Abstract
Large-scale chromatin decondensation has been observed after the targeting of certain acidic activators to heterochromatic chromatin domains. Acidic activators are often modular, with two or more separable transcriptional activation domains. Whether these smaller regions are sufficient for all functions of the activators has not been demonstrated. We adapted an inducible heterodimerization system to allow systematic dissection of the function of acidic activators, individual subdomains within these activators, and short acidic-hydrophobic peptide motifs within these subdomains. Here, we demonstrate that large-scale chromatin decondensation activity is a general property of acidic activators. Moreover, this activity maps to the same acidic activator subdomains and acidic-hydrophobic peptide motifs that are responsible for transcriptional activation. Two copies of a mutant peptide motif of VP16 (viral protein 16) possess large-scale chromatin decondensation activity but minimal transcriptional activity, and a synthetic acidic-hydrophobic peptide motif had large-scale chromatin decondensation activity comparable to the strongest full-length acidic activator but no transcriptional activity. Therefore, the general property of large-scale chromatin decondensation shared by most acidic activators is not simply a direct result of transcription per se but is most likely the result of the concerted action of coactivator proteins recruited by the activators' short acidic-hydrophobic peptide motifs.
Several transcriptional activators contain two or more distinct subdomains that can individually activate transcription in transient transcription assays (4, 15, 38, 39, 46). Furthermore, acidic activators' subdomains have been narrowed down to a short motif consisting mainly of acidic and hydrophobic residues (40). In the case of the strong viral transcriptional activator VP16 (viral protein 16), the motif from the N-terminal half of the C-terminal acidic activation domain has been carefully dissected to a short 11-amino-acid (11-aa) region, DALDDFDLDML (aa 437 to 447), whose activity can be diminished by an F442P mutation (8, 41). Single copies of the motif do not have strong activity, but two or more copies induce transcription even up to the level of the full activation domain (aa 413 to 490) (41). The 11-aa activation domain has been further simplified to a simple repetitive sequence: four copies of DDFDL (42). Similar regions have been noted in VP16 (aa 467 to 479), Fos (aa 267 to 277), and Gal4 (aa 861 to 873) (29, 40). Short KIX binding peptides, which interact with the KIX domain of p300 and CBP, have also been shown to have transcriptional activity on their own (13).
Transcriptional activators have historically been classified by the prevalence of certain amino acids in their activation domains, including acidic-, glutamine-, and proline-rich classes (31). This classification system is likely somewhat artificial, since it has been shown that the most prevalent amino acids can be dispensable for transcriptional activation (44). In light of these ambiguities, it would be useful to characterize transcriptional activators and their subdomains functionally, for example, by the ability to activate in certain contexts or by the ability to alter chromatin structure. While the transcriptional activating abilities of a few activators have been studied in the context of a stably integrated reporter gene (e.g., VP16) (47), systematic studies of several acidic activators' subdomains and the acidic-hydrophobic motifs have been performed only with transiently transfected reporter plasmids or have been conducted with Saccharomyces cerevisiae, where most of the genome is transcriptionally active, intergenic regions are small, and position effects are presumably minimal relative to the situation in mammalian cells. For example, an early systematic analysis of transcriptional ability was conducted with mammalian cells with a transiently transfected reporter gene (41), which typically displays some, but not all, of the characteristics of DNA embedded in the genome. This study revealed that while all activators tested can activate transcription when recruited to sites proximal to the TATA box (i.e., at a promoter position), acidic activators also had strong activity when bound to sites distal from the TATA box (i.e., at an enhancer position), whereas proline-rich activators had weak activity and glutamine-rich activators at distal locations had no activity (41). Studies at yeast promoters integrated into the genome showed similar results, except that the proline-rich activators were unable to activate from distal sites, and glutamine-rich activators were unable to activate even from proximal sites (23). Also in yeast, a short acidic-hydrophobic peptide was able to activate transcription from a promoter integrated into the genome and to alter the histone content of targeted promoters (12).
To activate transcription from genes in their natural chromatin location, transcriptional activators affect the local chromatin structure surrounding a target gene via a variety of proteins, including histone-modifying proteins (e.g., acetylases) and chromatin-remodeling complexes, which reposition nucleosomes (3, 18). The effects of transcriptional activators on higher levels of chromatin structure are just beginning to be characterized. To directly determine the effects of proteins on large-scale chromatin structure, a promoter can be integrated in multiple copies in the genome of cultured mammalian cells, and/or a protein can be targeted to an engineered chromatin site via a DNA binding domain (2). By targeting green fluorescent protein to the same chromatin region, changes in chromatin structure can be observed by fluorescence microscopy. In this way, several proteins were discovered to have large-scale chromatin-unfolding ability, including the VP16 acidic activation domain (48); BRCA1, COBRA1, E2F1, and p53 (54); the glucocorticoid receptor (32); and the estrogen receptor (5, 36). Accordingly, an in vivo system recently revealed an increase in mRNA transcripts simultaneous with large-scale chromatin unfolding after targeting VP16 to an engineered, heterochromatic site (20). By using fluorescence in situ hybridization, decondensation of chromatin was recently shown for the first time at an endogenous mammalian locus (7).
Demonstrating the causality and mechanism of this relationship between transcription and large-scale chromatin unfolding has proven perplexing and may reflect differences between different promoters and transcriptional activators. Careful studies of a mouse mammary tumor virus promoter array showed strong correlations between the level of transcription and the extent of large-scale chromatin unfolding from cell to cell as well as over time, during steroid induction and downregulation (32). This study also indicated that transcription is necessary to produce and maintain an unfolded chromatin state, as treatment of arrays with a transcriptional inhibitor (alpha-amanitin) or an inhibitor of RNA polymerase I elongation (DRB) reduced the initial chromatin unfolding and was able to reduce the size of already unfolded arrays (32). In contrast, the initial unfolding of a heterochromatic lac operator array by VP16 occurs even in the presence of alpha-amanitin, although maintenance of the unfolded structure may be partially inhibited by long-term alpha-amanitin treatment (48). Subdomains of the estrogen receptor have been identified which do not activate transcription but are fully capable of unfolding large-scale chromatin structure, further indicating that transcription is unnecessary in some contexts to produce unfolded chromatin (5). Precedent exists for chromatin changes being mechanistically decoupled from transcription itself. For example, histone modifications and decondensed chromatin produce a poised state that precedes actual transcriptional activation (7).
The precise relationship between transcriptional activity and large-scale chromatin decondensation activity has therefore been unclear. It was also unknown whether large-scale chromatin unfolding is a general property of all transcriptional activators and whether their subdomains or even shorter short acidic-hydrophobic motifs are sufficient for the unfolding. In this work, we demonstrate that the ability to unfold large-scale chromatin structure is a general property of acidic activators. Moreover, both transcriptional activity and the capability of unfolding large-scale chromatin structure are produced by the short acidic-hydrophobic peptide motifs contained within these activators.
MATERIALS AND METHODS
Plasmids, cell culture, transfections, and luciferase assays.
Plasmids constructed for the rapamycin recruitment system are described elsewhere (35). The direct fusions of the VP16 wild-type and mutant peptides to the nuclear localization sequence (NLS)-EYFP-lac repressor (NLS-EYFP-lac rep, called NYE127) were constructed by ligating the following blunted fragments into the EcoICRI-digested NYE127 vector: for NLS-EYFP-lac rep-VP16(437-448) (YFP-lac rep-monomer, called SEVI1), NYE105 was digested with EcoRV and SalI (filled in) to produce the 65-bp VP16(437 to 448) fragment. For NLS-EYFP-lac rep-VP16(437-448)x2 (YFP-lac rep-dimer, called SEVI2), NYE109 was digested with EcoRV and SalI (filled in) to produce the 104-bp VP16(437-448)x2 fragment. For NLS-EYFP-lac rep-VP16(437-448)F442P (YFP-lac rep-mutant monomer, called SEVI3), NYE129 was digested with EcoRV and SalI (filled in) to produce the 57-bp VP16(437-448)F442P fragment. For NLS-EYFP-lac rep-VP16(437-448)F442Px2 (YFP-lac rep-mutant dimer, called SEVI4), two oligonucleotides were annealed: VP16(F442P)2For (5′-GGG ACG CGC TAG ACG ATC CCG ATC TGG ACA TGT TGG GAT CTG ACG CGC TAG ACG ATC CCG ATC TGG ACA TGT TGG GAT CT-3′) and VP16(F442P)2Rev (5′-AGA TCC CAA CAT GTC CAG ATC GGG ATC GTC TAG CGC GTC AGA TCC CAA CAT GTC CAG ATC GGG ATC GTC TAG CGC GTC CC-3′). The insertions and junctions of these constructs were verified by sequencing.
Rapamycin was used at a final concentration of 100 nM (Sigma, St. Louis, Mo.). A03_1 cells (24) were cultured at 37°C with 5% CO2 in F-12 Ham's medium without hypoxanthine and thymidine (Invitrogen, Carlsbad, Calif.), with 10% dialyzed fetal bovine serum (HyClone Labs, Logan, Utah), and 0.3 μM methotrexate. Chinese hamster ovary cells (CHO-K1) were cultured at 37°C with 5% CO2 in F-12 Ham's medium (Invitrogen) with 10% fetal bovine serum (HyClone Labs). 2A5a cells and the stable luciferase reporter clones F3, G3, and H6 were cultured similarly but with 7.5 μg of puromycin/ml (6, 35).
Transfections on coverslips were performed with FuGENE 6 reagent (Roche, Indianapolis, Ind.) according to the manufacturer's instructions with 6 to 8 μl of reagent per 35-mm plate. Fresh medium was added 16 h after transfection. For fixed cells, 48 to 72 h after transfection, cells were rinsed in calcium and magnesium-free phosphate-buffered saline, fixed in magnesium-free phosphate-buffered saline with 1.6% formaldehyde (Polysciences, Warrington, Pa.), and mounted with ProLong Antifade reagent (Molecular Probes, Eugene, Oreg.).
Transfections for transient transcription assays used 350 ng of each effector, 900 ng of luciferase reporter, 100 ng of cytomegalovirus-beta-galactosidase reporter (Clontech, Palo Alto, CA), and 8 μl of FuGENE6. Fresh medium was added 16 h posttransfection, and cells were harvested and lysed with Passive Lysis buffer (Promega, Madison, Wis.) 48 h posttransfection. Luciferase assays, normalized for beta-galactosidase expression, were performed with Luciferase Assay reagent (Promega) and a Luminoskan luminometer (Thermo LabSystems, Vantaa, Finland).
Imaging and image analysis.
The traditional microscope (36) and automated microscope (5) have been previously described. We developed automated microscope software (RRnet36) for this work to collect images in three channels (cyan fluorescent protein [CFP], yellow fluorescent protein [YFP], and 4′,6-diamidino-2-phenylindole dihydrochloride [DAPI]). This software disregards cells with low CFP or YFP fluorescence, thereby excluding untransfected cells and making the assay independent of the percent of cells transiently transfected. Also, overlap of YFP fluorescence at the CFP array was measured by recording the average YFP intensity at the chromatin array (the region defined by CFP) and dividing this number by the average YFP intensity within the remainder of the nucleus (the region defined by DAPI excepting the chromatin array). The recruitment ratio is typically greater than 1, even if no recruitment is occurring, because a larger amount of fluorescent protein is present towards the interior of the nucleus, due to its three-dimensional shape. Editing several image sets to exclude images where the nucleus was imprecisely identified did not significantly alter the recruitment ratios, so we omitted this time-consuming editing step.
For chromatin-unfolding assays, the initial experiment is shown; a duplicate experiment showed similar results unless otherwise noted. The numbers below the plots are based on both experiments. The percentage of arrays that were unfolded uses 525 pixels as a threshold (5.25 μm2). The mean for each sample is the unweighted arithmetic mean of the means from the two experiments. The P values are step-down Bonferroni adjusted, were obtained with SAS PROC MIXED, and are based on each of the two experiments producing one measurement, the average of 50 to 100 cells so that n = 2 for each sample, rather than counting each cell on the slide as independent (28). Using the large data set generated in this work, there was no consistent correlation between nucleus size and chromatin array size (data not shown), so we did not normalize array size to nucleus size.
The automated microscope was also used for live imaging of cells in a Bioptechs FCS2 closed chamber, which maintains temperature and pH. At each time point, five 50-ms images were acquired in the CFP channel at various Z positions and the plane best in focus was used to collect 100-ms images in the CFP and YFP channels at several chosen stage locations. From these images, the CFP-labeled chromatin arrays were measured as previously described (5). The recruitment ratio was calculated by dividing the average pixel intensity of YFP at the chromatin array (the region defined by CFP fluorescence) by the average pixel intensity in the ring-shaped region surrounding the array (from the 4th to the 24th pixel in all directions from the edge of the array).
RESULTS
Construction and characterization of the rapamycin recruitment system.
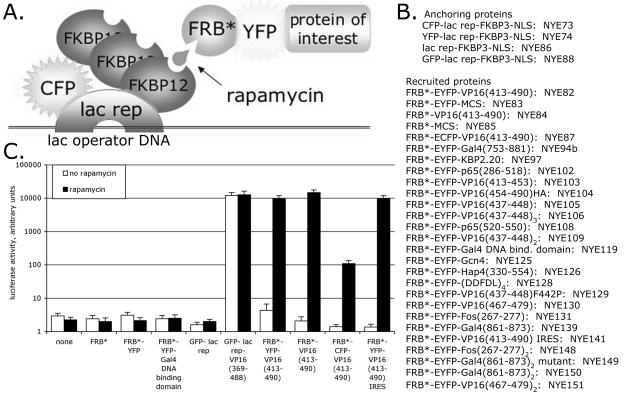
We adapted the FKBP12/FRB* rapamycin-inducible heterodimerization system (26) to allow inducible recruitment of a protein of interest to lac operator DNA sequences (Fig. 1A and B). This allows testing transcriptional activity using reporter plasmids with lac operator sites upstream of a core promoter. It also allows testing proteins for their ability to unfold large-scale chromatin structure with engineered cell lines with a condensed region of heterochromatin that contains lac operators (6, 48). The FKBP12 protein was present in three copies (designated FKBP3), and it interacted with the FRB* domain of mTOR/FRAP/RAFT1/RAPT1 in the presence of rapamycin.
FIG. 1.
Heterodimerization system for inducible protein targeting to lac operator arrays. (A) Schematic of the system. (B) Fusion proteins are listed with their names. MCS, multiple cloning site (polylinker); NLS, nuclear localization signal. (C) Results of transient transcription assays, where an 8-lac operator-E1b TATA-luciferase reporter plasmid was cotransfected with CFP-lac rep-FKBP3 plus each expression plasmid listed below the chart into CHO-K1 cells. Error bars show standard errors of the mean (SEM).
The system was tested by transient transfection assays with an 8-lac operator-E1b TATA-luciferase reporter plasmid (Fig. 1C). The reporter plasmid together with plasmid constructs for the anchoring protein, CFP-lac rep-FKBP3, and the recruited protein, FRB*-YFP-VP16 (aa 413 to 490) (hereafter called FRB*-YFP-VP16), were transfected into CHO-K1 cells which were then treated with rapamycin. The several-thousand-fold transcriptional activation observed with this inducible system was comparable to that observed with a direct GFP-lac rep-VP16 fusion protein. Rapamycin treatment and the FRB*-YFP-VP16 protein were required for this strong activation, and cotransfecting CFP-lac rep-FKBP3 with FRB*, FRB*-YFP-MCS (hereafter called FRB*-YFP), or FRB*-YFP-Gal4 DNA binding domain did not activate transcription. For unknown reasons, an FRB*-CFP-VP16 plasmid identical to FRB*-YFP-VP16, except for the amino acids specific to the fluorescent proteins, was not as active, possibly due to lower expression levels of the CFP fusion protein. We also constructed lac-FKBP3, YFP-lac rep-FKBP3, and GFP-lac rep-FKBP3. Although not tested as extensively as CFP-lac rep-FKBP3, they behaved similarly in several transient transcription assays (data not shown).
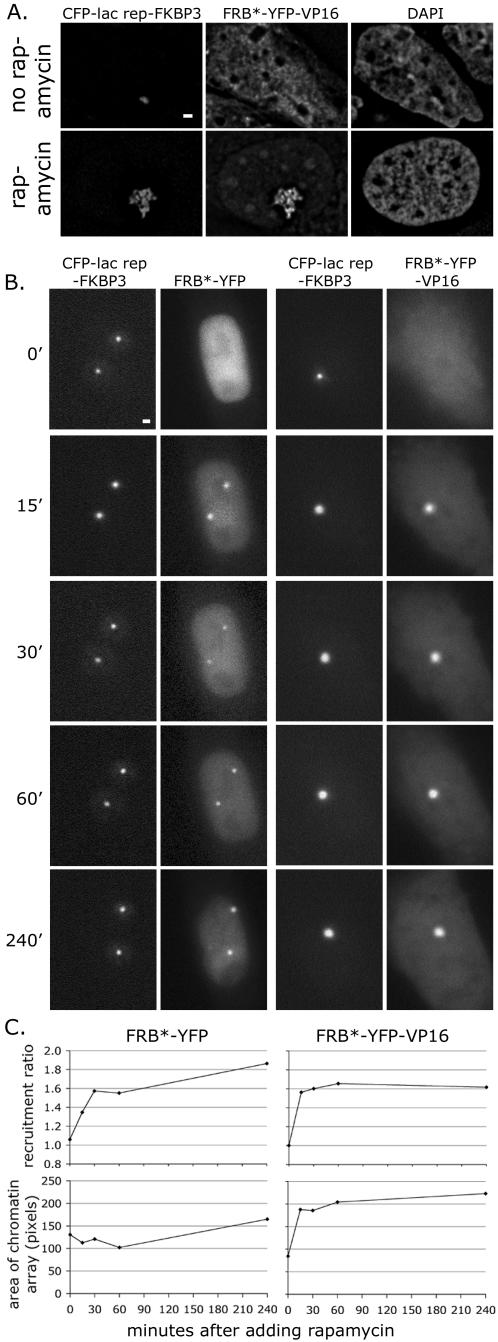
As observed by fluorescence microscopy, treatment of cells with rapamycin for 48 h induced recruitment of FRB*-YFP-VP16 to CFP-lac rep-FKBP3 at the heterochromatic lac operator chromatin array in A03_1 cells (Fig. 2A). In addition, this combination dramatically unfolds the normally condensed array, as was previously shown for GFP-lac rep-VP16 (48). We next observed the kinetics of recruitment and unfolding in living cells (Fig. 2B and C). The 2A5a cells used in these experiments, a subclone of clone 2A5, contain copies of a plasmid with 256 lac operators and the vitellogenin B1 TATA promoter driving expression of a CFP-peroxisome-targeting signal reporter gene (pSP21) (6). This engineered chromatin formed one or two condensed chromatin arrays per cell which, like the array in A03_1 cells, unfolded in response to VP16, although typically less dramatically (6). 2A5a cells were transiently transfected with the FRB*-YFP-VP16 and CFP-lac rep-FKBP3 constructs. Rapamycin was added 48 h after transfection. By 15 min after rapamycin addition, recruitment of FRB*-YFP-VP16 to the CFP-lac rep-FKBP3-bound 2A5a chromatin arrays reached a plateau and the chromatin arrays increased in size, indicating unfolding. Recruitment of FRB*-YFP occurred similarly, but without the accompanying unfolding of chromatin.
FIG. 2.
Rapid targeting of VP16 and inducible large-scale chromatin decondensation with heterodimerization system. (A) VP16 unfolds chromatin when inducibly targeted to a heterochromatic chromatin array. A03_1 cells which contain a compact heterochromatic lac operator array were transfected with CFP-lac rep-FKBP3 (NYE73) and FRB*-YFP-VP16 (NYE82) and treated with rapamycin 48 h prior to fixation. Single deconvolved optical sections are shown. Scale bar, 1 μm. (B) Unfolding of chromatin in living 2A5a cells. Cells were transfected with CFP-lac rep-FKBP3 and FRB*-YFP or FRB*-YFP-VP16 and incubated for 48 to 72 h. Cells were then maintained live on a microscope stage and imaged in single optical sections with short exposure times. Time points indicate minutes after rapamycin was added. Scale bar, 1 μm. (C) Quantitation of recruitment and chromatin unfolding in living 2A5a cells. Two independent experiments were carried out as above (B) for each condition. In each experiment, approximately 10 cells were imaged at time points before and after rapamycin was added. Each data point shown is the mean of about 20 cells.
Transcriptional activity and large-scale chromatin decondensation activity of inducible acidic activators.
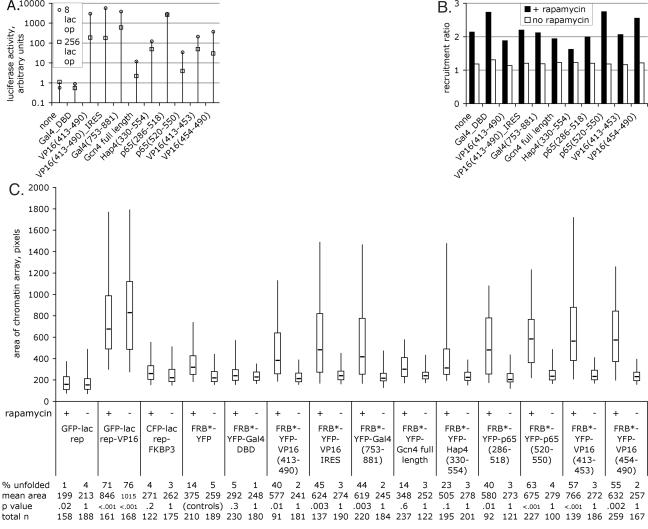
We fused several acidic activators to FRB*-YFP, and we conducted transient transcription assays using a luciferase reporter plasmid with either 8 or 256 lac operators upstream of an E1b TATA box (Fig. 3A). All activators activated transcription in this context, which lacked some features of genomic chromatin. The 256-lac operator plasmid was consistently less active than the 8-lac operator plasmid, as previously observed (35). We also tested these acidic activators in a more natural chromatin context using three independent stable cell lines containing an 8-lac operator-E1b TATA-luciferase reporter plasmid integrated into the genome (6, 35). The reporter gene in the stable cell lines showed a similar response to each acidic activator as in the transient transcription assays, except that induction was lower overall (data not shown).
FIG. 3.
Transcriptional activity and large-scale chromatin decondensation activity of inducible acidic activators. (A) The transient transcription assay used an 8- or 256-lac operator-E1b TATA-luciferase reporter plasmid in CHO-K1 cells. Fold induction is shown, meaning activity in the presence of rapamycin divided by activity in the absence of rapamycin. (B) Recruitment of each FRB*-YFP fusion protein to the chromatin array; see Materials and Methods for details. The median is shown. (C) Chromatin-unfolding assay. Chromatin arrays were measured for each FRB*-YFP fusion protein in the presence and absence of rapamycin. CFP-lac rep-FKBP3 was cotransfected with all FRB*-YFP fusion proteins. Tails of the box plots mark the 5th and 95th percentiles, boxes mark the 25th and 75th percentiles, and the line in the box marks the median for each sample. See Materials and Methods for details.
To rapidly assess the recruitment and chromatin unfolding activity of each FRB*-YFP fusion protein, we adapted our previously developed automated microscope software (5) to collect and analyze large numbers of images of A03_1 cells cotransfected with each FRB*-YFP fusion protein along with CFP-lac rep-FKBP3. Recruitment of each FRB*-YFP protein to the CFP-lac rep-FKBP3-labeled chromatin array was dependent on rapamycin, as expected (Fig. 3B). Measurements of chromatin arrays revealed that the negative controls, CFP-lac rep-FKBP3 alone and cotransfected with FRB*-YFP-Gal4 DNA binding domain, did not dramatically unfold large-scale chromatin structure, although the mean area of the chromatin array in these cases was somewhat larger than GFP-lac rep (Fig. 3C). None of the FRB*-YFP fusion proteins further unfolded chromatin in the absence of rapamycin (i.e., when not recruited to DNA). In the presence of rapamycin, FRB*-YFP showed slight chromatin-unfolding activity. While it is possible that this domain of the mTOR protein contains a chromatin-unfolding domain (see Fig. 6), the negative control FRB*-YFP-Gal4 DNA binding domain fusion contains the same region but did not exhibit chromatin-unfolding activity.
FIG. 6.
Large-scale chromatin-unfolding motifs. Hydrophobic, FMILVCW (red); acidic, DE (green); basic, KR (blue) (11). Protein segments listed have all been shown to unfold large-scale chromatin structure (this study and references 5, 36, and 54). The amino acids of each segment are shown in parentheses. The regions underlined are hypothesized to be responsible for the unfolding activity. The highlighted A in BRCA1 indicates the position where the cancer-predisposing mutation A1708E unfolds chromatin more effectively than the wild type in the context of the full-length protein (54). The highlighted Y in Hap4 indicates a discrepancy between the protein we used (34) and the National Center for Biotechnology Information sequence (accession no. X16727), which indicates an N at this position.
Most of the acidic activators tested were capable of dramatically unfolding large-scale chromatin structure well beyond these slight chromatin alterations, with 30 to 60% of the cells possessing a significantly unfolded array (versus 12% for FRB*-YFP) (Fig. 3C). Furthermore, individual activation domains, such as the two activation domains of p65 and of VP16, were capable of independently unfolding large-scale chromatin structures. The yeast activator Hap4 was not as dramatic, with 23% of arrays unfolded. The yeast activator Gcn4 was the only acidic activator that did not appear to unfold chromatin more than FRB*-YFP alone. We note that this was the weakest activator in transcriptional assays.
Transcriptional activity and large-scale chromatin decondensation activity of inducible acidic activation motifs.
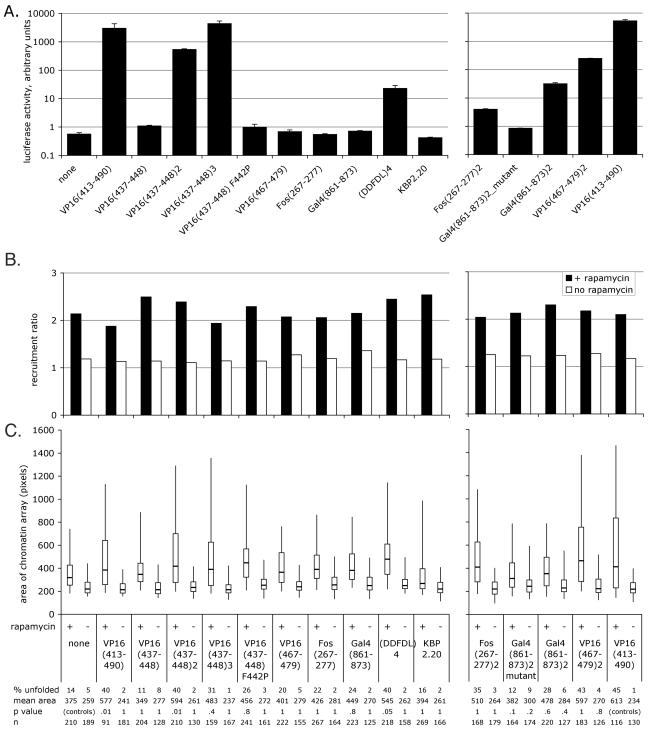
Several acidic activators share a common motif with transcriptional activity in transient transcription assays (40). We therefore constructed and tested these motifs for transcriptional activation on transfected and integrated reporter genes and for chromatin-unfolding ability. We also included a protein arising from a spontaneous mutation during PCR, the FRB*-YFP-Gal4(861-873)2 mutant, which contains one wild-type activation domain (MDDVYNYLFDDED), followed by one frameshift mutant domain (MDDVYNLSIR). In transient transcription assays, the fusions with one copy of each activation motif provided negligible activation, and two or more copies of the motifs [including (DDFDL)4] activated transcription (Fig. 4A). To our knowledge, this is the first demonstration that the motifs identified by inspection, VP16(467-479), Fos(267-277), and Gal4(861-873) (40), indeed are capable of transcriptional activation. Negligible activation was observed from the mutant version of FRB*-YFP-Gal4(861-873)2. Previous work indicated that KBP2.20, a KIX binding peptide, activated transcription about 10-fold less efficiently than VP16 (13). In our study, however, this peptide did not measurably activate transcription, so we conclude that in this protein context, KBP2.20 is not functional. Given the reduction in activity seen for the full-length acidic activators, we did not expect to see transcription induced by most of the motif activators in our stable transcription assays, since these motifs are much weaker. This was indeed the case: activation was observed only for the strongest two- and three-copy motifs: VP16(437-448)2, VP16(437-448)3, and VP16 (467-479)2 (data not shown). Recruitment of the FRB*-YFP-motif fusion proteins was as expected (Fig. 4B).
FIG. 4.
Transcriptional activity and large-scale chromatin decondensation activity of acidic activator motifs, all fused to FRB*-YFP. none, FRB*-YFP with no protein fused to it. See the legend to Fig. 3 for details. Error bars show SEMs. (A) Transient transcription assay. (B) Recruitment to the chromatin array. Sample labels are the same as for panels A and C. (C) Chromatin-unfolding assay. The initial experiment is shown. A subsequent experiment yielded similar results, except that the area of Gal4(861-873)2 was larger in the presence of rapamycin in the second experiment, and the area of VP16(437-448) was smaller in the absence of rapamycin and larger in the presence of rapamycin in the second experiment than in the first. The samples in the left and right charts were prepared separately and therefore statistically analyzed separately; note that the P values for the right side are compared to those of the positive control. The samples shown on the left (B and C) were prepared and analyzed at the same time as the samples in Fig. 3, so “none” and “VP16(413-490)” are the same data sets as shown in Fig. 3B and C.
Most of the two-copy motif fusion proteins, including VP16(437-448)2, (DDFDL)4, Fos(267-277)2, Gal4(861-873)2, and VP16(467-479)2, exhibited chromatin-unfolding activity not statistically significantly different from that of the full-length VP16 acidic activation domain, FRB*-YFP-VP16(413-490) (Fig. 4C, right). The one exception, with a P value of just under 0.1, is the one wild type, one mutant fusion of Gal4(861-873)2, which is less effective at unfolding chromatin than the fully wild-type version. Chromatin-unfolding ability, beyond that produced by FRB*-YFP alone, was unclear but probable for most of the one-copy motifs. While not statistically significant by a very stringent statistical test, most of these proteins increased the mean chromatin array area and unfolded 15 to 25% of the arrays, compared to 14% for FRB*-YFP alone (Fig. 4C, left). The mutant one-copy VP16 motif, VP16(437-448) F442P, was even more effective at unfolding chromatin than its wild-type counterpart. The KBP2.20 motif resembled the FRB*-YFP control, consistent with our hypothesis that this motif was not functional in this protein context.
Transcriptional activity and large-scale chromatin decondensation activity of acidic activation motifs directly bound to DNA.
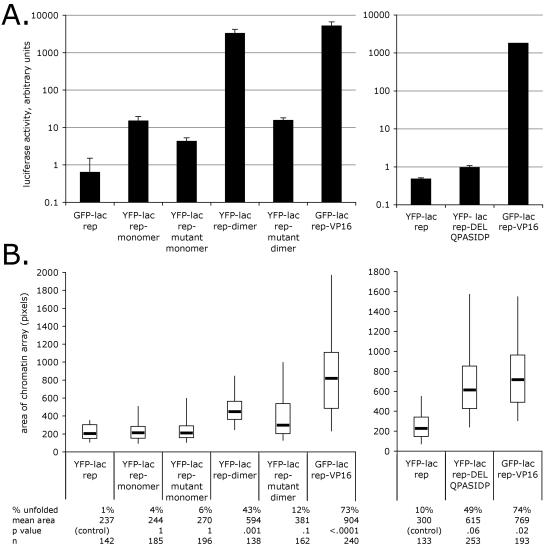
To confirm that the chromatin-unfolding activity of acidic activators revealed with the rapamycin recruitment system was not due to some unusual property of the rapamycin recruitment strategy or the FRB* domain, we fused several motifs directly to the lac repressor. These motifs included VP16(437-448) monomers and dimers, the F442P mutant form of each, and an acidic-hydrophobic sequence (DELQPASIDP) conveniently produced by a cloning strategy. Consistent with the results from the rapamycin recruitment system, the VP16 motif dimer activated transcription in transient assays much more strongly than the monomer form, and the F442P mutant forms showed reduced transcriptional activity (Fig. 5A, left). As in the rapamycin recruitment system, the wild-type dimer formed unfolded chromatin dramatically and the mutant dimer, while greatly diminished, still retained some unfolding activity (Fig. 5B, left). The wild-type and mutant monomers did not significantly unfold chromatin (Fig. 5B, left). Interestingly, the DELQPASIDP protein did not activate transcription (Fig. 5A, right) but showed strong large-scale chromatin unfolding activity (Fig. 5B, right).
FIG. 5.
Transcriptional activity and chromatin-unfolding ability of VP16(437-448) motifs and the F442P mutant thereof (left) and DELQPASIDP motif (right) recruited directly by the lac repressor. See the legend to Fig. 3 for details. The samples in the left and right charts were prepared separately and were therefore statistically analyzed separately. (A) Transient transcription assay. Note the log scale. Error bars show SEMs. (B) Chromatin-unfolding assay. The initial experiment is shown. A subsequent experiment yielded similar results, except that the distribution of areas showed more spread for the YFP-lac rep-dimer and less spread for the YFP-lac rep-mutant dimer in the second experiment.
2A5 cells (Fig. 2B) have a condensed lac operator chromatin array like A03_1 cells but also contain integrated CFP-peroxisome-targeting signal reporter genes. Consistent with the results in A03_1 cells, preliminary microscopy indicated that the VP16 F442P mutant motif dimer could unfold chromatin but did not detectably activate the reporter gene in 2A5 cells, while the wild-type VP16 motif dimer unfolded chromatin and strongly activated the reporter (data not shown).
Proteins which did not unfold large-scale chromatin structure.
We previously identified a number of transcription-related proteins which do not unfold a large-scale chromatin structure in this type of assay, including large portions of the estrogen receptor (36). In the course of this work, we also fused several glutamine- and proline-rich activators to FRB*-YFP, including Oct1 (aa 175 to 269) (41), Oct2 (aa 99 to 161) (41), Sp1 (aa 149 to 473) (A and B activation domains) (25), AP-2 (aa 31 to 76) (41), CTF (CAAT box transcription factor; aa 399 to 499) (41), 10 prolines, 10 glutamines, and the repressor Eed (bp 142 to 1,788) (9). We also fused several control proteins: 10 glycines, 10 cysteines, and the 963-bp full-length human cDNA for fibrillarin (1) (for construct details and further experiments, see reference 35). These proteins did not unfold chromatin except for a few examples when the DELQPASIDP linker was present in the fusion protein. None of these proteins affected transcription in transient assays (data not shown), although at least in the case of the relatively weak glutamine- and proline-rich activators we suspect the assay was not sensitive enough. Since the functionality of each protein could not be confirmed, we cannot conclude that these proteins do not have large-scale chromatin unfolding activity in their normal context, but these experiments did demonstrate that not every transcription-related protein recruited to DNA unfolds large-scale chromatin structure.
DISCUSSION
It was previously unclear whether all acidic activators, their subdomains, and their short motifs are capable of activation only on transiently transfected reporter plasmids or whether they contain all the necessary structural elements to unfold large-scale chromatin structure and activate transcription in a more natural chromatin environment. We systematically tested several full acidic activation domains (VP16, Gal4, Hap4, and Gcn4), several independent activation domains (from p65 and VP16), and several 10- to 20-aa motifs from acidic activators either alone or multimerized (VP16, Fos, and Gal4). The subdomains and some dimerized ∼10-aa motifs of acidic activators showed detectable transcriptional activity in the context of chromatin. Contrary to a hypothesis where different activities of the activator are carried out by different domains of the protein, it is particularly notable that activators with very simple sequence content, such as VP16(437-448)2, are able to activate genes within chromatin.
Whether large-scale chromatin unfolding is a general property of acidic activators was also unknown. While CFP-lac rep-FKBP3 alone and CFP-lac rep-FKBP3 with FRB*-YFP and rapamycin alter large-scale chromatin structure slightly when targeted to the heterochromatin array, it is clear that acidic activators were able to unfold chromatin beyond that produced by FRB*-YFP alone. In addition, we tested VP16 activation motifs in an entirely different protein context, direct fusion to a lac repressor, with similar results. Prior to this work, the acidic activators VP16 and p53, as well as BRCA1, E2F1, COBRA1, the estrogen receptor, and the glucocorticoid receptor, were known to possess large-scale chromatin-unfolding activity (32, 36, 48, 54). We add to this list the acidic activator proteins p65, Gal4, and Hap4. The only acidic activator tested that did not dramatically unfold chromatin was Gcn4; it appears likely that this fusion protein was not entirely functional, because it gave low levels of transcriptional activity in transient assays.
VP16 and p65 each have two distinct activation domains (4, 15, 38, 39, 46), and each of these domains was able to unfold large-scale chromatin structure, consistent with the identification of several chromatin-unfolding domains within the ligand-inducible activation function 2 of the estrogen receptor (5) and BRCA1 (54). It is also clear from this work that the same small motifs from acidic activators which recruit proteins to activate transcription are also sufficient to recruit proteins that unfold large-scale chromatin structure. This was seen for VP16(437-448)2, VP16(437-448)3, VP16(467-479)2, (DDFDL)4, Fos(267-277)2, and Gal4(861-873)2.
Based on the work presented here and previous studies (42), two ∼10-aa motifs on the same protein chain appear to be necessary to obtain detectable transcriptional activity and chromatin unfolding. The disproportionate strength of two copies of a motif versus one is still poorly understood even for the simplest case of transcription from a transiently transfected reporter plasmid, which was reported more than a decade ago (41). Interestingly, having many copies of a single ∼10-aa motif recruited to the same promoter appears to be insufficient. A single copy of the motif embedded within its normal protein context is sufficient to bind transcription-related proteins (49). If indeed these motifs function by binding a particular protein(s) (see further discussion below), the simplest explanation is that the ∼10-aa motif is the minimal binding unit, but that two copies allow the protein(s) to bind cooperatively or in a functionally productive way. While possible, it seems less likely that a novel motif has been formed by the junction between the two motifs, because the natural activator VP16 has two motifs separated by many amino acids and every two-motif protein we engineered shows the same behavior, despite the fact that their junctions were not designed in any particular systematic way. In any case, the ∼20-aa two-copy sequence is a remarkably short and simple motif to function in transcriptional activation and chromatin unfolding.
In fact, all proteins discovered so far to unfold chromatin have a region of acidic-hydrophobic amino acid residues, consistent with the hypothesis that this region interacts with a protein or proteins that can unfold chromatin (Fig. 6). Small activation motifs, particularly those containing hydrophobic residues, are found not only in acidic activators but also in other types of activators (16, 45). These motifs are apparently capable of binding a number of different proteins, at least in vitro, including histone acetylation complexes, chromatin-remodeling complexes, and general transcription factors (17, 21). The work presented here indicates that these or novel proteins have large-scale chromatin-unfolding activity. The fact that large-scale chromatin structure began unfolding within minutes after rapamycin was added in the live cell studies described here, as was previously seen with injection of lac-VP16 proteins into nuclei (48), provides further evidence for an active mechanism behind chromatin unfolding, rather than a passive blocking of chromatin assembly.
The mechanism by which these short motifs recruit so many proteins is still controversial. The predominant model suggests that these motifs can support transient, weak, direct interactions with a variety of proteins (37). A recent study suggests that activation motifs are inherently “sticky,” with hydrophobic residues that are capable of binding a variety of proteins interspersed with either acidic or at least hydrophilic residues that force the motif to remain accessible in aqueous solution (30). An intriguing alternate model suggests that acidic-hydrophobic activation motifs may act by interacting directly with histones, possibly distorting them structurally and thereby marking them for the recruitment of other proteins (12).
Overall, these motifs are better understood in terms of developing models of functional intrinsically disordered proteins rather than the traditional lock-and-key model of structure-function relationship (10, 51, 52). The intrinsic disorder of short protein binding peptide motifs may be critical for function by minimizing steric hindrances in the early stages of molecular recognition, such that the motif becomes structured only after the binding event has initiated (43, 49). The lack of precise sequence conservation among acidic-hydrophobic motifs and the low specificity of acidic activation domains for various binding partners is consistent with speculation that the intrinsic disorder of a protein can contribute to function by allowing independent binding of several different target proteins (10, 51).
In agreement with the first study of large-scale chromatin decondensation of an endogenous mammalian locus (7) and our previous work on the estrogen receptor (5), the DELQPASIDP motif's ability to unfold chromatin in the absence of transcriptional ability lends further support to the hypothesis that large-scale chromatin unfolding is unlikely to be a direct result of a physical disruption of chromatin by transcription itself but rather is likely due to proteins recruited by these short motifs. While a large number of proteins are recruited by these small motifs, this study provides a route to gain insight into which of these proteins are involved in regulating large-scale chromatin structure. For example, the F442P mutation in VP16 is reported to significantly reduce binding of several interacting proteins (14, 15, 19, 27, 50, 53), yet a dimer of a motif with this mutation can still unfold chromatin. More dramatically, the DELQPASIDP motif unfolds chromatin but does not activate transcription. The behavior of these motifs provides further evidence consistent with the hypothesis that large-scale chromatin unfolding is necessary but not sufficient to activate transcription (36).
Previous studies have examined the in vitro binding of a variety of transcription-related proteins to several versions of VP16, including truncations and mutations thereof (14, 15, 19, 22, 27, 33, 34, 50, 53). The reported binding of none of these proteins neatly correlates with the large-scale chromatin-unfolding activity discovered in this study. There are several possible explanations. First, whether in vitro protein-protein binding assays reflect physiological interactions is uncertain, and we intend to use the lac operator-repressor system in vivo to address this issue. Second, it is possible, perhaps even likely, that different proteins are involved in large-scale chromatin unfolding in different cellular contexts (i.e., redundancy exists). Third, it is possible that the proteins recruited by acidic activation domains which unfold large-scale chromatin structure have not yet been discovered. The DELQPASIDP motif is particularly useful to identify such proteins, since the motif apparently binds large-scale chromatin-unfolding proteins but does not detectably activate transcription. Identifying the proteins responsible for large-scale chromatin unfolding, whether previously known or novel, should shed light on its mechanism.
Acknowledgments
This work was supported by grants from the National Institutes of Health to A.S.B. (R01-GM58460 and R01-GM42516). A.E.C. was a Howard Hughes Medical Institute Predoctoral Fellow.
We appreciate the technical assistance of Anousheh Ashouri. Statistical analysis was provided by Paul Holmes and Susanne Aref of the Illinois Statistics Office. We gratefully acknowledge the gift of plasmids from Stuart Schreiber, Karol Bomsztyk, Rong Li, and Walter Schaffner.
REFERENCES
- 1.Aris, J. P., and G. Blobel. 1991. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc. Natl. Acad. Sci. USA 88:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belmont, A. S. 2001. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 11:250-257. [DOI] [PubMed] [Google Scholar]
- 3.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. L., W. D. Cress, A. Cress, S. J. Triezenberg, and L. Guarente. 1990. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: evidence for transcriptional adaptors. Cell 61:1199-1208. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter, A. E., A. Ashouri, and A. S. Belmont. 2004. Automated microscopy identifies estrogen receptor subdomains with large-scale chromatin structure unfolding activity. Cytometry 58A:157-166. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter, A. E., and A. S. Belmont. 2004. Direct visualization of transcription factor induced chromatin remodeling and cofactor recruitment in vivo. Methods Enzymol. 375:366-381. [DOI] [PubMed] [Google Scholar]
- 7.Chambeyron, S., and W. A. Bickmore. 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18:1119-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cress, W. D., and S. J. Triezenberg. 1991. Critical structural elements of the VP16 transcriptional activation domain. Science 251:87-90. [DOI] [PubMed] [Google Scholar]
- 9.Denisenko, O. N., and K. Bomsztyk. 1997. The product of the murine homolog of the Drosophila extra sex combs gene displays transcriptional repressor activity. Mol. Cell. Biol. 17:4707-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunker, A. K., J. D. Lawson, C. J. Brown, R. M. Williams, P. Romero, J. S. Oh, C. J. Oldfield, A. M. Campen, C. M. Ratliff, K. W. Hipps, J. Ausio, M. S. Nissen, R. Reeves, C. Kang, C. R. Kissinger, R. W. Bailey, M. D. Griswold, W. Chiu, E. C. Garner, and Z. Obradovic. 2001. Intrinsically disordered protein. J. Mol. Graph. Model. 19:26-59. [DOI] [PubMed] [Google Scholar]
- 11.Engelman, D. M., T. A. Steitz, and A. Goldman. 1986. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 15:321-353. [DOI] [PubMed] [Google Scholar]
- 12.Erkine, A. M., and D. S. Gross. 2003. Dynamic chromatin alterations triggered by natural and synthetic activation domains. J. Biol. Chem. 278:7755-7764. [DOI] [PubMed] [Google Scholar]
- 13.Frangioni, J. V., L. M. LaRiccia, L. C. Cantley, and M. R. Montminy. 2000. Minimal activators that bind to the KIX domain of p300/CBP identified by phage display screening. Nat. Biotechnol. 18:1080-1085. [DOI] [PubMed] [Google Scholar]
- 14.Ge, H., and R. G. Roeder. 1994. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 78:513-523. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich, J. A., T. Hoey, C. J. Thut, A. Admon, and R. Tjian. 1993. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell 75:519-530. [DOI] [PubMed] [Google Scholar]
- 16.Gugneja, S., C. M. Virbasius, and R. C. Scarpulla. 1996. Nuclear respiratory factors 1 and 2 utilize similar glutamine-containing clusters of hydrophobic residues to activate transcription. Mol. Cell. Biol. 16:5708-5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, D. B., and K. Struhl. 2002. The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. J. Biol. Chem. 277:46043-46050. [DOI] [PubMed] [Google Scholar]
- 18.Hassan, A. H., K. E. Neely, M. Vignali, J. C. Reese, and J. L. Workman. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6:D1054-D1064. [DOI] [PubMed] [Google Scholar]
- 19.Ingles, C. J., M. Shales, W. D. Cress, S. J. Triezenberg, and J. Greenblatt. 1991. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature 351:588-590. [DOI] [PubMed] [Google Scholar]
- 20.Janicki, S. M., T. Tsukamoto, S. E. Salghetti, W. P. Tansey, R. Sachidanandam, K. V. Prasanth, T. Ried, Y. Shav-Tal, E. Bertrand, R. H. Singer, and D. L. Spector. 2004. From silencing to gene expression: real-time analysis in single cells. Cell 116:683-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein, J., M. Nolden, S. L. Sanders, J. Kirchner, P. A. Weil, and K. Melcher. 2003. Use of a genetically introduced cross-linker to identify interaction sites of acidic activators within native transcription factor IID and SAGA. J. Biol. Chem. 278:6779-6786. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, N., T. G. Boyer, and A. J. Berk. 1995. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol. Cell. Biol. 15:6465-6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunzler, M., G. H. Braus, O. Georgiev, K. Seipel, and W. Schaffner. 1994. Functional differences between mammalian transcription activation domains at the yeast GAL1 promoter. EMBO J. 13:641-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, G., G. Sudlow, and A. S. Belmont. 1998. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning. J. Cell Biol. 140:975-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, R. 1999. Stimulation of DNA replication in Saccharomyces cerevisiae by a glutamine- and proline-rich transcriptional activation domain. J. Biol. Chem. 274:30310-30314. [DOI] [PubMed] [Google Scholar]
- 26.Liberles, S. D., S. T. Diver, D. J. Austin, and S. L. Schreiber. 1997. Inducible gene expression and protein translocation using nontoxic ligands identified by a mammalian three-hybrid screen. Proc. Natl. Acad. Sci. USA 94:7825-7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, Y. S., I. Ha, E. Maldonado, D. Reinberg, and M. R. Green. 1991. Binding of general transcription factor TFIIB to an acidic activating region. Nature 353:569-571. [DOI] [PubMed] [Google Scholar]
- 28.Littell, R. C., G. A. Milliken, W. W. Stroup, and R. D. Wolfinger. 1996. SAS system for mixed models. SAS Institute, Inc., Cary, N.C.
- 29.Ma, J., and M. Ptashne. 1987. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48:847-853. [DOI] [PubMed] [Google Scholar]
- 30.Melcher, K. 2000. The strength of acidic activation domains correlates with their affinity for both transcriptional and non-transcriptional proteins. J. Mol. Biol. 301:1097-1112. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell, P. J., and R. Tjian. 1989. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245:371-378. [DOI] [PubMed] [Google Scholar]
- 32.Muller, W. G., D. Walker, G. L. Hager, and J. G. McNally. 2001. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J. Cell Biol. 154:33-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nedialkov, Y. A., and S. J. Triezenberg. 2004. Quantitative assessment of in vitro interactions implicates TATA-binding protein as a target of the VP16C transcriptional activation region. Arch. Biochem. Biophys. 425:77-86. [DOI] [PubMed] [Google Scholar]
- 34.Neely, K. E., A. H. Hassan, C. E. Brown, L. Howe, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 22:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nye, A. C. 2003. Effects of transcriptional activators on large-scale chromatin structure. Ph.D. thesis. University of Illinois, Urbana-Champaign.
- 36.Nye, A. C., R. R. Rajendran, D. L. Stenoien, M. A. Mancini, B. S. Katzenellenbogen, and A. S. Belmont. 2002. Alteration of large-scale chromatin structure by estrogen receptor. Mol. Cell. Biol. 22:3437-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ptashne, M., and A. Gann. 1997. Transcriptional activation by recruitment. Nature 386:569-577. [DOI] [PubMed] [Google Scholar]
- 38.Regier, J. L., F. Shen, and S. J. Triezenberg. 1993. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 90:883-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitz, M. L., and P. A. Baeuerle. 1991. The p65 subunit is responsible for the strong transcription activating potential of NF-kappa B. EMBO J. 10:3805-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitz, M. L., M. A. dos Santos Silva, H. Altmann, M. Czisch, T. A. Holak, and P. A. Baeuerle. 1994. Structural and functional analysis of the NF-κB p65 C terminus. An acidic and modular transactivation domain with the potential to adopt an alpha-helical conformation. J. Biol. Chem. 269:25613-25620. [PubMed] [Google Scholar]
- 41.Seipel, K., O. Georgiev, and W. Schaffner. 1992. Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions. EMBO J. 11:4961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seipel, K., O. Georgiev, and W. Schaffner. 1994. A minimal transcription activation domain consisting of a specific array of aspartic acid and leucine residues. Biol. Chem. Hoppe-Seyler 375:463-470. [DOI] [PubMed] [Google Scholar]
- 43.Shen, F., S. J. Triezenberg, P. Hensley, D. Porter, and J. R. Knutson. 1996. Transcriptional activation domain of the herpesvirus protein VP16 becomes conformationally constrained upon interaction with basal transcription factors. J. Biol. Chem. 271:4827-4837. [DOI] [PubMed] [Google Scholar]
- 44.Sullivan, S. M., P. J. Horn, V. A. Olson, A. H. Koop, W. Niu, R. H. Ebright, and S. J. Triezenberg. 1998. Mutational analysis of a transcriptional activation region of the VP16 protein of herpes simplex virus. Nucleic Acids Res. 26:4487-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka, M., and W. Herr. 1994. Reconstitution of transcriptional activation domains by reiteration of short peptide segments reveals the modular organization of a glutamine-rich activation domain. Mol. Cell. Biol. 14:6056-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Triezenberg, S. J., R. C. Kingsbury, and S. L. McKnight. 1988. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 2:718-729. [DOI] [PubMed] [Google Scholar]
- 47.Tsukamoto, T., N. Hashiguchi, S. M. Janicki, T. Tumbar, A. S. Belmont, and D. L. Spector. 2000. Visualization of gene activity in living cells. Nat. Cell Biol. 2:871-878. [DOI] [PubMed] [Google Scholar]
- 48.Tumbar, T., G. Sudlow, and A. S. Belmont. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uesugi, M., O. Nyanguile, H. Lu, A. J. Levine, and G. L. Verdine. 1997. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science 277:1310-1313. [DOI] [PubMed] [Google Scholar]
- 50.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 51.Uversky, V. N. 2002. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11:739-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wright, P. E., and H. J. Dyson. 1999. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 293:321-331. [DOI] [PubMed] [Google Scholar]
- 53.Xiao, H., A. Pearson, B. Coulombe, R. Truant, S. Zhang, J. L. Regier, S. J. Triezenberg, D. Reinberg, O. Flores, and C. J. Ingles. 1994. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol. Cell. Biol 14:7013-7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye, Q., Y. F. Hu, H. Zhong, A. C. Nye, A. S. Belmont, and R. Li. 2001. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J. Cell Biol. 155:911-921. [DOI] [PMC free article] [PubMed] [Google Scholar]