the N terminus of MOZ. Similarly, full-length MOZ, which does not contain LXXLL motifs required for NR binding (23), showed no interaction with the ERα or RXRα LBD (Fig. 1B). These results indicate that MOZ and MOZ-TIF2 do not interact directly with the LBDs of the NRs tested. In contrast, MOZ-TIF2 was found to interact strongly with a GST fusion protein containing the minimal SID of CBP (54) (GST-SID) and with GST-CBP full-length protein, but not with GST alone (Fig. 1C). Although the GST-CBP full-length protein was expressed at very low levels in Escherichia coli extracts, we were able to detect specific binding of MOZ-TIF2 to this protein. Weak binding of MOZ to GST-SID was detected upon prolonged exposures, but in contrast to the case for MOZ-TIF2, this binding was disrupted under more stringent binding conditions (data not shown). Thus, MOZ-TIF2 interacts with CBP but not NRs in vitro.
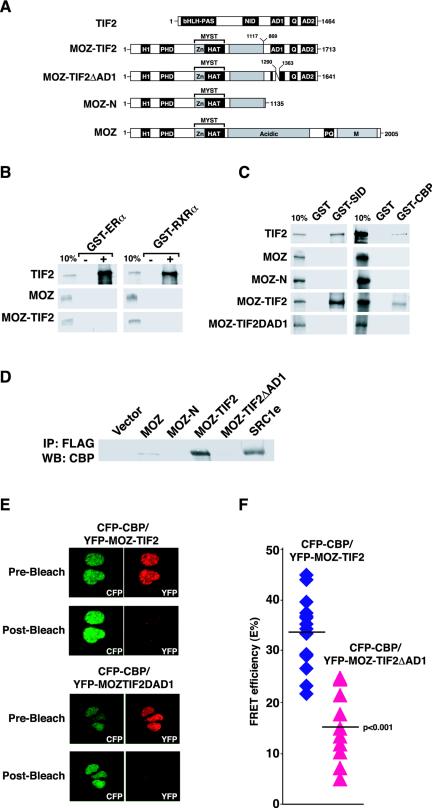
FIG. 1.
MOZ-TIF2 interacts with CBP via the AD1 domain in vitro and in vivo. (A) Schematic representations of domains in TIF2, MOZ-TIF2, MOZ, and derivatives. bHLH-PAS, basic helix-loop-helix/Per-ARNT-Sim domain; NID, nuclear receptor interaction domain; Q, glutamine-rich domain; H1, histone H1/5-like domain, PHD, plant homeodomain; Zn, C2HC zinc finger. Residue-rich domains, acidic, proline and glutamine, and methionine. Residue numbers 1117 and 869 in MOZ-TIF2 indicate the amino acid breakpoints in MOZ and TIF2, respectively. Numbers 1290 and 1363 in MOZ-TIF2ΔAD1 indicate amino acid boundaries of the deleted AD1 domain. (B) In vitro-translated TIF2, MOZ, and MOZ-TIF2were tested for interaction with GST proteins or GST fused to the LBDs ERα or RXRα in the presence (+) or absence (−) of ligand. One-tenth of the total input of labeled protein is shown for comparison (10%). (C) Interactions of TIF2, MOZ, MOZ-N, MOZ-TIF2, and MOZ-TIF2ΔAD1 with GST, GST-CBP-SID (residues 2058 to 2130), and full-length GST-CBP as in (B). (D) Coimmunoprecipitation data showing in vivo interactions between ectopically expressed CBP and FLAG fusion proteins in HEK293 cells. (E) FRET analyses between CFP-CBP and YFP-MOZ-TIF2 or YFP-MOZ-TIF2ΔAD1. Representative nuclei showing fluorescence images pre- and post-YFP bleach are shown. (F) Percent FRET efficiencies (E%) for nuclei (15 and 10 nuclei, respectively). The line indicates the average E% value for each group. The differences in E% between the two groups were analyzed by using a two-tailed type 2 t test, and the P value is indicated.
The interaction of the p160 proteins TIF2, SRC1, and ACTR with CBP is mediated by a short sequence, termed AD1, that contains leucine-rich amphipathic α-helices (17, 39, 54). In order to investigate whether AD1 is required for the CBP binding and other functions of MOZ-TIF2, the deletion mutants MOZ-N (amino acids 1 to 1135) and MOZ-TIF2ΔAD1 (deletion of amino acids 1290 to 1363) were generated (Fig. 1A). MOZ-N, representing the truncated MOZ sequence contained within MOZ-TIF2, did not bind CBP in vitro (Fig. 1C). The deletion construct MOZ-TIF2ΔAD1 also failed to interact with GST-CBP proteins under similar conditions (Fig. 1C). Thus, the AD1 sequence is essential for the interaction of MOZ-TIF2 and CBP proteins in vitro.
To investigate whether MOZ-TIF2 and CBP proteins can associate in vivo, coimmunoprecipitation experiments were performed. FLAG-tagged MOZ, MOZ-N, MOZ-TIF2, MOZ-TIF2ΔAD1, SRC1 (TIF2 homolog), or empty vector was cotransfected with CBP. Whole-cell extracts were examined for the interaction of FLAG-purified proteins with CBP. These experiments confirmed that both SRC1 and MOZ-TIF2 proteins associate strongly with CBP, whereas MOZ showed only weak binding (Fig. 1D). Moreover, deletion of the TIF2 moiety (MOZ-N) or the AD1 sequence (MOZ-TIF2ΔAD1) resulted in a failure to coimmunoprecipitate CBP (Fig. 1D), supporting the conclusion that the AD1 domain mediates the interaction of MOZ-TIF2 with CBP. In similar experiments, we confirmed that the HA-tagged GRIP1 protein (the mouse homolog of TIF2) coimmunoprecipitated with CBP (data not shown).
To assess whether the MOZ-TIF2 and CBP proteins interact directly in vivo, FRET experiments were carried out with CFP-CBP as the donor protein and YFP-MOZ-TIF2 or YFP-MOZ-TIF2ΔAD1 as the acceptor protein. The expression of full-length fusion proteins in transfected cells was confirmed by Western blotting (data not shown). FRET interactions were assessed on an LSM510 META microscope (Zeiss), using the donor-dequenching method (55) on fixed cells. The FRET was determined as an increase in donor-emitted fluorescence after photobleaching of the acceptor and was expressed as FRET efficiency (E%). As controls, cells expressing CFP-CBP alone were bleached with light at a wavelength of 514 nm to confirm the absence of nonspecific increases in CFP fluorescence due to the bleaching protocol (data not shown). Transfected cells coexpressing both CFP and YFP (as confirmed by their emission spectra) were selected, and YFP in regions of interest was photobleached (Fig. 1E, compare pre- and postbleach). Concomitantly, CFP fluorescence intensities were monitored pre- and postbleach. In cells expressing CFP-CBP and YFP-MOZ-TIF2, strong FRET efficiencies were observed (E% = 33.7% ± 7%), indicating a significant degree of physical association of the CBP and MOZ-TIF2 proteins in vivo (Fig. 1F). In contrast, in cells expressing CFP-CBP and YFP-MOZ-TIF2ΔAD1 (Fig. 1F), the FRET efficiencies detected were significantly reduced (E% = 15.2% ± 6.9%; P < 0.001). Taken together, these data confirm that MOZ-TIF2 interacts directly with CBP both in vivo and in vitro and that this interaction is facilitated by the AD1 sequence.
MOZ-TIF2 inhibits transcription mediated by nuclear receptors and p53.
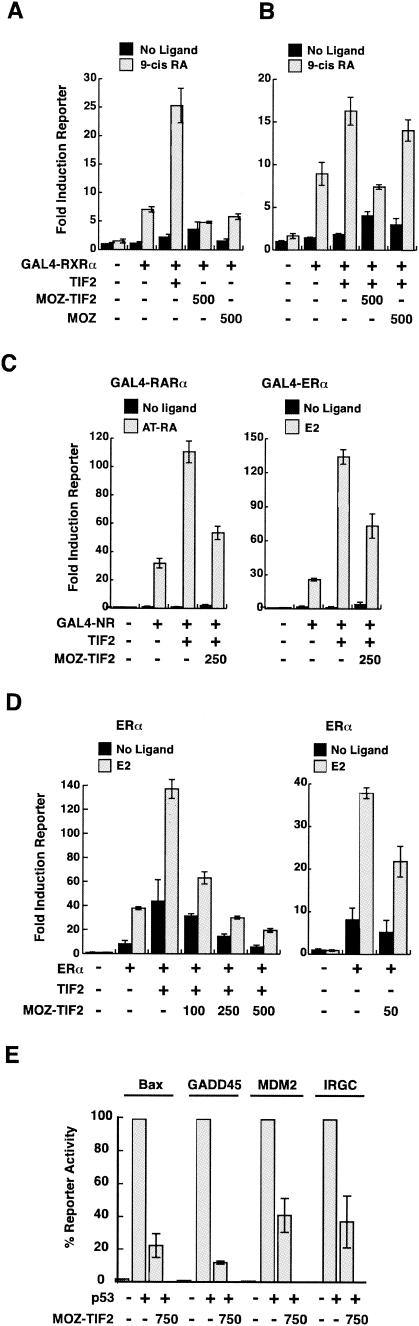
Having established that MOZ-TIF2 binds to CBP but not NRs, we assessed the impact of exogenous TIF2, MOZ, and MOZ-TIF2 proteins on ligand-dependent reporter activation by GAL4-NR-LBD proteins in COS-1 cells. Substantial ligand-dependent activation of the reporter was observed for GAL4-RXRα, GAL4-RARα, and GAL4-ERα in the absence of other exogenous factors (7-, 32-, and 26-fold, respectively) (Fig. 2A and C), as expected. Coexpression with the TIF2 coactivator further enhanced reporter activation by GAL4-RXRα, GAL4-RARα, and GAL4-ERα to 25-, 111-, and 134-fold above the basal level, respectively (Fig. 2A and C). In contrast, neither MOZ nor MOZ-TIF2 enhanced GAL4-NR-LBD activity, and moreover, we noted reduced ligand-dependent reporter activation by GAL4-RXRα in the presence of MOZ-TIF2 (Fig. 2A). This inhibitory effect of MOZ-TIF2, but not MOZ, on the activation of the reporter was even more striking when GAL4-NRs were coexpressed with TIF2 (Fig. 2B and C and data not shown). The GAL4-RARα reporter activation data from Fig. 2C were combined with data from five independent experiments (data not shown) for statistical analysis. Analysis of variance showed significant overall group differences (F6,101 = 83.211; P < 0.0001). Post hoc t tests with Bonferroni correction confirmed a highly significant difference between the groups GAL4-RARα + TIF2 and GAL4-RARα + TIF2 + MOZ-TIF2 (P < 0.0001). Consistency between experiments was confirmed by analysis of variance, which demonstrated no significant effect of experimental replication on reporter activity across the experimental groups tested.
FIG.2.
MOZ-TIF2 inhibits transcription activation by nuclear receptors and by p53. (A to D) GAL4-NR- and full-length-ERα-mediated luciferase assays from transiently transfected COS-1 cells. Cognate ligand (shaded bars) (9-cis-retinoic acid [9-cis RA], all-trans-retinoic acid [AT-RA], or 17β-estradiol [E2]) or an equal volume of vehicle (black bars) was added. Reporter activation is represented as fold induction over the control value (reporters in the absence of NRs, coactivators, or ligand), and results from representative experiments are shown. (E) p53-mediated luciferase assays from transiently transfected SaOs-2 cells with different p53-responsive reporter plasmids. The reporter activation is shown as a percentage relative to the activation of the reporter gene in the presence of p53, which is set to 100%. The quantity of DNA (nanograms) transfected is indicated, with + indicating 100 ng and − indicating no DNA. The data are presented as averages and standard errors of the means from triplicate samples.
To determine whether MOZ-TIF2 had a similar effect on full-length NRs, we assessed its effect on activation of the p(ERE)3-tk-Luc reporter by ERα coexpressed with TIF2. In agreement with the results with GAL4-fused NRs, MOZ-TIF2 inhibited activation of an estrogen-responsive reporter by ERα/TIF2 in a dose-dependent manner (Fig. 2D). ERα-mediated reporter activation was also reduced by MOZ-TIF2 in the absence of coexpressed TIF2 (Fig. 2D, right panel), suggesting that MOZ-TIF2 inhibits the activity of ERα mediated by endogenous coactivators. Consistent with this, MOZ-TIF2 inhibited reporter activation by GAL4-RARα/SRC1 (data not shown), indicating that the activities of different p160/NR complexes are inhibited by MOZ-TIF2. In conclusion, these results indicate that MOZ-TIF2 acts as a dominant inhibitor of ligand-dependent transcription by NRs.
To determine whether MOZ-TIF2 interferes with the activity of other CBP-dependent activators, we assayed the activity of the tumor suppressor protein p53 in the presence and absence of MOZ-TIF2. The p53-deficient cell line SaOS2 was transfected with a p53 expression vector in combination with reporters containing promoter sequences derived from a range of p53-responsive genes, including those for Bax, GADD45, and MDM2, or the synthetic IRGC element. Each of the four reporters displayed significant induction in the presence of p53, which was inhibited by coexpression with MOZ-TIF2 (Fig. 2E). In contrast, wild-type MOZ did not inhibit p53 activity on these promoters (data not shown). Thus, our results show that MOZ-TIF2 acts as a dominant inhibitor of the transcriptional activities of a number of different CBP-dependent activators in vivo.
The AD1 domain is essential for inhibition of NR and p53 activities by MOZ-TIF2.
To confirm that the addition of YFP and CFP tags did not alter the dominant inhibitory properties of MOZ-TIF2 used in FRET assays, we assessed the effects of YFP-MOZ-TIF2 and CFP-MOZ-TIF2 in reporter assays. As shown in Fig. 3A, YFP- and CFP-tagged MOZ-TIF2 proteins inhibited the ligand-dependent activity of GAL4-RARα/TIF2, whereas YFP-MOZ and CFP-MOZ did not. To test whether the AD1 domain is required for MOZ-TIF2 inhibition of NR activity, we compared the effects of MOZ-TIF2, MOZ-N, and MOZ-TIF2ΔAD1 on GAL4-RARα/TIF2 reporter activation (Fig. 3B). In contrast to MOZ-TIF2, MOZ-TIF2ΔAD1 did not inhibit GAL4-RARα/TIF2 reporter activation, indicating that interaction with CBP via the AD1 domain is essential for inhibition of NR activity. Similarly, MOZ-N had little effect on the ligand-dependent reporter activation by GAL4-RARα/TIF2, suggesting that the expression of a truncated MOZ N-terminal sequence is not sufficient to inhibit NR activity. Consistent with these results, expression of CBP in conjunction with MOZ-TIF2 almost fully rescued the GAL4-RARα/TIF2 reporter activation to levels observed in the absence of MOZ-TIF2 (Fig. 3C).
We next assessed the effects of MOZ-TIF2 and MOZ-TIF2ΔAD1 on reporter activation by endogenous NRs and coactivators in the hematopoietic cell line U937, which has a differentiation status similar to that of M5-type AML cells (3, 57). These cells were electroporated with luciferase reporter plasmids responsive to either retinoic acid receptors (RARE-Luc) or peroxisome proliferator-activated receptors (PPRE-Luc), and we confirmed that the reporter activities were induced by addition of cognate ligands to the culture medium (retinoic acid or rosiglitazone, respectively) (Fig. 3D). However, cotransfection of U937 cells with MOZ-TIF2 expression plasmid decreased the ligand-induced activation of RARE- and PPRE-luciferase reporter genes in a dose-dependent manner (Fig. 3D), indicating that MOZ-TIF2 inhibits the activity of endogenous PPAR and retinoid receptors in hematopoietic cells. In contrast, no decrease in the ligand-dependent reporter activity was observed in U937 cells expressing MOZ-TIF2ΔAD1 (Fig. 3D).
We also assessed whether CBP binding by MOZ-TIF2 was required for the inhibition of p53 activity. Our reporter assays in SaOS-2 cells revealed that the AD1 domain is also necessary for MOZ-TIF2 inhibition of p53 activity, as MOZ-TIF2ΔAD1 failed to reduce p53-mediated activation of IRGC-luc and GADD45-luc (Fig. 3E). As observed with NRs, exogenous CBP achieved a partial rescue of MOZ-TIF2 inhibition of p53 activity (Fig. 3F). Taken together, our results suggest that interaction of the AD1 domain of MOZ-TIF2 with CBP is necessary for its transcriptional inhibitory effects in vivo.
Distinct subnuclear localization of MOZ and MOZ-TIF2 proteins.
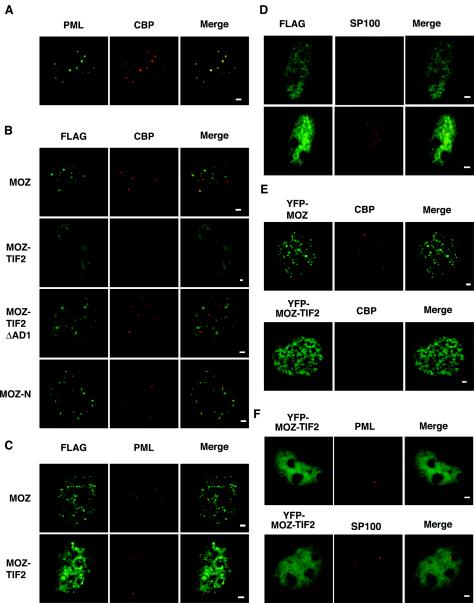
To investigate the subcellular localization of MOZ proteins in different cell lines, we carried out indirect immunofluorescence experiments with antibodies raised against MOZ (Autogen Bioclear). Endogenous MOZ proteins were detected in the nuclei of the adherent cell lines COS-1 (kidney), U-2Os (bone), and HEK293 (kidney) and the nonadherent leukemic cell lines HL60 and U937 (Fig. 4A). Similar staining patterns were detected in Calu-1 (lung), N-Tera (testis), HeLa (cervix), and KMH2 (B-lymphocyte) cells, indicating that MOZ is expressed in cell lines derived from a variety of different tissues (data not shown). In most cell types, MOZ was partially concentrated in subnuclear foci or speckles and was excluded from nucleoli (Fig. 4A). A similar subcellular distribution was observed for recombinant FLAG-tagged MOZ proteins expressed ectopically in COS-1 (Fig. 4B) and HEK293 (data not shown) cells as detected with anti-FLAG and anti-MOZ antibodies. Costaining for other nuclear proteins revealed that MOZ speckles were distinct from splicing speckles, Cajal bodies, paraspeckles (data not shown), and PML bodies (Fig. 5C). Thus, MOZ appears to occupy a distinct nuclear subdomain.
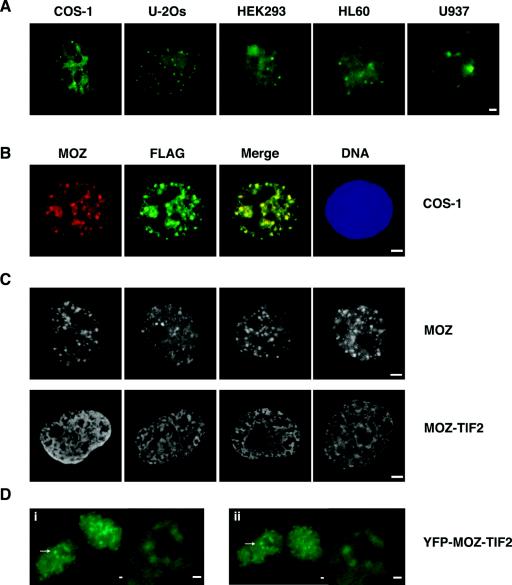
FIG. 4.
Subcellular localization of endogenous and exogenously expressed MOZ proteins. (A) Epifluorescence microscope images of endogenous MOZ proteins for COS-1, U-2Os, HEK293, HL60, and U937 cells. (B) FLAG-MOZ proteins detected with anti-FLAG (green) and anti-MOZ (red) antibodies in COS-1 cells. Yellow indicates colocalization. (C) Distinct nuclear localization of epitope-tagged MOZ (upper panels) and MOZ-TIF2 proteins (lower panels), showing punctate and mesh-like staining, respectively. (D) Representative time lapse images of YFP-MOZ-TIF2 proteins expressed in HEK293 cells. Images i and ii are still pictures of the same cells captured 25 min apart. The arrows indicate perinucleolar regions of interest highlighted at higher magnification in the insets. Bars, 1 μm.
FIG. 5.
Subcellular localization of MOZ, MOZ-TIF2, MOZ-N, and MOZ-TIF2ΔAD1 and their effect on the nuclear distribution of endogenous CBP and PML in COS-1 cells. (A) Staining of COS-1 cells for endogenous CBP and PML. (B) Detection of the indicated FLAG-MOZ fusion proteins and endogenous CBP. (C) Costaining of FLAG-tagged MOZ proteins with endogenous PML. (D) Costaining of FLAG-MOZ-TIF2 for endogenous SP100. (E) Detection of YFP-MOZ and YFP-MOZ-TIF2 proteins with staining for endogenous CBP. (F) Staining of cells expressing YFP-MOZ-TIF2 for endogenous PML. Red and green channels are shown in the Merge panels; yellow indicates colocalization. Bar, 1 μm.
In contrast to FLAG-MOZ proteins, FLAG-MOZ-TIF2 displayed a mesh-like nuclear distribution that was distinct from that of MOZ in approximately 80% of transiently transfected COS-1 (Fig. 4C) and HEK293, U-2Os, and U937 (data not shown) cells. We frequently observed an increased concentration of FLAG-MOZ-TIF2 at the peripheries of nucleolar centers, and occasionally speckles could be clearly observed in these regions (Fig. 4C). These speckles may represent nucleolar caps, which are structures formed at the nucleolar periphery in the vicinity of perinucleolar heterochromatin (A. Lamond, personal communication). This localization was also apparent in time-lapse imaging of live HEK293 cells expressing YFP-MOZ-TIF2 (Fig. 4D and data not shown). However, the majority of cells show clearly distinct subnuclear distribution profiles for MOZ and MOZ-TIF2.
MOZ-TIF2 depletes CBP from PML bodies.
CBP and PML proteins colocalize to subnuclear structures termed PML bodies or nuclear bodies (see reference 66 for a review). We confirmed that endogenous CBP and PML proteins colocalized to PML bodies in COS-1 cells (Fig. 5A). Although MOZ proteins were also detected in nuclear speckles, these were clearly distinct from PML bodies (Fig. 5B and C, merged images). The endogenous CBP distribution in COS-1 cells expressing exogenous MOZ, MOZ-N, or MOZ-TIF2ΔAD1 proteins was indistinguishable from that in mock-transfected controls, in that endogenous CBP staining was diffuse nuclear and concentrated in foci, which are likely to represent PML bodies (Fig. 5B). The association of CBP foci with PML bodies was confirmed by costaining with anti-PML antibodies (data not shown). However, we noted that in a large proportion of cells expressing MOZ-TIF2, we were unable to detect endogenous CBP in PML bodies (Fig. 5B), despite confirming that PML bodies were intact by using antibodies raised against PML (Fig. 5C) or SP100 (Fig. 5D). Similarly, COS-1 cells expressing YFP-MOZ-TIF2 showed a reduced staining of CBP in PML bodies (Fig. 5E and 6B) compared to cells expressing YFP-MOZ. We also confirmed that PML bodies were intact in these cells by costaining for PML and SP100, respectively (Fig. 5F). This result suggests that expression of MOZ-TIF2 alters the subcellular localization of a major proportion of CBP proteins in these cells, while leaving PML bodies undisturbed. In contrast, endogenous CBP showed typical subcellular distributions (i.e., association with PML bodies) in cells expressing MOZ-TIF2ΔAD1 (Fig. 5B), YFP-MOZ-TIF2ΔAD1 (Fig. 6B), YFP-MOZ (Fig. 5E), or TIF2/GRIP1 (data not shown). Similarly, others have reported that ectopic expression of TIF2/GRIP1 did not disrupt the association of endogenous CBP with PML bodies, although CBP was observed to be recruited into additional non-PML speckles (5). Our results indicate that the integrity of the AD1 domain in MOZ-TIF2, and the CBP interaction it mediates, is necessary for CBP depletion from PML bodies in addition to the inhibition of NR transcriptional activity.
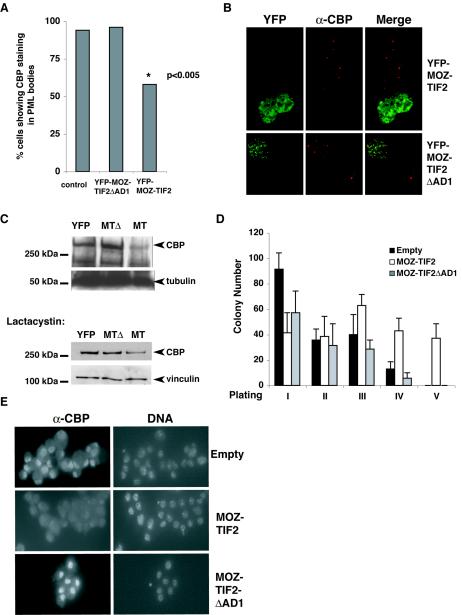
FIG. 6.
Effect of MOZ-TIF2 on cellular CBP protein levels and on the proliferation of hematopoietic stem cells in vitro. (A) Quantitative analysis of CBP-containing speckles (PML bodies) in control cells (n = 300) and YFP-MOZ-TIF2- and YFP-MOZ-TIF2ΔAD1-expressing cells (n = 50). (B) Representative images of COS-1 cells, showing the effect of YFP-MOZ-TIF2 and YFP-MOZ-TIF2ΔAD1 expression on detection of endogenous CBP speckles. (C) Western blot analysis of whole-cell extracts from YFP+ sorted HEK293 cells expressing YFP, YFP-MOZ-TIF2,and YFP-MOZ-TIF2ΔAD1 cultured in the absence (upper panels) or presence (lower panels) of lactacystin. The band corresponding to full-length CBP is indicated. The loading controls α-tubulin and vinculin are also indicated. (D) Serial replating assays to assess the proliferative potential of GFP+ Lin− cells transduced with the indicated vectors in methylcellulose medium containing SCF, IL-3, and Il-6. Error bars indicate standard errors of the means. (E) Anti-CBP staining of sorted GFP+ Lin− cells transduced with the indicated retroviral vectors. Images were taken at identical exposure times. DNA was counterstained with 0.1 μg of DAPI per ml.
MOZ-TIF2 reduces cellular levels of CBP.
To assess the effect of MOZ-TIF2 on CBP localization to PML bodies in a quantitative manner, we employed YFP-MOZ-TIF2 or YFP-MOZ-TIF2ΔAD1 DNA in immunofluorescence studies. COS-1 cells expressing YFP fusion proteins were stained for endogenous CBP (Fig. 6B) or PML and scored for the presence of CBP speckles resembling PML bodies. CBP speckles showing a pattern typical for PML bodies were detected in 94% of 300 randomly chosen nontransfected control cells and in 96% of 50 transfected cells expressing YFP-MOZ-TIF2ΔAD1 (Fig. 6A). However, in cells expressing YFP-MOZ-TIF2 (n = 50), the number of nuclei displaying CBP speckles was significantly reduced, to 58% of the cells counted (Fig. 6A) [chi-square analysis, χ2 (2) = 13.12; P < 0.005]. In contrast, intact PML bodies were detected in cells expressing YFP-MOZ-TIF2ΔAD1 or YFP-MOZ-TIF2 and in control cells by using an anti-PML antibody (data not shown), as was also found for the FLAG-tagged MOZ and MOZ-TIF2 (Fig. 5C). Thus, our results indicate that MOZ-TIF2 depletes CBP from PML bodies in vivo.
In addition to the loss of CBP staining at PML bodies, we observed that COS-1 cells expressing MOZ-TIF2 showed an overall reduction in the level of endogenous CBP, as indicated by the intensity of staining with anti-CBP antibodies (Fig. 5B). This was also observed in COS-1 and HEK293 cells expressing YFP-MOZ-TIF2 (Fig. 5E and 6B and data not shown). To confirm that endogenous CBP levels were reduced in the presence of MOZ-TIF2, transiently transfected HEK293 cells expressing YFP, YFP-MOZ-TIF2, or YFP-MOZ-TIF2ΔAD1 were sorted by FACS for YFP-positive (YFP+) cells. After equalization of cell numbers, whole-cell extracts were subjected to Western blotting with CBP antibodies. As shown in Fig. 6C, the level of CBP in YFP+-sorted cells expressing MOZ-TIF2 was reduced by approximately twofold compared to that in control YFP cells, whereas CBP levels were increased in YFP+-sorted cells expressing MOZ-TIF2ΔAD1. Equal loading of the samples was confirmed by using antitubulin antibodies (upper panel). To explore the molecular mechanism of the observed CBP degradation, transiently transfected HEK293 cells were incubated in the presence of the proteasome-specific inhibitor lactacystin, and Western blot analysis was performed as described above with CBP antibodies. Vinculin antibodies were used to assess equal loading. As shown in the lower panel of Fig. 6C, the level of CBP in cells expressing MOZ-TIF2 was still reduced compared to that in control YFP cells, despite the presence of the proteasome inhibitor lactacystin, thereby indicating that the mechanism involved is proteasome independent. Therefore, we have conclusively shown that expression of MOZ-TIF2 leads to CBP depletion, which is not dependent on the proteasome.
CBP depletion and transformation of myeloid progenitors by MOZ-TIF2 are dependent on the AD1 domain.
To test the effects of MOZ-TIF2 on proliferation and CBP expression in hematopoietic progenitor cells, lineage-negative (Lin−) cells were purified from the bone marrow of 8- to 10-week-old 129SvEv mice. To facilitate introduction of MOZ-TIF2 cDNAs by retroviral transduction, MOZ-TIF2 and MOZ-TIF2ΔAD1 cDNAs were subcloned into the pMIE (MSCV-IRES-EGFP) vector, which contains an internal ribosome entry sequence driving translation of the enhanced GFP (19). Transfection of COS-1 cells with these vectors confirmed that MOZ-TIF2 and MOZ-TIF2ΔAD1 proteins expressed from pMIE displayed typical nuclear staining phenotypes (data not shown). In addition, reporter assays confirmed that pMIE-MOZ-TIF2, but not pMIE-MOZ-TIF2ΔAD1, downregulated NR transcriptional activity (data not shown). Ecotropic Phoenix cells were transfected with pMIE constructs to produce high titers of retroviruses, which were used to infect Lin− cells as previously described (41). After infection, GFP-positive (GFP+) cells were separated by FACS and tested for colony formation in methylcellulose medium containing appropriate cytokines by serial replating assays (35, 41). Our results indicate that only GFP+ Lin− cells expressing MOZ-TIF2 gave rise to colonies after multiple rounds of replating, in contrast to empty vector or MOZ-TIF2ΔAD1 (Fig. 6D). This clearly demonstrates an increase in the proliferative potential of hematopoietic progenitor cells mediated by MOZ-TIF2, which is dependent on the AD1 domain. Our results are in agreement with those of Deguchi et al. (16), showing that MOZ-TIF2 but not a derivative containing a mutation in the CBP-binding domain can transform murine Lin− cells and induce the development of AML in mice.
To determine the effect of MOZ-TIF2 proteins on endogenous CBP levels in Lin− cells, transduced FACS-sorted GFP+ cells were fixed and subjected to immunofluorescence staining with anti-CBP antibodies. As shown in Fig. 6E, CBP staining in the nuclei of Lin− cells expressing MOZ-TIF2 was reduced in comparison to that in control or Lin− cells expressing MOZ-TIF2ΔAD1, as previously observed in COS-1 and HEK293 cells. Interestingly, we note that the intensity of CBP staining is increased in cells expressing MOZ-TIF2ΔAD1, although the molecular mechanism for this is unclear. Our results demonstrate that MOZ-TIF2 reduces the levels of endogenous CBP and that this decrease is dependent on interaction with CBP via the AD1 domain. Thus, we have demonstrated that the biological activity of MOZ-TIF2, including transcriptional inhibition and transformation of Lin− progenitor cells, is dependent on an interaction with endogenous CBP, leading to depletion of this global coactivator in myeloid progenitors.
DISCUSSION
The acquisition of MOZ-TIF2 and MOZ-CBP reciprocal translocations in hematopoietic stem cells or myeloid progenitors leads to myelodysplastic disease and AML (1, 8, 9, 42). MOZ-CBP and MOZ-TIF2 translocations are likely to affect cell function in a number of ways, including (i) a reduction of the normal cellular complement of parental proteins and (ii) aberrant function of resultant fusion proteins, including both dominant negative and gain-of-function effects. Due to the function of TIF2, CBP, and possibly MOZ in transcriptional regulation and chromatin modification, it is likely that MOZ-CBP and MOZ-TIF2 fusion proteins affect gene expression networks important in cell differentiation and proliferation. Our results provide novel insight into the molecular mechanisms of MOZ-TIF2 action and indicate that CBP function is compromised in cells expressing this protein.
MOZ-TIF2 and MOZ-CBP translocations are associated with similar AML phenotypes, and it has been suggested that the MOZ-TIF2 fusion protein may mimic MOZ-CBP through interaction with CBP (9, 26). Our results confirm that MOZ-TIF2 interacts with CBP directly both in vitro and in vivo, as demonstrated by GST pull-down (Fig. 1C), coimmunoprecipitation (Fig. 1D), and FRET (Fig. 1E and F) experiments, and they establish that this interaction is dependent on the AD1 sequence. In contrast, MOZ-TIF2 showed no direct binding to NRs in our assays, as it lacks the TIF2 NR interaction domain (Fig. 1B). Moreover, in contrast to the case for TIP60, which interacts with NR LBDs via a single LXXLL motif and stimulates NR-mediated transcription (20), we did not detect any interaction of MOZ with the LBDs of steroid or retinoid receptors, in the presence or absence of ligand (Fig. 1B). Consistent with this, coexpression of MOZ did not stimulate the transcriptional activities of NRs in our experiments (Fig. 2A and B and 3A and B and data not shown); thus, wild-type MOZ protein does not appear to directly regulate NR function. However, it has been reported that MORF and CBP are components of a PPARα-containing complex (58), whereas MOZ, MORF, and CBP have been detected in AML1-containing complexes (32). Thus, it remains possible that MOZ may contribute to the regulation of certain NR target genes by indirect recruitment via other activators.
We have demonstrated here that as a direct consequence of the MOZ-TIF2/CBP interaction, the transcriptional activities of a number of CBP-dependent activators are compromised (Fig. 2), including that of NRs, which are important for normal hematopoiesis (34, 59). Our results show clearly that ligand-dependent transcription by ectopically expressed RARα, RXRα, and ERα is inhibited by the MOZ-TIF2 protein (Fig. 2 and 3). In addition, the activity of endogenous retinoid receptors and PPARs in U937 cells is inhibited by expression of MOZ-TIF2 (Fig. 3). Retinoic acid, vitamin D, and PPARγ ligands have been reported to inhibit cell growth and induce lineage-specific differentiation of myeloid progenitors, leukemic blasts, and hematopoietic cell lines such as U937 and HL-60 (14, 27, 45, 50, 59). In acute promyelocytic leukemia (APL) (the M3 subtype of AML), retinoid differentiation therapy is an important treatment for patients expressing the PML-RARα fusion protein, and mutations in the RARα gene are also associated with non-APL forms of myeloid leukemia (49). Thus, nuclear receptor target genes are important for normal hematopoiesis and represent one set of genes whose expression may be compromised in MOZ-TIF2-associated AML.
It has been shown that p53 requires CBP as a transcriptional coactivator (37, 53) and that this involves a specific interaction of p53 with CBP SID (38, 39). Here we have shown that the transcriptional activity of p53 is reduced in cells expressing MOZ-TIF2 protein, in a CBP-binding-dependent manner (Fig. 2 and 3). Several other leukemic fusion proteins have been shown to disrupt p53 function. MLL-ELL inhibits p53 function by disruption of its interaction with CBP/p300, leading to a reduction in p53 acetylation in vivo (63). The PML-RAR fusion protein also causes deacetylation and degradation of p53, which results in the downregulation of p53 transcriptional activity and protection from p53-dependent responses (25). Thus, while p53 gene mutations are less frequent in hematological malignancies than in solid tumors (61), it is possible that p53 function may be disrupted in AML, through loss of CBP. Similarly, MOZ-CBP expressed in SaOS-2 cells was found to inhibit the transcriptional activity of AML1, another key regulator of hematopoiesis (32). Given the role of CBP as a global coactivator, it is likely that the functions of many other activators expressed in myeloid progenitor cells will also be compromised.
To examine other molecular consequences of MOZ-TIF2 expression in cells, we compared the subcellular localizations of MOZ and MOZ-TIF2 proteins. Analysis of endogenous or overexpressed MOZ proteins in a variety of cell lines by indirect immunofluorescence or YFP tagging indicated that MOZ is a predominantly nuclear protein concentrated in subnuclear foci distinct from PML bodies (Fig. 4 and 5B, C, and E). In contrast, MOZ-TIF2 displayed a distinct mesh-like nuclear stain (Fig. 4C and D, 5B to F, and 6B). Both MOZ and MOZ-TIF2 proteins showed staining at structures which may represent nucleolar caps (Fig. 4 C and D, which are formed at the nucleolar periphery in the vicinity of perinucleolar heterochromatin (Lamond, personal communication). A high proportion of cells expressing MOZ-TIF2 (but not MOZ) displayed mislocalization and depletion of endogenous CBP from its normal association with PML bodies (Fig. 5B and E and 6B). This effect was dependent on the CBP interaction sequence AD1, as MOZ-N and MOZ-TIF2ΔAD1 staining patterns resembled that of wild-type MOZ (Fig. 5B). However, MOZ-TIF2 did not appear to disrupt the integrity of PML bodies (Fig. 5C, D, and F). Although the consequence of depletion of CBP from PML bodies is not known, it has been reported that PML binds CBP and enhances NR transcriptional activity (18, 66). Interestingly, mislocalization of CBP is a feature of both Huntington's and Kennedy's diseases, due to sequestration of CBP by polyglutamine expansions (40, 43). Similarly, the NUP98-HoxA9 translocation in AML has been proposed to result in sequestration of CBP via binding to the NUP98 FG repeats (30).
Our experiments also demonstrate that endogenous CBP protein levels were decreased in MOZ-TIF2-expressing cells and that this depletion is dependent on the AD1 domain and independent of the proteasome (Fig. 6B and C). Similarly, reduced CBP staining was observed in murine bone marrow Lin− cells transduced with retroviruses expressing MOZ-TIF2 (Fig. 6E), but not MOZ-TIF2ΔAD1 or vector controls, indicating again that this decrease was dependent on the AD1 domain. Loss of CBP and p300 expression in human disease and animal models provides evidence that these proteins may act as tumor suppressors. Rubenstein-Taybi syndrome results from a loss of one allele of CBP, and, in addition to severe developmental defects, patients show an increased incidence of tumors. Mice heterozygous for the CBP gene exhibit hematopoietic diseases (33) and reduced self-renewal of hematopoietic stem cells (51), whereas null embryos present with defective hematopoiesis (44, 65). In addition, it was recently reported that loss of CBP in thymocytes accelerates the development of T-cell lymphoma in mice defective for p27Kip (29). CBP and p300 are cellular targets of transforming viruses via the action of viral proteins such as adenoviral protein E1A, human T-cell leukemia virus Tax protein and simian virus 40 T antigen, which compete with cellular factors for binding to the CBP SID domain, the docking site for TIF2 and other p160s (38, 39, 52). Thus, CBP may be a common target for somatically induced leukemogenesis and cell transformation by viral proteins.
Experiments with mice have confirmed that MOZ-TIF2 causes AML in a bone marrow transplant assay (16). The induction of AML was observed to be dependent on the integrity of two functional domains, namely, the CBP-binding domain and a C2HC zinc finger in the MYST domain that has been proposed to act as a nucleosome-binding motif in the Drosophila Mof protein (2). It has been reported that replacement of cysteine at position 543 in this zinc finger or of glycine 657 in the HAT domain leads to inhibition of MOZ-TIF2 HAT activity, indicating that HAT activity may be another important feature of MOZ-TIF2 function in AML (16). In this study we have investigated the molecular mechanisms underlying the requirement for the CBP-binding (AD1) sequence and have established that MOZ-TIF2 interaction with CBP leads to mislocalization of CBP from PML bodies and depletion of cellular levels of CBP via a proteasome-independent mechanism. This results in a concomitant reduction in transcriptional activity of CBP-dependent activators such as NRs and p53, which are important in cell differentiation pathways. MOZ-TIF2 displays an abnormal subnuclear distribution, distinct from that of either parental protein, which appears to be dependent on the AD1 CBP-binding sequence. We have found a strong correlation between the CBP binding and the cell transformation and transcriptional inhibitory properties of MOZ-TIF2. Therefore, we conclude that the MOZ-TIF2 translocation leads to impairment of CBP, a key player in chromatin modification and transcriptional regulation. Further experiments to address the importance of the MYST domain for MOZ-TIF2 action are ongoing.
Our results, coupled with the studies investigating effects on AML1 transcriptional activity (32), indicate that translocations involving MOZ-TIF2 and MOZ-CBP disrupt the action of transcription factors such as NRs, p53, and Runx proteins, which regulate myeloid cell differentiation. This report provides novel insights into molecular mechanisms of MOZ-TIF2 action and indicates that CBP function is compromised in cells expressing this protein. Future studies will focus on transcriptional profiling of Lin− hematopoietic precursors expressing MOZ fusion proteins.
Acknowledgments
We thank S. Ali, C. Bevan, M. Carapeti, K. Chatterjee, P. Chambon, N. Cross, C. J. Di Como, A. Dejean, D. G. Gilliland, R. Goodman, A. Harel-Bellan, T. G. Hofmann, X. Liu, J. Norman, M. Parker, M. Stallcup, and A. Zelent for gifts of plasmids or antibodies. We thank E. Louis, K. Straatman, and N. Royle for access to microscope facilities, C. Guerin for assistance with the FRET experiments, R. Snowdon and G. Cohen for cell sorting, and P. Moran for statistical analyses. We thank A. Lamond for useful discussions. We are also grateful to A. Tilley, C. Wright, and P. Simpson (Improvision) for advice on cell imaging and microscopy.
This work was funded by grants from the Wellcome Trust and Cancer Research UK. P.J.F.T. was supported by a Ph.D. stipend from the Medical Research Council. E.K. was supported by a fellowship from the Royal Netherlands Academy of Science. P.S. and C.B. were supported by the Medical Research Council.
REFERENCES
- 1.Aguiar, R. C., A. Chase, S. Coulthard, D. H. Macdonald, M. Carapeti, A. Reiter, J. Sohal, A. Lennard, J. M. Goldman, and N. C. Cross. 1997. Abnormalities of chromosome band 8p11 in leukemia: two clinical syndromes can be distinguished on the basis of MOZ involvement. Blood 90:3130-3135. [PubMed] [Google Scholar]
- 2.Akhtar, A., and P. B. Becker. 2001. The histone H4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition. EMBO Rep. 2:113-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alcalay, M., N. Meani, V. Gelmetti, A. Fantozzi, M. Fagioli, A. Orleth, D. Riganelli, C. Sebastiani, E. Cappelli, C. Casciari, M. T. Sciurpi, A. R. Mariano, S. P. Minardi, L. Luzi, H. Muller, P. P. Di Fiore, G. Frosina, and P. G. Pelicci. 2003. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J. Clin. Investig. 112:1751-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. [DOI] [PubMed] [Google Scholar]
- 5.Baumann, C. T., H. Ma, R. Wolford, J. C. Reyes, P. Maruvada, C. Lim, P. M. Yen, M. R. Stallcup, and G. L. Hager. 2001. The glucocorticoid receptor interacting protein 1 (GRIP1) localizes in discrete nuclear foci that associate with ND10 bodies and are enriched in components of the 26S proteasome. Mol. Endocrinol. 15:485-500. [DOI] [PubMed] [Google Scholar]
- 6.Billio, A., E. J. Steer, G. Pianezze, M. Svaldi, M. Casin, B. Amato, P. Coser, and N. C. Cross. 2002. A further case of acute myeloid leukaemia with inv(8)(p11q13) and MOZ-TIF2 fusion. Haematologica 87:ECR15. [PubMed] [Google Scholar]
- 7.Blobel, G. A. 2000. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood 95:745-755. [PubMed] [Google Scholar]
- 8.Borrow, J., V. P. Stanton, Jr., J. M. Andresen, R. Becher, F. G. Behm, R. S. Chaganti, C. I. Civin, C. Disteche, I. Dube, A. M. Frischauf, D. Horsman, F. Mitelman, S. Volinia, A. E. Watmore, and D. E. Housman. 1996. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat. Genet. 14:33-41. [DOI] [PubMed] [Google Scholar]
- 9.Carapeti, M., R. C. Aguiar, J. M. Goldman, and N. C. Cross. 1998. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood 91:3127-3133. [PubMed] [Google Scholar]
- 10.Chaffanet, M., L. Gressin, C. Preudhomme, V. Soenen-Cornu, D. Birnbaum, and M. J. Pebusque. 2000. MOZ is fused to p300 in an acute monocytic leukemia with t(8;22). Genes Chromosomes Cancer 28:138-144. [DOI] [PubMed] [Google Scholar]
- 11.Champagne, N., N. R. Bertos, N. Pelletier, A. H. Wang, M. Vezmar, Y. Yang, H. H. Heng, and X. J. Yang. 1999. Identification of a human histone acetyltransferase related to monocytic leukemia zinc finger protein. J. Biol. Chem. 274:28528-28536. [DOI] [PubMed] [Google Scholar]
- 12.Champagne, N., N. Pelletier, and X. J. Yang. 2001. The monocytic leukemia zinc finger protein MOZ is a histone acetyltransferase. Oncogene 20:404-409. [DOI] [PubMed] [Google Scholar]
- 13.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 14.Collins, S. J., K. A. Robertson, and L. Mueller. 1990. Retinoic acid-induced granulocytic differentiation of Hl-60 myeloid-leukemia cells is mediated directly through the retinoic acid receptor (RAR-alpha). Mol. Cell. Biol. 10:2154-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coulthard, S., A. Chase, K. Orchard, A. Watmore, A. Vora, J. A. M. Goldman, and D. M. Swirsky. 1998. Two cases of inv(8)(p11q13) in AML with erythrophagocytosis: a new cytogenetic variant. Br. J. Haematol. 124:561-563. [DOI] [PubMed] [Google Scholar]
- 16.Deguchi, K., P. M. Ayton, M. Carapeti, J. L. Kutok, C. S. Snyder, I. R. Williams, N. C. Cross, C. K. Glass, M. L. Cleary, and D. G. Gilliland. 2003. MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell 3:259-271. [DOI] [PubMed] [Google Scholar]
- 17.Demarest, S. J., M. Martinez-Yamout, J. Chung, H. Chen, W. Xu, H. J. Dyson, R. M. Evans, and P. E. Wright. 2002. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 415:549-553. [DOI] [PubMed] [Google Scholar]
- 18.Doucas, V., M. Tini, D. A. Egan, and R. M. Evans. 1999. Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling. Proc. Natl. Acad. Sci. USA 96:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du, C., R. L. Redner, M. P. Cooke, and C. Lavau. 1999. Overexpression of wild-type retinoic acid receptor alpha (RARalpha) recapitulates retinoic acid-sensitive transformation of primary myeloid progenitors by acute promyelocytic leukemia RARalpha-fusion genes. Blood 94:793-802. [PubMed] [Google Scholar]
- 20.Gaughan, L., M. E. Brady, S. Cook, D. E. Neal, and C. N. Robson. 2001. Tip60 is a co-activator specific for class I nuclear hormone receptors. J. Biol. Chem. 276:46841-46848. [DOI] [PubMed] [Google Scholar]
- 21.Giordano, A., and M. L. Avantaggiati. 1999. p300 and CBP: partners for life and death. J. Cell. Physiol. 181:218-230. [DOI] [PubMed] [Google Scholar]
- 22.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 23.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 24.Hong, H., K. Kohli, A. Trivedi, D. L. Johnson, and M. R. Stallcup. 1996. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl. Acad. Sci. USA 93:948-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Insinga, A., S. Monestiroli, S. Ronzoni, R. Carbone, M. Pearson, G. Pruneri, G. Viale, E. Appella, P. Pelicci, and S. Minucci. 2004. Impairment of p53 acetylation, stability and function by an oncogenic transcription factor. EMBO J. 23:1144-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson, S., and L. Pillus. 1999. Modifying chromatin and concepts of cancer. Curr. Opin. Genet. Dev. 9:175-184. [DOI] [PubMed] [Google Scholar]
- 27.James, S. Y., M. A. Williams, A. C. Newland, and K. W. Colston. 1999. Leukemia cell differentiation: cellular and molecular interactions of retinoids and vitamin D. Gen. Pharmacol. 32:43-154. [DOI] [PubMed] [Google Scholar]
- 28.Kalkhoven, E., J. E. Valentine, D. M. Heery, and M. G. Parker. 1998. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 17:232-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang-Decker, N., C. Tong, F. Boussouar, D. J. Baker, W. Xu, A. A. Leontovich, W. R. Taylor, P. K. Brindle, and J. M. van Deursen. 2004. Loss of CBP causes T cell lymphomagenesis in synergy with p27Kip1 insufficiency. Cancer Cell. 5:177-189. [DOI] [PubMed] [Google Scholar]
- 30.Kasper, L. H., P. K. Brindle, C. A. Schnabel, C. E. Pritchard, M. L. Cleary, and J. M. van Deursen. 1999. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol. Cell. Biol. 19:764-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, M. Y., S. J. Hsiao, and W. L. Kraus. 2001. A role for coactivators and histone acetylation in estrogen receptor alpha-mediated transcription initiation. EMBO J. 20:6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitabayashi, I., Y. Aikawa, L. A. Nguyen, A. Yokoyama, and M. Ohki. 2001. Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein. EMBO J. 20:7184-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kung, A. L., V. I. Rebel, R. T. Bronson, L. E. Ch'ng, C. A. Sieff, D. M. Livingston, and T. P. Yao. 2000. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 14:272-277. [PMC free article] [PubMed] [Google Scholar]
- 34.Kuwata, T., I. M. Wang, T. Tamura, R. M. Ponnamperuma, R. Levine, K. L. Holmes, H. C. Morse, L. M. De Luca, and K. Ozato. 2000. Vitamin A deficiency in mice causes a systemic expansion of myeloid cells. Blood 95:3349-3356. [PubMed] [Google Scholar]
- 35.Lavau, C., J. M. Heard, O. Danos, and A. Dejean. 1996. Retroviral vectors for the transduction of the PML-RARalpha fusion product of acute promyelocytic leukemia. Exp. Hematol. 24:544-551. [PubMed] [Google Scholar]
- 36.Liang, J., L. Prouty, B. J. Williams, M. A. Dayton, and K. L. Blanchard. 1998. Acute mixed lineage leukemia with an inv(8)(p11q13) resulting in fusion of the genes for MOZ and TIF2. Blood 92:2118-2122. [PubMed] [Google Scholar]
- 37.Lill, N. L., S. R. Grossman, D. Ginsberg, J. DeCaprio, and D. M. Livingston. 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature 387:823-827. [DOI] [PubMed] [Google Scholar]
- 38.Livengood, J. A., K. E. S. Scoggin., K. Van Orden, S. J. McBryant, R. Edayathumangalam, P. J. Laybourn, and J. K. Nyborg. 2002. p53 Transcriptional activity is mediated through the SRC1-interacting domain of CBP/p300. J. Biol. Chem. 277:9054-9061. [DOI] [PubMed] [Google Scholar]
- 39.Matsuda, S., J. C. Harries, M. Viskaduraki, P. J. F. Troke, K. B. Kindle, C. M. Ryan, and D. M. Heery. 2004. A conserved alpha-helical motif mediates the binding of diverse nuclear proteins to the SRC1 interaction domain of CBP. J. Biol. Chem. 279:14055-14064. [DOI] [PubMed] [Google Scholar]
- 40.McCampbell, A., J. P. Taylor, A. A. Taye, J. Robitschek, M. Li, J. Walcott, D. Merry, Y. H. Chai, H. Paulson, G. Sobue, and K. H. Fischbeck. 2000. CREB-binding protein sequestration by expanded polyglutamine. Hum. Mol. Genet. 9:2197-2202. [DOI] [PubMed] [Google Scholar]
- 41.Minucci, S., S. Monestiroli, S. Giavara, S. Ronzoni, F. Marchesi, A. Insinga, D. Diverio, P. Gasparini, M. Capillo, E. Colombo, C. Matteucci, F. Contegno, F. Lo-Coco, E. Scanziani, A. Gobbi, and P. G. Pelicci. 2002. PML-RAR induces promyelocytic leukemias with high efficiency following retroviral gene transfer into purified murine hematopoietic progenitors. Blood 100:2989-2995. [DOI] [PubMed] [Google Scholar]
- 42.Murati, A., J. Adelaide, C. Popovici, M. J. Mozziconacci, C. Arnoulet, M. Lafage-Pochitaloff, D. Sainty, D. Birnbaum, and M. Chaffanet. 2003. A further case of acute myelomonocytic leukemia with inv(8) chromosomal rearrangement and MOZ-NCOA2 gene fusion. Int. J. Mol. Med. 12:423-428. [PubMed] [Google Scholar]
- 43.Nucifora, F. C., M. Sasaki, M. F. Peters, H. Huang, J. K. Cooper, M. Yamada, H. Takahashi, S. Tsuji, J. Troncoso, V. L. Dawson, T. M. Dawson, and C. A. Ross. 2001. Interference by huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science 291:2423-2428. [DOI] [PubMed] [Google Scholar]
- 44.Oike, Y., N. Takakura, A. Hata, T. Kaname, M. Akizuki, Y. Yamaguchi, H. Yasue, K. Araki, K. Yamamura, and T. Suda. 1999. Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood 93:2771-2779. [PubMed] [Google Scholar]
- 45.Olsson, I., U. Gullberg, I. Ivhed, and K. Nilsson. 1983. Induction of differentiation of the human histiocytic lymphoma cell-line U-937 by 1-alpha,25-dihydroxycholecalciferol. Cancer Res. 43:5862-5867. [PubMed] [Google Scholar]
- 46.Onate, S. A., S. Y. Tsai, M. J. Tsai and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 47.Panagopoulos, I., M. R. Teixeira, F. Micci, J. Hammerstrom, M. Isaksson, B. Johansson, F. Mitelman, and S. Heim. 2000. Acute myeloid leukemia with inv(8)(p11q13). Leuk. Lymphoma 39:651-656. [DOI] [PubMed] [Google Scholar]
- 48.Panagopoulos, I., T. Fioretos, M. Isaksson, U. Samuelsson, R. Billstrom, B. Strombeck, F. Mitelman, and B. Johansson. 2001. Fusion of the MORF and CBP genes in acute myeloid leukemia with the t(10;16)(q22;p13). Hum. Mol. Genet. 10:395-404. [DOI] [PubMed] [Google Scholar]
- 49.Parrado, A., C. Chomienne, and R. A. Padua. 2000. Retinoic acid receptor alpha (RAR alpha) mutations in human leukemia. Leuk. Lymphoma 39:271-282. [DOI] [PubMed] [Google Scholar]
- 50.Pizzimenti, S., S. Laurora, F. Briatore, C. Ferretti, M. U. Dianzani, and G. Barrera. 2002. Synergistic effect of 4-hydroxynonenal and PPAR ligands in controlling human leukemic cell growth and differentiation. Free Radic. Biol. Med. 32:33-245. [DOI] [PubMed] [Google Scholar]
- 51.Rebel, V. I., A. L. Kung, E. A. Tanner, H. Yang, R. T. Bronson, and D. M. Livingston. 2002. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc. Natl. Acad. Sci. USA 99:14789-14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scoggin, K. E. S., A. Ulloa, and J. K. Nyborg. 2001. The oncoprotein tax binds the SRC1-interacting domain of CBP/p300 to mediate transcriptional activation. Mol. Cell. Biol. 21:5520-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scolnick, D. M., N. H. Chehab, E. S. Stavridi, M. C. Lien, L. Caruso, E. Moran, S. L. Berger, and T. D. Halazonetis. 1997. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 57:3693-3696. [PubMed] [Google Scholar]
- 54.Sheppard, H. M., J. C. Harries, S. Hussain, C. Bevan, and D. M. Heery. 2001. Analysis of the steroid receptor coactivator 1 (SRC1)-CREB binding protein interaction interface and its importance for the function of SRC1. Mol. Cell. Biol. 21:39-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siegel, R. M., F. K. Chan, D. A. Zacharias, R. Swofford, K. L. Holmes, R. Y. Tsien, and M. J. Lenardo. 2000. Measurement of molecular interactions in living cells by fluorescence resonance energy transfer between variants of the green fluorescent protein. Sci. STKE 38:PL1. [DOI] [PubMed] [Google Scholar]
- 56.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sundstrom, C., and K. Nilsson. 1976. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 17:565-577. [DOI] [PubMed] [Google Scholar]
- 58.Surapureddi, S., S. Yu, H. Bu, T. Hashimoto, A. V. Yeldandi, P. Kashireddy, M. Cherkaoui-Malki, C. Qi, Y. J. Zhu, M. S. Rao, and J. K. Reddy. 2002. Identification of a transcriptionally active peroxisome proliferator-activated receptor alpha-interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proc. Natl. Acad. Sci. USA 99:11836-11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tenen, D. G., R. Hromas, J. D. Licht, and D. E. Zhang. 1997. Transcription factors, normal myeloid development, and leukemia. Blood 90:489-519. [PubMed] [Google Scholar]
- 60.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 61.Trecca, D., L. Longo, A. Biondi, L. Cro, R. Calori, F. Grignani, A. T. Maiolo, P. G. Pelicci, and A. Neri. 1994. Analysis of p53 gene mutations in acute myeloid leukemia. Am. J. Hematol. 46:304-309. [DOI] [PubMed] [Google Scholar]
- 62.Voegel, J. J., M. J. Heine, M. Tini, V. Vivat, P. Chambon, and H. Gronemeyer. 1998. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 17:507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiederschain, D., H. Kawai, J. Gu, A. Shilatifard, and Z. M. Yuan. 2003. Molecular basis of p53 functional inactivation by the leukemic protein MLL-ELL. Mol. Cell. Biol. 23:4230-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, X. J. 2004. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32:959-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361-372. [DOI] [PubMed] [Google Scholar]
- 66.Zhong, S., P. Salomoni, P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell. Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]